Abstract
Streptococcus pneumoniae is a major cause of morbidity and mortality worldwide. Its polysaccharide capsule causes resistance to phagocytosis and interferes with the innate immune system's ability to clear infections at an early stage. Nevertheless, we found that encapsulated pneumococci are sensitive to killing by a human neutrophil granule extract. We fractionated the extract by high-performance liquid chromatography and identified α-defensins by mass spectrometry as the proteins responsible for killing pneumococci. Analysis of sensitivity to the commercial α-defensins human neutrophil proteins 1 to 3 (HNP1-3) confirmed these findings. We analyzed the sensitivities of different pneumococcal strains to HNP1-3 and found that encapsulated strains are efficiently killed at physiological concentrations (7.5 μg/ml). Surprisingly, nonencapsulated, nonvirulent pneumococci were significantly less sensitive to α-defensins. The proposed mechanisms of α-defensin resistance in nonencapsulated pneumococci is surface charge modification, e.g., by introduction of positive charge by d-alanylation of surface-exposed lipoteichoic acids. These mechanisms are surmounted by the presence of the capsule, which we hypothesize is masking these charge modifications. Hence, α-defensins in the phagolysosome of neutrophils possibly contribute to intracellular killing after antibody-mediated opsonophagocytosis of encapsulated pneumococci.
Streptococcus pneumoniae (the pneumococcus) is a major human pathogen and the cause of community-acquired pneumonia, sinusitis, otitis media, and meningitis. Despite access to antibiotic treatment, pneumococci remain a major cause of morbidity and mortality worldwide. Pneumococci colonize the nasopharynx of up to 60% of healthy children (21) and the microbial and host factors that determine if carriage leads to disease or not need to be better characterized. The role of the effectors of the innate immune system, such as neutrophils and antimicrobial peptides (AMPs), which could affect early clearance of pneumococci, is not well understood.
Neutrophils are professional phagocytes that have cytoplasmic granules (azurophilic, specific, and small storage) that contain AMPs (see below) and enzymes (e.g., neutrophil elastase and cathepsin G). Upon encountering microbes, neutrophils are activated and kill bacteria by phagocytosis (22, 24). In this process, microbes are engulfed into a phagosome that fuses with granules where reactive oxygen species are produced and AMPs and enzymes are present. Neutrophils also degranulate, killing microbes extracellularly through the release of AMPs from granules that fuse with the cytoplasmic membrane. Finally, neutrophils can form neutrophil extracellular traps (NETs), extracellular structures consisting of DNA, histones, and granule proteins that capture and kill microbes by means of a high local concentration of AMPs and histones (3, 35).
Typically, AMPs are small (12- to 100-amino-acid), amphiphilic, and cationic proteins. Two major groups of mammalian AMPs are the cathelicidins and the defensins. Defensins are small (15- to 20-residue), cysteine-rich cationic peptides with a characteristic β-sheet-rich fold. They constitute 30 to 50% of the content of azurophilic granules and are active against a wide range of bacteria, fungi, and enveloped viruses. Due to their net positive charge and hydrophobicity, defensins as well as other AMPs are thought to exert their antimicrobial effects by permeabilizing the bacterial cytoplasmic membrane (15). There are three subfamilies of defensins described for mammals, namely, α, β, and θ, which were classified based on their cysteine pairing and structure (9, 15). Human neutrophil proteins 1 to 4 (HNP1-4) are α-defensins, expressed exclusively by neutrophils.
Gram-positive pathogens can be resistant to phagocytosis, AMPs, and NETs, preventing efficient clearance by innate immune mechanisms. The pneumococcal polysaccharide capsule, of which there are more than 90 different types, makes pneumococci resistant to complement-mediated opsonophagocytosis (5). The capsule also hampers trapping by NETs (34). Since most AMPs are cationic, the introduction of positive charge via d-alanylation of surface-exposed lipoteichoic acids (LTAs) (13, 17, 18, 25) makes microbes resistant to AMPs. Indeed, we recently showed that d-alanylation in nonencapsulated pneumococci rendered them resistant to killing by antimicrobial components present in NETs (34). The combination of capsule, DNase expression, and d-alanylation of LTA makes pneumococci resistant to NETs (1, 34, 35).
Other surface proteins modifying pneumococcal surface charge and thereby potentially the susceptibility to positively charged AMPs include LytA and PgdA. The murein hydrolase LytA, a choline-binding protein bound to phosphocholine residues on LTA, is involved in autolysis and drug-induced lysis (32). Pneumococcal strains lacking choline-binding proteins, such as LytA, are more net-negatively charged than wild-type strains (30). PgdA removes negatively charged acetyl groups from the GlcNAc sugar moieties on cell wall peptidoglycan (33). The inactivation of pgdA is therefore expected to increase the net negative surface charge.
