Abstract
Francisella tularensis infects wild animals and humans to cause tularemia. This pathogen targets the cytosol of macrophages, where it replicates using the genes in the Francisella pathogenicity island (FPI). Virulence gene regulation in Francisella is complex, but transcriptional regulators MglA and SspA have been shown to regulate the expression of approximately 100 genes, including the entire FPI. We utilized a Francisella novicida transposon mutant library to identify additional regulatory factors and identified five additional genes that are essential for virulence gene expression. One regulatory gene, FTN_0480 (fevR, Francisella effector of virulence regulation), present in all Francisella species, is required for expression of the FPI genes and other genes in the MglA/SspA regulon. The expression of fevR is positively regulated by MglA. However, constitutive expression of fevR in an mglA mutant strain did not restore expression of the MglA/SspA regulon, demonstrating that mglA and fevR act in parallel to positively regulate virulence gene expression. Virulence studies revealed that fevR is essential for bacterial replication in macrophages and in mice, where we additionally show that fevR is required for the expression of genes in the MglA/SspA regulon in vivo. Thus, fevR is a crucial virulence gene in Francisella, required for the expression of virulence factors known to be essential for this pathogen's subversion of host defenses and pathogenesis in vivo.
Francisella tularensis is a gram-negative facultative intracellular pathogen that causes the zoonotic disease tularemia. F. tularensis can be contracted from ticks, contaminated water, or infected mammals, particularly rabbits. Since F. tularensis can survive in a wide range of environments, the bacterium must be able to sense and respond appropriately to each niche by altering expression of survival genes. Indeed, it is known that stresses, such as nutrient and iron limitation, influence the expression of Francisella virulence genes (7, 12, 20). During infections, F. tularensis replicates in host cells, and the virulence of the pathogen is linked to its ability to replicate intracellularly (1). Intracellular bacteria initially reside within a vacuole that stains with cellular markers typical of a late endosome/early lysosome, but the vacuole does not acidify appreciably and does not contain cathepsin D, suggesting that the vacuole is modified (10, 15, 26, 27). F. tularensis escapes this vacuole within 5 h and replicates to high numbers in the cytosol (11, 15). After replication in the cytosol, F. tularensis is found in an autophagic vacuole that fuses with lysosomes, though without harm to the bacteria (9). Thus, it seems likely that F. tularensis senses specific phases of the intracellular life cycle and alters its gene expression accordingly.
Francisella tularensis subsp. tularensis is the most virulent subspecies of Francisella and is found throughout North America and in parts of Europe. Francisella novicida, a close relative of F. tularensis, causes severe disease in immunocompromised humans but rarely does so in immunocompetent individuals (24). However, F. novicida has the same families of virulence genes as F. tularensis, causes a similar disease in mice, and thereby serves as a good experimental model with which to study Francisella pathogenesis (18, 23).
The transcriptional regulator macrophage growth locus A (mglA) was discovered in F. novicida (4). mglA is upregulated inside macrophages 1.5 h postinfection (3), suggesting that the MglA regulon represents a subset of Francisella genes involved in interactions with macrophages. Indeed, MglA regulates the expression of 102 genes, several of which are necessary for intracellular replication, including a large cluster of 17 genes called the Francisella pathogenicity island (FPI) (6, 19). MglA interacts with stringent starvation protein A (SspA) to form a heterodimer that binds to RNA polymerase, consistent with a direct role in transcriptional regulation (8). In addition, it was recently reported that the two-component regulator pmrA regulates the expression of the FPI and other virulence genes (22). However, MglA does not regulate pmrA, nor does PmrA regulate MglA. During growth in broth, mglA and sspA mRNAs are expressed during the lag phase and levels decrease over time (3, 6, 8). These kinetics are opposite to those of MgA/SspA-regulated transcripts, whose levels start low and increase over time. Taken together, these results suggest that additional factors or signals are necessary for regulon expression.
We devised a genetic screen using an F. novicida transposon library (33) to identify genes involved in the expression of the MglA/SspA regulon. We report here that one novel gene, FTN_0480 (fevR, for Francisella effector of virulence regulation), which is positively regulated by MglA and SspA (6, 8), is required for replication in macrophages and for virulence in mice. We also demonstrate that fevR functions in mice to regulate the known virulence determinants in the MglA/SspA regulon. Thus, we have identified fevR, a novel regulator of virulence genes which is essential for Francisella pathogenesis.
MATERIALS AND METHODS
Bacterial strains and growth.
Wild-type F. novicida strain U112 and an isogenic mglA point mutant (GB2) have been described previously (4). Bacteria were grown in tryptic soy broth (TSB; Difco Laboratories, Detroit, MI) supplemented with 0.2% cysteine or plated on tryptic soy agar (Becton Dickinson, Sparks, MD) supplemented with 0.1% cysteine (Sigma, St. Louis, MO) or modified Mueller-Hinton (MH) agar (Difco) supplemented with 0.025% ferric pyrophosphate (Sigma), 0.02% IsoVitaleX (Becton Dickinson), 0.1% glucose, and 0.025% calf serum (Gibco, Carlsbad, CA). Kanamycin (15 μg/ml), chloramphenicol (3 μg/ml), and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 50 μg/ml) (Sigma) were added to the growth media when appropriate. Escherichia coli strain DH12S was used for cloning and grown in Luria broth (Difco Laboratories, Detroit, MI).
Screen to identify transcriptional regulators.