Since many pneumococcal infections are controlled by the innate immune system, we set out to identify individual AMPs from neutrophils that kill pneumococci. We fractionated human neutrophil granule extracts (hNGE) by high-performance liquid chromatography (HPLC) and tested the effect on different pneumococcal serotypes and mutants.
MATERIALS AND METHODS
hNGE isolation.
Neutrophils were purified from human blood from healthy donors as approved by the local ethics committee. A protocol using 3% dextran (MP Biomedicals, Illkirch, France) and Ficoll-Hypaque (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) was used as described in reference 1. The isolation of hNGE was performed as previously described (6, 23).
Chromatography.
hNGE was fractionated with a C4 reverse-phase HPLC column (Vydac protein C4 column; column length of 250 mm, with particle size of 5 μm). Proteins were eluted with a gradient of increasing concentrations of acetonitrile containing 0.1% (vol/vol) trifluoroacetic acid (TFA) at a flow rate of 1 ml/min. Fractions were lyophilized, dissolved in 20 mM sodium acetate buffer (pH 4.0), and tested for antimicrobial activity (see below). Separation was performed with a Waters 626 LC system connected to a photodiode array detector (Waters, Milford, MA).
Mass spectrometry.
The identity and purity of the antimicrobial component were analyzed by matrix-assisted laser desorption ionization mass spectrometry (Proteomics 4700 workstation; Applied Biosystems, Foster City, CA). The lyophilized sample was digested with 50 mM NH4HCO3, 5% acetonitrile, 2% (wt/vol) trypsin (sequencing-grade modified trypsin [Promega, Madison, WI]), and 0.15 M dithiothreitol for 4 h at 37°C. The reaction was stopped with 0.2% TFA and the reaction mixture was further mixed with matrix alpha-cyano-4-hydroxycinnamic acid (CHCA) solubilized in 50% acetonitrile-0.3% TFA at a concentration of 5 mg/ml.
Analysis of peptide mass fingerprints was obtained with the following parameters: reflectron mode, 20-kV accelerating voltage, and a low mass gate of 800 Da. Tandem mass spectrometry spectra were obtained without collision gas. The parameters for database searches (MASCOT; Matrix Science) were as follows: 30-ppm peptide mass tolerance for peptide mass fingerprint and 0.3 Da for tandem mass spectrometry spectra. The uncleaved protein was analyzed in linear mode with CHCA as the matrix with an internal marker (Mr, 2465.21).
Bacterial strains used.
Pneumococci were grown on blood agar plates overnight at 37°C in 5% CO2. The bacteria were subcultured in semisynthetic medium c+y (19) to an optical density at 620 nm of 0.5 and diluted in Dulbecco's phosphate-buffered saline (PBS) without Ca2+ and Mg2+ (Invitrogen, Lidingö, Sweden). The following strains were used: the serotype 1 strain BHN32, the serotype 2 strain D39, the serotype 4 strain TIGR4 (31), and the serotype 9V strain I95 (8), as well as their nonencapsulated derivatives, type 1R (34), type 2R (R6) (14), type 4R (TIGR4R) (8), and type 9VR (8), respectively. The TIGR4 ΔlytA mutant (with an erythromycin cassette replacing lytA) was obtained from E. Tuomanen (12). The TIGR4R ΔlytA mutant was created by transforming TIGR4R with genomic DNA from the TIGR4 ΔlytA mutant and selecting for nonencapsulated mutants on erythromycin plates. We used PCR ligation mutagenesis (20, 29) to inactivate dltA (34) or pgdA in TIGR4 and TIGR4R. For pgdA, primers used for construction and screening of deletion alleles were 5′-taagactttctttcctgctg-3′ and 5′-TTGGGCCCggtctgttagatatttgacag-3′ flanked with ApaI for the upstream fragment and 5′-gactatccaacagagaggag-3′ and 5′-TTGGATCCgcaatccacaattcctctag-3′ flanked with BamHI for the downstream fragment (lowercase letters indicate the gene sequence, and underlining indicates the restriction enzyme site). The PCR products were ligated to an erythromycin cassette (GenBank; AB057644) containing ApaI and BamHI sites and transformed into the recipient pneumococcal strain as previously described (2). The resulting transformants were selected on blood agar plates containing erythromycin (1 μg/ml) and confirmed by PCR and sequencing of the insertion area.
Shigella flexneri (strain M90T) and Escherichia coli (Top10; Invitrogen, Lidingö, Sweden) were grown in tryptic soy broth and LB broth, respectively, overnight at 37°C and subcultured at a 1/100 dilution on the day of the experiment.