An MglA-regulated β-galactosidase reporter plasmid was constructed by fusing the promoter of pepO to lacZY (pepO::lacZY). The region 5′ to the pepO open reading frame (FTN_1186) was amplified using pepO F and pepO R (see Table S1 in the supplemental material), introducing BglI and XhoI sites, respectively. lacZY was amplified from pCE37 using lacZ F and lacY R (see Table S1 in the supplemental material), introducing XhoI and PstI sites, respectively. PCR products were digested and ligated to pDSC (6) digested with BlgI and PstI. The ligations were transformed into E. coli DH12S. Chloramphenicol-resistant colonies were sequenced, and plasmid DNA was isolated using a DNA midiprep kit (Qiagen) and transformed into U112 by chemical transformation. Among the 15 chloramphenicol-resistant U112 colonies, only 2 turned blue on MH agar supplemented with X-Gal (termed U112-B1 and U112-B2). To test whether the MglA pathway was still functional in U112-B1, we deleted mglA. The mglA mutant in the U112-B1 strain background did not express β-galactosidase and the colonies were white on indicator plates, consistent with the role of MglA in regulating pepO::lacZY expression. Thus, we performed our genetic screen with the U112-B1(pDSC3) strain. To create a library of mutants in the U112-B1 background, genomic DNA was isolated from the previously described U112 transposon library (33). Genomic DNA (gDNA; 0.65 μg) from the total library was chemically transformed into U112-B1, and transformants were plated on MH agar (3 μg/ml chloramphenicol, 15 μg/ml kanamycin, and X-Gal). White colonies were picked 2 days after plating and verified to have only one transposon insertion by Southern blotting (data not shown). Candidate transposon mutants were additionally screened by quantitative reverse transcription-PCR (RT-PCR). The wild type and transposon mutants were subcultured to an optical density (OD) of 0.03. RNA was isolated from transposon mutants grown for 7 h to an OD of 2.0, on average.
Mutagenesis, epitope-tagged clonings, and complementation.
F. novicida deletion and complemented mutants were made as described previously using the primers listed in Table S1 in the supplemental material (6). The groES::fevR constructs, C-terminal epitope-tagged constructs FevR-glutathione S-transferase (GST), MglA-hemagglutinin (HA), and SspA-His, were made by overlapping PCR using the primers listed in Table S1 in the supplemental material. Chemically competent F. novicida was used in transformations (2). Mutants were assessed by PCR to verify homologous recombination and sequenced.
Identification of transposon insertion.
The sites of transposon insertion were identified by either inverse PCR or direct sequencing of gDNA. gDNA was isolated from the mutants using DNeasy (Qiagen). Next the gDNA was either sent for sequencing using the transposon primer TnF or cut with XbaI and ligated in a large volume to promote self-ligation of DNA fragments. The ligations were purified using Zymo DNA cleaner and concentrator, used as the template in a PCR with TnF and TnR (primers that extended out from the transposon), and then sequenced using primer TnF.
Time courses and RNA isolation.
Overnight cultures of F. novicida strains were grown in TSB-0.2% cysteine at 37°C with shaking and subcultured to an OD at 600 nm (OD600) of 0.01 in 200 ml. Samples were taken at various time points throughout the 8-h time course for RNA isolation and determination of the OD600. Five samples were taken during the time course at 1, 3, 5, and 7.5, and 10 h for characterization of single mutants or at 2, 4, 6, and 8 h in gro::fevR bypass experiments. RNA was harvested as previously described (6).
Preparation of cDNA probes, hybridization, and analysis.
The Francisella microarray and hybridization conditions were described previously (6). Normalized data were collected using the Stanford Microarray Database (30). Spots were excluded from analysis due to obvious abnormalities, a regression correlation of <0.6, or a Cy3 net mean intensity of <100. Only spots with at least 70% good data across the experiment were included for analysis. The normalized log2 (Cy5/Cy3) ratio was used for hierarchical clustering using the CLUSTER program (14). Results were visualized using the TREEVIEW program (14). Significant differences in gene transcript between the mglA and fevR mutant growth curves were determined using Significance Analysis for Microarrays program, version 1.21 (32), with the two-class statistical analysis tool and a calculated false-discovery rate of 0. Microarray results are available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/).
Quantitative RT-PCR.
Quantitative RT-PCR for bacteria grown in broth was performed as previously described (6). To quantify transcripts from infected mice, cDNA was created using the high-capacity cDNA kit and quantified using Power Sybr green PCR master mix, both from Applied Biosystems. The gene-specific message was calculated by plotting a standard curve for each primer set using a dilution series of RNA from a sample known to contain the message of interest. To compare transcript abundances for the wild type and mutants, values were normalized to that for DNA helicase uvrD (FTT0121) to obtain relative quantities of message.
Macrophage replication assay.
Bone marrow-derived macrophages were prepared as described elsewhere (28). Bone marrow-derived macrophages were seeded in 24-well plates at a density of 2.5 × 105 macrophages per well and incubated at 37°C and 5% CO2 overnight. The macrophages were then infected at a multiplicity of infection of 25:1 (bacteria-to-macrophage ratio) and centrifuged at 730 × g for 15 min to mediate attachment. The macrophages were incubated (time zero) for 1 h and washed three times with warm media. Macrophages were lysed 0.5, 4, 8, and 12 h postinfection with 1% saponin in water for 5 min and then diluted in phosphate-buffered saline and plated on tryptic soy agar with 0.1% cysteine.
Mouse infections.
Six- to 8-week-old, female wild-type C57BL/6 (Jackson Laboratories, Bar Harbor, ME) mice were kept under specific-pathogen-free conditions in filter top cages. Mice were provided with sterile water and food ad libitum. Competitive-index (CI) experiments were performed as previously described (6). CI values were calculated as the ratio of mutant to wild-type output, normalized for the input, and significance was calculated by comparing the log of the CI to 0. Standard errors were calculated, and significance of results was determined by application of the Mann-Whitney statistical test on the log10 value of the CI. For RNA isolation from the skin, hair was removed from the mouse abdomen and mice were infected subcutaneously with 107 CFU in two spots. After 24 h, skin was harvested and one side was taken for bacterial enumeration, while the other was placed immediately in 1 ml of Trizol and homogenized. RNA was then extracted as described previously (6). All animal infection experiments were approved by the Institutional Animal Care and Use Committee and the Institutional Biosafety Committee of Stanford University.
Coimmunoprecipitations.
MglA-HA/SspA-His-, MglA-HA/FevR-GST-, and SspA-His/FevR-GST-tagged strains were grown in 100 ml at 37°C until an OD of 1.0. Bacteria were pelleted and resuspended in 5 ml of Bugbuster (Novagen) and 80 μl of protease inhibitor cocktail (Novagen). Cell extracts were sonicated four times with 15-second bursts at a 70 output setting on a Kontes ultrasonic cell disruptor to decrease viscosity. Cell extracts were spun for 1 h at ∼150,000 × g in a Beckman ultracentrifuge to separate soluble and insoluble fractions. To purify SspA-His- and SspA-interacting partners, 1.5 ml of the soluble fraction was incubated with 80 μl of His-Bind resin (Novagen) overnight at 4°C and purified by following the manufacturer's instructions. To purify MglA-HA and MglA-HA binding partners, 5 μl of anti-HA antibody (Sigma; H3663) was noncovalently attached to 80 μl protein G plus agarose (Calbiochem) by following Sigma's instructions. The soluble fraction (1.5 ml) was incubated overnight at 4°C with the HA antibody and beads. To purify FevR-GST and copurifying partners, 1.5 ml of the soluble fraction was added to 80 μl BaculoGold glutathione-agarose beads (BD Biosciences) by following the manufacturer's instructions and incubated overnight at 4°C. The HA- and glutathione-containing beads were washed five times with 500 μl of radioimmunoprecipitation assay (RIPA) buffer. Fifty microliters of sodium dodecyl sulfate sample buffer was then added to the beads, and 15 μl of sample was run on a 12% acrylamide sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and transferred to nitrocellulose. Immunoblots were blocked overnight with Odyssey blocking buffer (LI-COR Biosciences) and probed with anti-HA antibody at 1:2,000 dilution (Roche), anti-GST antibody at 1:4,000 dilution (Sigma; G1160), and anti-His antibody at 1:2,000 dilution (R&D Systems; MAB050).
RESULTS
Genetic screen to identify genes that affect F. novicida virulence gene expression.
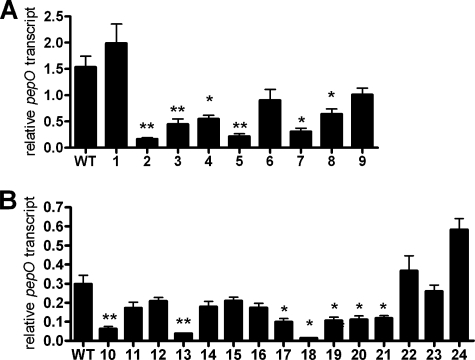
Since MglA and SspA were known to regulate the expression of many virulence genes in F. tularensis, including the FPI (6, 8, 19), we devised a screen to identify additional regulators involved in the MglA/SspA pathway using an MglA-regulated β-galactosidase reporter vector. The pepO promoter, which is expressed roughly 50-fold higher in the wild-type strain than in an mglA mutant (6), was fused to lacZY (pDSC3) and introduced into our F. novicida transposon library, which represents eightfold coverage of the nonessential genome (33). Transposon mutants with wild-type expression of pepO formed blue colonies on plates containing the indicator X-Gal, due to pepO promoter-dependent expression of lacZY. Mutants defective in the induction of the pepO promoter formed white colonies. Approximately 40,000 colonies were screened, representing approximately 25-fold coverage of the genome; 24 white colonies were isolated and designated W1 to W24. All candidate mutant clones were additionally screened by quantitative RT-PCR for decreased pepO expression to eliminate false positives. Thirteen of the 24 isolates contained significantly lower levels of pepO transcript than the wild type (Fig. 1A and B). Thus, this genetic screen led to the identification of genes required for the expression of the MglA-regulated gene pepO.
FIG. 1.
Screen for F. novicida transposon mutants that regulate pepO. (A and B) RNA was isolated from transposon mutants grown in broth at 37°C to early stationary phase. pepO mRNA levels were determined by quantitative RT-PCR and normalized to that for the uvrD transcript. Thirteen of 24 transposon mutants had significantly lower levels of pepO transcript than the wild type (WT). Samples were obtained in triplicate. Experiments were performed at least three times. Means and standard deviations from a representative experiment are shown. *, P < 0.05; **, P < 0.01.
fevR positively regulates expression of the MglA/SspA regulon in vitro.
The transposon insertion sites were determined for the 13 mutants identified in the screen and were mapped to five distinct genes: two hypothetical genes (FTN_0480 and FTN_1069), caiC (encoding AMP binding protein), uhpC (encoding a major facilitator superfamily transport protein), and cphA (encoding cyanophycin synthetase) (Table 1). We did not identify mglA or sspA in our screen; thus, our results may represent an underestimate of genes involved in the regulation of MglA/SspA-dependent gene expression. However, we focused our efforts on hypothetical gene FTN_0480 for several reasons: (i) it encodes a novel protein; (ii) its transcript is positively regulated by MglA and SspA, which are required for virulence (6, 8); and (iii) it is induced by iron depletion, a condition which induces the expression of critical virulence genes in many bacterial pathogens (6, 8, 12).
TABLE 1.