Killing assays.
The bactericidal activities of hNGE, HPLC fractions, and HNP1-3 (Hycult Biotechnology b.v., Uden, The Netherlands), all of them in sodium acetate buffer (pH 4.0), were determined by incubating bacteria (1.0 × 106/ml) in Dulbecco's PBS (yielding a pH of 7.0) at 37°C with the indicated doses for 1 h. Survivors were counted by plating serial dilutions on blood agar plates. Bacterial killing was measured as percentages of control values (obtained using bacteria incubated with sodium acetate buffer and PBS alone). Low-binding, siliconized tubes and pipette tips (Biozym, Hessich Oldendorf, Germany) were used for handling HNP1-3, hNGE, and the individual HPLC fractions. Experiments were performed at least three independent times.
Defensin binding assay.
Pneumococci were grown to mid-logarithmic phase and diluted to gain a 1.7 × 106/ml solution and incubated together with HNP1-3 (HyCult Biotechnology) at concentrations of 0.15, 1.5, and 15 μg/ml for 1 to 2 min. After centrifugation, the pellet was lysed using lysozyme (10 mg/ml) for 30 min at 37°C. An HNP1-3 enzyme-linked immunosorbent assay (ELISA) (HyCult Biotechnology) was performed with dilution series of the lysed bacteria as outlined in the manufacturer's protocol. Binding of defensins (femtograms per single bacterium) was calculated. Bacteria incubated with sodium acetate buffer were used as the control.
Statistical analyses.
The nonparametric Mann-Whitney U test was used for analysis. A P value of ≤0.05 was considered significant.
RESULTS
Killing of pneumococci by hNGE.
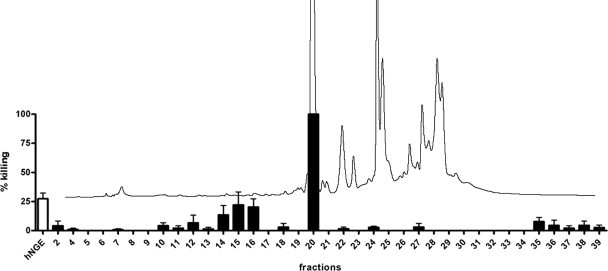
To determine the effect of neutrophil granule components on pneumococci, we prepared a crude extract from human neutrophils (hNGE) and tested its activity on the encapsulated S. pneumoniae TIGR4 strain (Fig. 1). After 1 h, a 1/10 dilution of the 6.4-mg/ml protein-containing hNGE fraction killed 27% of the pneumococcal culture. These data demonstrate that encapsulated pneumococci are sensitive to components present in human neutrophil granules.
FIG. 1.
Killing of encapsulated pneumococci by a hNGE and its individual HPLC fractions. The antimicrobial activity in hNGE (white bar) was determined by incubating encapsulated TIGR4 pneumococci with a 1/10 dilution of hNGE in sodium acetate buffer and determining survivors by plating serial dilutions. The extract was fractionated on a C4 reverse-phase HPLC column into 40 fractions and incubated with pneumococci to determine killing of individual fractions (black bars). The mean percentage of killing plus standard error of the mean (SEM) for each fraction is shown. The elution profile at A280 from the HPLC is shown (black curves). Encapsulated TIGR4 pneumococci are killed by the hNGE and efficiently by fraction 20.
Identification of active substances in hNGE.
To identify the components in hNGE active against pneumococci, we fractionated this extract with C4 HPLC columns into 40 fractions. The killing activity of each fraction was tested (Fig. 1) by incubation with TIGR4 pneumococci as described in Materials and Methods. Sodium acetate buffer, the vehicle in the fractions, was used as a control. Killing was calculated as the percent CFU recovered after incubation with individual fractions compared to that obtained after incubation with buffer alone. The elution profile (Fig. 1) at A280 shows the elution of major protein peaks in fractions 20 to 30. Fraction 20, with a protein concentration of 1.6 mg/ml, showed the most pronounced antimicrobial effect (Fig. 1). Mass spectrometry analysis showed that this fraction contained α-defensins HNP1 (gi:228797) and HNP3 (gi:229858). We also observed antimicrobial activity in fractions 10 to 16, but this was not further investigated.
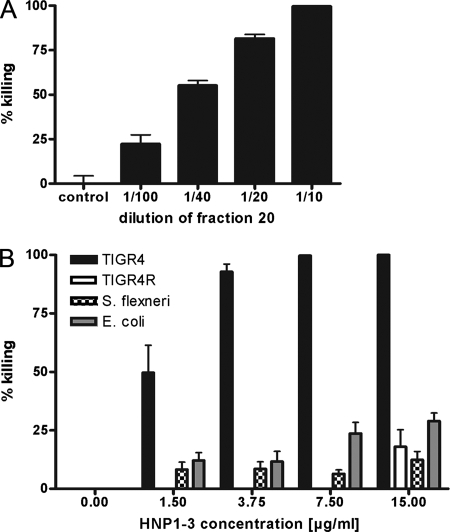
The killing activity of fraction 20 on pneumococci was dose dependent (Fig. 2A). A 1/10 dilution of the 1.6-mg/ml protein-containing fraction killed 100% of the inoculum, while a 1/100 dilution resulted in more than 20% killing. We used commercial HNP1-3 to confirm that α-defensins kill encapsulated pneumococci (Fig. 2B). A concentration as low as 3.75 μg/ml killed 90% of the TIGR4 inoculum, and this activity was also dose dependent. The gram-negative bacteria Shigella flexneri and Escherichia coli were significantly less susceptible to HNP1-3 than were encapsulated pneumococci. Fifteen micrograms of HNP1-3 per milliliter killed only 25% of either of these two gram-negative organisms, a sensitivity similar to that seen for the nonencapsulated derivative TIGR4R strain (Fig. 2B). The nonencapsulated TIGR4R strain was less sensitive not only to HNP1-3 but also to hNGE (data not shown). We conclude that human HNP1-3 is the most effective component of hNGE against TIGR4 pneumococci, and the presence of the type 4 capsule confers this high sensitivity.
FIG. 2.
Killing of pneumococci and gram-negative bacteria by defensins. Bacterial killing was determined by incubating bacteria with increasing concentrations of fraction 20, which contains defensins (A), or with commercial HNP1-3 (B) and enumerating survivors. The mean values plus SEM are shown. (A) Killing of encapsulated TIGR4 by fraction 20 diluted 1/10 to 1/100 in sodium acetate buffer. Sodium acetate buffer only was used as a control. (B) Killing of encapsulated TIGR4 and nonencapsulated TIGR4R pneumococci, Shigella flexneri, and Escherichia coli by HNP1-3. Effective concentrations ranged from 0 to 15.0 μg/ml of HNP1-3. Encapsulated pneumococci are more sensitive to HNP1-3 than are nonencapsulated pneumococci or gram-negative bacteria.
Differences in pneumococcal sensitivity to HNP1-3 depend on capsule and charge.
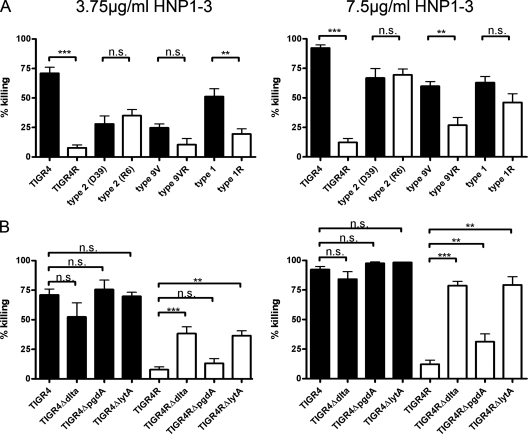
To test if capsular types other than type 4 sensitize pneumococci to HNP1-3, we incubated sets of encapsulated (serotypes 1, 2, 4, and 9V) and the corresponding nonencapsulated (“rough”-morphology serotypes 1R, 2R, R6, 4R, and 9VR) strains with HNP1-3 at 3.75 (Fig. 3, left) and 7.5 (Fig. 3, right) μg/ml. Killing was determined for either HNP1-3 concentration after a 1-hour incubation period at 37°C. Controls were incubated with sodium acetate buffer alone and were used as a reference to determine the percentage of killing. All strains were killed in a dose-dependent manner, and encapsulated strains were (with the exception of serotype 2 [see below]) killed more efficiently (Fig. 3A). We conclude that pneumococcal strains expressing capsule polysaccharide are more efficiently killed by HNP1-3 than are their nonencapsulated derivatives. These results contradict the common view that the capsule is a mediator of AMP resistance.
FIG. 3.
Dose-dependent killing of different pneumococci by HNP1-3. Bacterial killing was determined by incubating bacteria with 3.75 μg/ml (left) and 7.5 μg/ml (right) of HNP1-3. The mean values plus SEM are shown. (A) Encapsulated strains (black bars) are more sensitive than the corresponding nonencapsulated strains (white bars) to HNP1-3. (B) The absence of LTA d-alanylation (ΔdltA) or of the LytA amidase (ΔlytA) sensitizes nonencapsulated pneumococci (white bars) but not encapsulated pneumococci (black bars) to HNP1-3. The absence of peptidoglycan deacetylation (ΔpgdA) has a less pronounced effect on determining α-defensin sensitivity in nonencapsulated pneumococci. **, P ≤ 0.01; ***, P ≤ 0.001.