Identity of transposon insertion sites in mutants with significantly lower levels of pepO
| Clone(s) | U112 designation | Gene | Description of product |
|---|---|---|---|
| 3, 4, 13 | FTN_0480 | fevR | Hypothetical protein |
| 2, 5, 7, 18, 21 | FTN_0604 | caiC | AMP-binding protein |
| 8 | FTN_1069 | Protein of unknown function | |
| 17, 19, 20 | FTN_1112 | cphA | Cyanophycin synthetase |
| 10 | FTN_1611 | uhpC | Major facilitator superfamily transport protein |
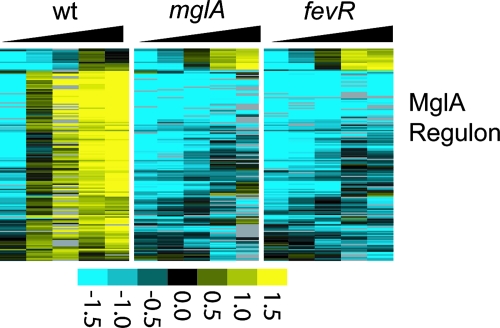
To determine the role of FTN_0480 in the regulation of F. novicida gene expression, we compared the transcriptomes of the wild type, an FTN_0480 deletion mutant, and an mglA point mutant (GB2) (4) by microarray analysis. RNA was isolated from the bacterial strains during a time course in broth at 37°C. Similar to our previous findings (6), the majority of MglA-regulated genes were downregulated in the mglA point mutant (92), while 10 genes were expressed at higher levels than in wild-type bacteria (Fig. 2). The ΔFTN_0480 mutant transcriptome was strikingly similar to that of the mglA mutant. Indeed, statistical analysis of the microarray data using Significance Analysis for Microarrays revealed that the only gene whose expression varied significantly between the mglA and ΔFTN_0480 mutant strains was FTN_0480 itself. Thus, under the conditions used, the mglA and ΔfevR mutant transcriptomes appear identical. This result demonstrates that expression of the genes within the FPI, all of which are required for Francisella virulence, is dependent on FTN_0480. Thus, we annotate the FTN_0480 locus as fevR.
FIG. 2.
The MglA/SspA regulon is not expressed in mglA and ΔfevR mutants grown in broth. RNA was isolated from wild-type, mglA, and ΔfevR mutants during growth in TSB at 37°C to compare transcriptomes by microarray. The levels of the gene transcripts of the previously published MglA regulon (6) are shown. The levels of these transcripts in mglA and fevR mutants are identical and demonstrate that these genes contribute to MglA-regulated gene expression in broth. Columns represent individual time points increasing from left to right during the time course at 1, 3, 5, 7, and 9 h. Rows represent individual genes.
mglA and fevR act in parallel to positively regulate virulence gene expression in vitro.
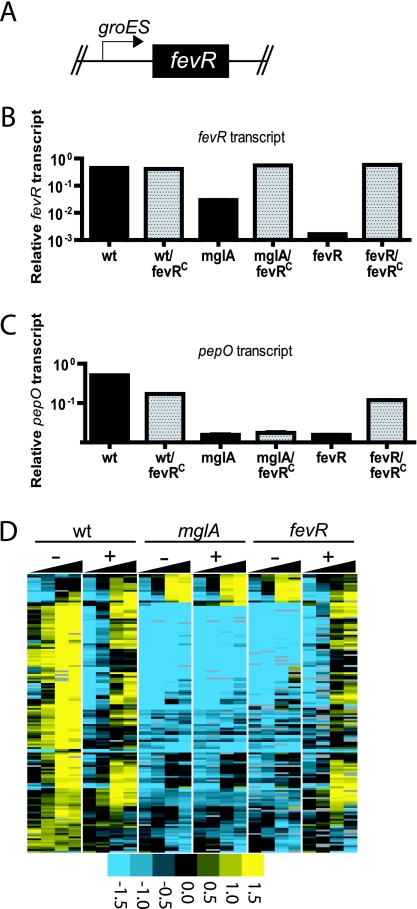
MglA and SspA positively regulate transcript expression of fevR (6, 8). mglA and sspA transcript levels are themselves unaffected in a ΔfevR mutant (Fig. 2), suggesting that fevR does not positively regulate the transcription of mglA and sspA. One hypothesis to explain this result is that FevR is a transcriptional activator that is downstream of MglA/SspA in a regulatory cascade and that FevR can act independently of MglA/SspA to induce the expression of the regulon. To test this model, we designed a strain that expresses fevR and contains a mutated mglA. Rescue of FPI expression in this strain lacking mglA would indicate that FevR does indeed act downstream and independently of MglA. We replaced the fevR promoter with that of groES, which is constitutively active in the wild-type and mglA and ΔfevR mutant backgrounds, resulting in a fevR(Con) strain (Fig. 3A). RNA was then isolated during growth, and the transcriptomes were analyzed by quantitative RT-PCR and microarray analysis. As expected, the fevR transcript is expressed in the mglA fevR(Con) and ΔfevR fevR(Con) mutant backgrounds, establishing that MglA no longer regulates expression of the fevR transcript in these strains (Fig. 3B). To test whether uncoupling fevR expression from MglA would result in expression of pepO and the MglA/SspA regulon, we measured gene expression by quantitative RT-PCR and microarray analysis, respectively. In the wild-type fevR fevR(Con) and ΔfevR fevR(Con) backgrounds the MglA/SspA regulon was expressed, albeit at lower levels than in the wild-type strain (Fig. 3C and D). The physiological cause of the decreased pepO transcript expression is unclear. The mglA fevR(Con) mutant, however, did not express pepO or the entire MglA regulon. Thus, fevR is not sufficient to induce the MglA/SspA regulon in the absence of mglA. Instead, these data suggest that MglA positively regulates the expression of fevR and that, once expressed, MglA and FevR are both required to initiate transcription of the genes in the MglA/SspA regulon.
FIG. 3.
Constitutive expression of an fevR transcript does not rescue expression of the MglA/SspA regulon in an mglA mutant. (A) The fevR promoter was replaced with the groES promoter and the resulting gene was inserted into the chromosome of wild-type (wt) and mglA and ΔfevR mutant backgrounds to create wild-type fevR(Con) (fevRC), mglA fevR(Con), and ΔfevR fevR(Con) mutants. Strains were grown in TSB at 37°C, and RNA was isolated at 2, 4, 6, and 8 h. Quantitative RT-PCR of fevR (B) and pepO (C) mRNA was performed at the 8-h time point, and levels were normalized to that for the uvrD control transcript. Although the mglA mutant containing the fevR(Con) construct expresses the fevR transcript, pepO expression is not rescued. Sampling was performed in triplicate. Means and standard deviations from a representative experiment are shown. (D) Microarray analysis of MglA regulon expression throughout the growth curves confirms the quantitative RT-PCR findings that both mglA and fevR are necessary for MglA/SspA regulon expression.
FevR does coprecipitate with MglA or SspA.
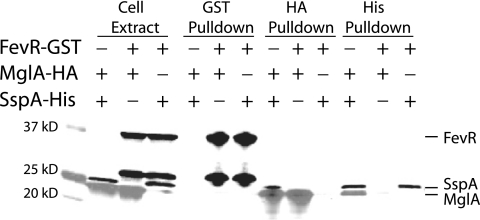
Since MglA and FevR are both required for expression of the MglA/SspA regulon and constitutive expression of fevR did not lead to MglA-independent virulence gene expression, we proposed that MglA and FevR may interact. To determine whether MglA and FevR directly interact, we constructed epitope-tagged versions of these proteins. The C terminus of endogenous MglA was tagged with HA2 and FevR was tagged with GST. As a positive control for coprecipitation, the C terminus of endogenous SspA was also tagged with an eight-histidine epitope (8). Insertion of the epitope tags did not disrupt regulatory functions, as MglA/SspA-regulated transcripts were still expressed in each of the strains (data not shown). Strains were grown to late exponential phase, and cell extracts were prepared under nondenaturing conditions (Fig. 4). Similar to published studies, MglA-HA coimmunoprecipated with SspA-His and vice versa, demonstrating that conditions were sufficient for identifying direct interactions within this complex (Fig. 4) (8). However, neither MglA-HA nor SspA-His copurified with FevR-GST. Likewise, FevR-GST did not copurify with MglA-HA or SspA-His. These data suggest that, while MglA and FevR function in parallel, FevR does not bind MglA or SspA to promote expression of the MglA/SspA regulon.
FIG. 4.
MglA and SspA do not coprecipitate with FevR. MglA-HA/SspA-His-, MglA-HA/FevR-GST-, and SspA-His/FevR-GST-tagged strains were created by placing the corresponding epitope tag at the C termini of the endogenous copies of products of mglA, sspA, and fevR. Cell extracts were prepared from double-tagged strains grown to late exponential phase. Each cell extract was used in coprecipitations with nickel beads, anti-HA antibody/protein G resin, or glutathione beads and immunoblotted to detect His, HA, and GST tags. The anti-GST antibody stained two bands, a high-molecular-weight band representing FevR-GST and a low-molecular-weight band likely to be a GST cleavage product.
fevR is required for intracellular growth in macrophages.
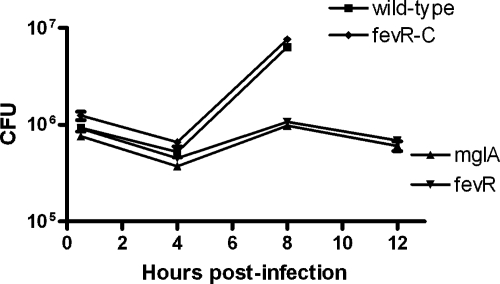
MglA and several MglA-regulated genes, including many FPI genes and a putative transglutaminase gene (FTN_0869), are necessary for replication of F. novicida in macrophages (6, 16, 19). Since fevR contributes to the expression of these genes in vitro, we predicted that a ΔfevR mutant would be unable to grow in macrophages. To test this, we infected bone marrow-derived macrophages with the wild-type, mglA, ΔfevR, and ΔfevR complemented strains (Fig. 5). The numbers of both the wild-type and the complemented ΔfevR mutant bacteria increased 10-fold inside macrophages over 8 h (Fig. 5A), resulting in the death of infected macrophages at 9 h postinfection (data not shown). In contrast, the numbers of mglA and ΔfevR mutant bacteria did not increase inside macrophages, nor did the mglA and ΔfevR mutant-infected macrophages die throughout the course of the experiment. These data demonstrate that fevR is required for Francisella replication within macrophages, a critical virulence trait of this pathogen.
FIG. 5.
mglA and fevR contribute to replication in macrophages. Bone marrow-derived macrophages were infected with either the wild-type, mglA, ΔfevR, or ΔfevR complemented (fevR-C) bacterial strain at a multiplicity of infection of 20 bacteria per macrophage. Macrophages were washed at 30 min postinfection, lysed at 2, 4, 8, and 12 h postinfection, and plated to enumerate the number of CFU. The means from a single experiment are shown and are representative of three independent experiments. The standard deviations are shown and are less than 10%.
fevR is required for virulence in a mouse model of infection.
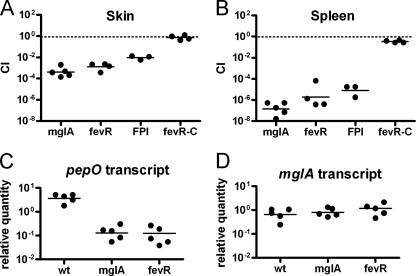
MglA positively regulates the expression of many virulence factors, including the FPI. Indeed, an mglA mutant strain is severely attenuated in a mouse model of infection (6, 22, 33). Likewise, if fevR is involved in regulating this pathway in vivo, we anticipate that a ΔfevR mutant would also be severely attenuated. To measure the contribution of fevR to virulence, we performed competition experiments in which mice were infected subcutaneously with a 1:1 mixture of wild-type bacteria and either the mglA or ΔfevR mutant bacteria. Forty-eight hours postinfection, the skin and spleens were collected and the numbers of wild-type and mutant bacteria in each sample were enumerated. The ΔfevR mutant exhibited a severe decrease in fitness compared to the wild type in both the spleen and the skin, with a CI similar to that of an mglA or Δfpi mutant (Fig. 6 A and B). The attenuation of the ΔfevR mutant could be rescued by adding back a wild-type copy of fevR. These data show that a ΔfevR mutant has one of the most severe attenuations described for Francisella and that therefore fevR is critical for F. novicida virulence.
FIG. 6.
mglA and fevR are necessary for virulence gene expression and survival in vivo. (A and B) Mice were infected subcutaneously with 5 × 104 CFU of wild-type bacteria and 5 × 104 CFU of mglA (point mutant), ΔfevR, ΔfevR complemented (fevR-C), and FPI deletion mutant bacteria, for a total of 105 bacteria/mouse. After 2 days, the skin (A) and the spleens (B) were taken for counts and the CI was calculated. Each dot represents the CI value for one mouse. Bars represent the geometric means. (C and D) Mice were infected subcutaneously in two different spots on the abdomen with 107 CFU of wild-type (wt) bacteria or mglA or ΔfevR mutants. After 24 h, one injection site was taken for counts, while the other was taken to isolate total RNA. Transcript levels of pepO (C) and mglA (D) were determined by quantitative RT-PCR and normalized to that for control transcript uvrD. Since the mglA mutant is a point mutant, the mglA transcript is still expressed in this strain. Each dot represents the average of quantitative PCR values from experiments performed triplicate for one mouse. Bars represent the geometric means.
Virulence gene expression in tissue requires mglA and fevR.
To conclusively demonstrate that both mglA and fevR are necessary for expression of MglA/SspA-regulated genes in vivo, we quantified the transcripts from MglA-regulated gene pepO during an infection in mice. Mice were injected subcutaneously in two spots on the abdomen with 107 CFU of the wild-type, mglA, or ΔfevR strain. At 24 h postinfection, one injection site was taken to quantify bacterial load, while the other was taken for total RNA isolation. The wild-type strain replicated to ∼109 bacteria per gram in infected skin in 24 h, whereas mglA and ΔfevR mutant counts were 100-fold lower. These single-infection results are in agreement with our data from CI experiments and conclusively demonstrate that fevR is required for F. novicida virulence in vivo. Quantitative RT-PCR was performed to determine the levels of pepO and mglA transcripts, which were normalized to those for control gene uvrD, which is expressed at similar levels in these strains. mglA was included as a control because microarray results suggested that the gene was not differentially regulated between the wild-type, mglA point mutant, and ΔfevR mutant bacteria. Indeed, the mglA transcript did not vary significantly between the strains (Fig. 6D). However, the expression of the pepO transcript was 10 times higher in the wild type than in the mglA and ΔfevR mutants (Fig. 6C). These results establish a function for fevR as a critical regulator of virulence gene expression in vivo.
DISCUSSION
Francisella is a highly infectious pathogen that possesses the ability to survive in various environments including freshwater, arthropods, and mammals, suggesting that this organism has the ability to regulate gene expression in response to various external signals. Nutrient and iron limitation has been shown to influence the expression of Francisella virulence genes (7, 12, 20). Several F. tularensis regulatory factors have been described. MglA and SspA act together to regulate the expression of at least 100 genes, including FPI genes, which are essential for virulence. In addition, the two-component regulator pmrA regulates the expression of the FPI and other virulence genes (22). mglA and pmrA are required for the ability of F. tularensis to survive within mammalian hosts, yet we do not understand how their gene products integrate to control virulence. These regulators likely play roles in F. tularensis subversion of the immune system, including growth within macrophages and dendritic cells and modulation of innate immunity signaling pathways within macrophages (1, 5, 25, 31).
To further elucidate the MglA/SspA signaling cascade, we screened for transposon mutants unable to induce transcription from the MglA-regulated promoter pepO. The majority of the transposon mutants we identified contained insertions in either FTN_0480 (fevR), caiC, or cphA. We have shown that fevR is required for the expression of the MglA/SspA regulon during growth in rich media and in tissue. In addition, we demonstrate that a ΔfevR mutant is unable to replicate within macrophages and exhibits one of the most severe attenuations described for Francisella during a mouse infection. Previously, all regulation studies of Francisella have utilized broth or in vitro tissue culture systems to track virulence regulation. Since we do not know what all of the environmental signals that integrate to control virulence are, it was important to address the regulation of known virulence factors, such as pepO, in the host. In this study, we demonstrate that fevR regulates known virulence factors in the environmental context of a natural infection.
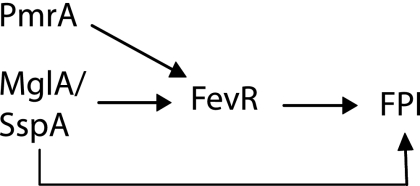
The expression of fevR is positively regulated by MglA and SspA, suggesting that FevR may be downstream of these proteins in a pathway. However, the requirement of MglA for expression of genes in the MglA/SspA regulon could not be bypassed by constitutively expressing an fevR transcript in an mglA mutant (Fig. 3), leading us to test if FevR may directly interact with the MglA/SspA complex. We created epitope-tagged versions of MglA, SspA, and FevR in F. novicida to assay for interactions between these proteins. Similar to results from previously published work for another organism (Francisella tularensis subspecies holarctica strain LVS) (8), MglA and SspA copurified. However, we could not demonstrate MglA or SspA protein interactions with FevR in coprecipitation assays, suggesting that MglA and SspA do not directly interact with FevR. Taken together our data indicate that MglA/SspA and FevR function in parallel to regulate the transcription of the MglA/SspA regulon (Fig. 7). This resembles a network motif called a feed forward loop (FFL), where one inducer activates the other and then the two inducers act together to activate the downstream genes (29). FFLs have been shown to accelerate the time for response to a given stimulus, as well as decrease responses to transient signals so that the bacterium responds only to persistent environmental cues (21). Previous work has also shown that the orphan response regulator PmrA contributes to expression of fevR (22), suggesting that expression of the fevR transcript requires the integration of multiple input signals and that the upregulation of fevR may represent a commitment step for FPI transcript expression. Thus, we have identified an essential novel regulator of Francisella virulence.
FIG. 7.
Model for the MglA/SspA pathway. MglA and SspA positively regulate fevR expression, and fevR regulates expression of the MglA/SspA regulon. Constitutive expression of the fevR transcript in an mglA mutant did not rescue MglA/SspA regulon expression, suggesting that MglA/SspA and FevR work in parallel to activate downstream genes. Furthermore, while MglA and SspA copurify with each other, neither copurifies with FevR-GST. These data suggest that the MglA/SspA regulon is under the control of a common network motif called an FFL.
The identification of fevR as a regulator may help to explain an interesting observation. When Francisella is grown in TSB with 0.2% cysteine at 37°C, mglA and sspA are maximally expressed during lag phase and the transcript levels decrease throughout the growth curve (3, 6). Curiously, MglA/SspA-regulated genes, including fevR, follow a pattern of gene expression that is opposite of mglA and sspA transcript expression, with maximal levels occurring during stationary phase. This suggests that MglA/SspA regulatory activity increases as levels of fevR transcript and presumably levels of FevR protein increase in the bacterium. Since mglA-sspA and fevR transcripts display different patterns of gene expression, it also suggests that their upregulation may result from different environmental cues. The future study of how F. tularensis incorporates these signals in order to regulate virulence gene expression during infection will be very instructive.
FevR is annotated as a hypothetical protein and shows little similarity to proteins from non-Francisella species. However, iterative Psi-BLAST searches indicate that FevR is weakly homologous to the MerR family of transcription factors. The observed similarity was highly driven by a putative DNA binding domain in FevR (see Fig. S1 in the supplemental material). The MerR family of transcription factors typically contain a conserved N terminus with a helix-turn-helix domain followed by a highly divergent C-terminal metal binding domain that regulates MerR function. FevR is 111 amino acids, which is smaller than typical MerRs, and alignments suggest that FevR may lack a metal binding domain (see Fig. S1 in the supplemental material). In fact, the similarity between FevR and MerR transcription factors is mostly contained within a 22-amino-acid region that encompasses the MerR helix-turn-helix domain (13). So, while FevR exhibits some degree of homology to MerRs in the helix-turn-helix region, the remainder of the FevR predicted protein appears divergent, and it is unclear if FevR would have a mechanism of action similar to that of MerRs. However, MerRs act by binding DNA independently of RNA polymerase, which may explain why we were unable to coprecipitate FevR with MglA or SspA. We are currently conducting experiments to test the model that FevR positively regulates gene expression in F. tularensis by binding DNA.
SspA homologs in Francisella are required for expression of the FPI and other virulence genes. SspA is highly conserved in gram-negative bacteria, including pathogenic Neisseria and Yersinia spp. and E. coli. Although SspA is involved in virulence gene regulation in Neisseria and Yersinia species, the specific mechanism of SspA-mediated gene regulation is not known. However, in E. coli SspA requires a coactivator for binding to phage promoters (17). Given the integration of SspA/MglA and FevR in Francisella, the work presented here supports the idea that SspA homologs require a coactivator for gene regulation. It is possible that Neisseria and Yersinia spp. encode a functional homolog of FevR and that this could be a conserved mechanism of SspA-mediated gene regulation in bacterial pathogens.
Our screen identified four other genes, uhpC, cphA, FTN_1069, and caiC, that are involved in the regulating the expression of the virulence factor pepO and possibly the MglA/SspA regulon. The further characterization of FevR and these additional genes will lead to further advances in understanding how F. tularensis regulates the expression of genes important for survival and virulence in response to various environmental cues. As we learn more about the complex signaling cascades that regulate gene expression, we may also gain further understanding of virulence regulation in related gram-negative pathogens.
Supplementary Material
Acknowledgments
We thank Elizabeth Joyce, Jeff Margolis, and David Weiss for careful reading of the manuscript and for their thoughtful discussions.
This work was supported by grants AI063302 and AI065359 from the NIH-NIAID to Denise Monack. A.B. was supported by a Cellular and Molecular Biology Training Program grant (NIH 5 T32 GM007276).
Editor: B. A. McCormick
Footnotes
Published ahead of print on 16 June 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Anthony, L. D., R. D. Burke, and F. E. Nano. 1991. Growth of Francisella spp. in rodent macrophages. Infect. Immun. 593291-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, L. S., M. Z. Gu, S. C. Cowley, W. W. Leung, and F. E. Nano. 1991. Transformation and allelic replacement in Francisella spp. J. Gen. Microbiol. 1372697-2703. [DOI] [PubMed] [Google Scholar]
- 3.Baron, G. S., and F. E. Nano. 1999. An erythromycin resistance cassette and mini-transposon for constructing transcriptional fusions to cat. Gene 22959-65. [DOI] [PubMed] [Google Scholar]
- 4.Baron, G. S., and F. E. Nano. 1998. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol. Microbiol. 29247-259. [DOI] [PubMed] [Google Scholar]
- 5.Bosio, C. M., and S. W. Dow. 2005. Francisella tularensis induces aberrant activation of pulmonary dendritic cells. J. Immunol. 1756792-6801. [DOI] [PubMed] [Google Scholar]
- 6.Brotcke, A., D. S. Weiss, C. C. Kim, P. Chain, S. Malfatti, E. Garcia, and D. M. Monack. 2006. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect. Immun. 746642-6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchan, B. W., M. K. McLendon, and B. D. Jones. 2008. Identification of differentially regulated Francisella tularensis genes using a newly developed Tn5-based transposon delivery system. Appl. Environ. Microbiol. 742637-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charity, J. C., M. M. Costante-Hamm, E. L. Balon, D. H. Boyd, E. J. Rubin, and S. L. Dove. 2007. Twin RNA polymerase-associated proteins control virulence gene expression in Francisella tularensis. PLoS Pathog. 3e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Checroun, C., T. D. Wehrly, E. R. Fischer, S. F. Hayes, and J. Celli. 2006. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. USA 10314578-14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemens, D. L., and M. A. Horwitz. 2007. Uptake and intracellular fate of Francisella tularensis in human macrophages. Ann. N. Y. Acad. Sci. 1105160-186. [DOI] [PubMed] [Google Scholar]
- 11.Clemens, D. L., B. Y. Lee, and M. A. Horwitz. 2004. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 723204-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, K., R. J. Blick, W. Liu, and E. J. Hansen. 2006. Identification of Francisella tularensis genes affected by iron limitation. Infect. Immun. 744224-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 185019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 9514863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golovliov, I., V. Baranov, Z. Krocova, H. Kovarova, and A. Sjostedt. 2003. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 715940-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray, C. G., S. C. Cowley, K. K. Cheung, and F. E. Nano. 2002. The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol. Lett. 21553-56. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, A. M., H. Lehnherr, X. Wang, V. Mobley, and D. J. Jin. 2003. Escherichia coli SspA is a transcription activator for bacteriophage P1 late genes. Mol. Microbiol. 481621-1631. [DOI] [PubMed] [Google Scholar]
- 18.Kieffer, T. L., S. Cowley, F. E. Nano, and K. L. Elkins. 2003. Francisella novicida LPS has greater immunobiological activity in mice than F. tularensis LPS, and contributes to F. novicida murine pathogenesis. Microbes Infect. 5397-403. [DOI] [PubMed] [Google Scholar]
- 19.Lauriano, C. M., J. R. Barker, S. S. Yoon, F. E. Nano, B. P. Arulanandam, D. J. Hassett, and K. E. Klose. 2004. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. USA 1014246-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenco, J., M. Hubalek, P. Larsson, A. Fucikova, M. Brychta, A. Macela, and J. Stulik. 2007. Proteomics analysis of the Francisella tularensis LVS response to iron restriction: induction of the F. tularensis pathogenicity island proteins IglABC. FEMS Microbiol. Lett. 26911-21. [DOI] [PubMed] [Google Scholar]
- 21.Mangan, S., and U. Alon. 2003. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA 10011980-11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohapatra, N. P., S. Soni, B. L. Bell, R. Warren, R. K. Ernst, A. Muszynski, R. W. Carlson, and J. S. Gunn. 2007. Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infect. Immun. 753305-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nano, F. E., N. Zhang, S. C. Cowley, K. E. Klose, K. K. Cheung, M. J. Roberts, J. S. Ludu, G. W. Letendre, A. I. Meierovics, G. Stephens, and K. L. Elkins. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 1866430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owen, C. R., E. O. Buker, W. L. Jellison, D. B. Lackman, and J. F. Bell. 1964. Comparative studies of Francisella tularensis and Francisella novicida. J. Bacteriol. 87676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reilly, T. J., G. S. Baron, F. E. Nano, and M. S. Kuhlenschmidt. 1996. Characterization and sequencing of a respiratory burst-inhibiting acid phosphatase from Francisella tularensis. J. Biol. Chem. 27110973-10983. [DOI] [PubMed] [Google Scholar]
- 26.Santic, M., M. Molmeret, and Y. Abu Kwaik. 2005. Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-gamma. Cell. Microbiol. 7957-967. [DOI] [PubMed] [Google Scholar]
- 27.Santic, M., M. Molmeret, K. E. Klose, S. Jones, and Y. A. Kwaik. 2005. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7969-979. [DOI] [PubMed] [Google Scholar]
- 28.Schaible, U. E., and S. H. E. Kaufmann. 2002. Studying trafficking of intracellular pathogens in antigen-presenting cells. Methods Microbiol. 31343-360. [Google Scholar]
- 29.Shen-Orr, S. S., R. Milo, S. Mangan, and U. Alon. 2002. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 3164-68. [DOI] [PubMed] [Google Scholar]
- 30.Sherlock, G., T. Hernandez-Boussard, A. Kasarskis, G. Binkley, J. C. Matese, S. S. Dwight, M. Kaloper, S. Weng, H. Jin, C. A. Ball, M. B. Eisen, P. T. Spellman, P. O. Brown, D. Botstein, and J. M. Cherry. 2001. The Stanford Microarray Database. Nucleic Acids Res. 29152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Telepnev, M., I. Golovliov, T. Grundstrom, A. Tarnvik, and A. Sjostedt. 2003. Francisella tularensis inhibits Toll-like receptor-mediated activation of intracellular signalling and secretion of TNF-alpha and IL-1 from murine macrophages. Cell. Microbiol. 541-51. [DOI] [PubMed] [Google Scholar]
- 32.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss, D. S., A. Brotcke, T. Henry, J. J. Margolis, K. Chan, and D. M. Monack. 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. USA 1046037-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.