Strain R6 is the exception to the rule outlined above (Fig. 3A). However, R6 is not an isogenic capsule mutant of parental strain D39; rather, it also harbors a spontaneous dltA mutation (17). To further investigate this relation, we analyzed surface charge-affected dltA (d-alanylation) mutants in the TIGR4 background (Fig. 3B). Deletion of dltA from the encapsulated TIGR4 ΔdltA mutant did not increase sensitivity to HNP1-3. However, upon inactivating dltA in the nonencapsulated TIGR4R background (the TIGR4R ΔdltA mutant), we observed a marked increase in sensitivity to HNP1-3, yielding a result similar to that for the R6 strain. Hence, modification in surface charge by abolishing the introduction of positively charged d-alanine to LTAs sensitizes nonencapsulated strains (R6, the TIGR4R ΔdltA mutant) to the positively charged HNP1-3.
We also used lytA and pgdA mutants to determine their effect on defensin-mediated killing (Fig. 3B). The presence or absence of LytA and PgdA had no effect on the killing of encapsulated TIGR4. However, the nonencapsulated TIGR4R ΔlytA mutant was more sensitive than the parental strain, TIGR4R. Also, the TIGR4R ΔpgdA mutant was killed more efficiently than the wild-type strain but not in a fashion as pronounced as that for the TIGR4R ΔdltA and TIGR4R ΔlytA mutants.
Taken together, these data suggest that the introduction of positive surface charge (via d-alanylation) or the reduction of negative surface charge (proteins decreasing exposure of choline residues or removal of acetyl groups) leads to resistance to HNP1-3. However, this mechanism is inactivated in the presence of the capsule.
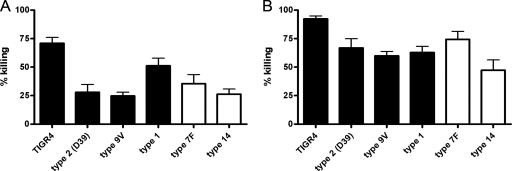
We did not find a correlation between negatively charged capsules (types 1, 2, 4, and 9V) and noncharged capsules (types 7F and 14) and susceptibility to HNP1-3 (Fig. 4). Hence, the exact mechanism by which the capsule interferes with protective surface charge modifications remains to be elucidated.
FIG. 4.
Dose-dependent HNP1-3-mediated killing of pneumococci with different capsular charges. Bacterial killing was determined by incubating bacteria with 3.75 μg/ml (A) and 7.5 μg/ml (B) of HNP1-3. The mean values plus SEM are shown. There was no correlation between negatively charged capsules (types 1, 2, 4, and 9V; black) or noncharged capsules (types 7F and 14; white) and susceptibility to HNP1-3.
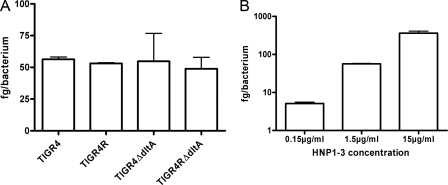
In order to determine whether differences in defensin killing are dependent on differences in binding, we analyzed the binding of defensins to different pneumococcal strains (Fig. 5A). For this, the TIGR4 strain, its isogenic capsular mutant, and the two dltA mutants were used and incubated shortly together with HNP1-3. After lysis, binding was determined by using a commercially available ELISA kit (see Materials and Methods). We did not observe any significant difference in HNP1-3 binding between the individual strains. However, incubating the TIGR4 strain with different concentrations of HNP1-3 clearly showed a dose-dependent binding (Fig. 5B).
FIG. 5.
Binding of HNP1-3 to different pneumococcal strains. Binding was determined using an ELISA, as outlined in Materials and Methods. Binding was calculated as femtograms of HNP1-3 per single bacterium. One representative experiment is shown. (A) Binding of HNP1-3 (1.5 μg/ml) to TIGR4 pneumococci and isogenic capsular and dltA mutants. No significant difference between the strains in the binding of defensins was detected. (B) Dose-dependent binding of HNP1-3 (0.15, 1.5, and 15 μg/ml) to TIGR4 pneumococci.
DISCUSSION
Pneumococci are major human pathogens that produce an antiphagocytic polysaccharide capsule. Nonencapsulated pneumococci are virtually avirulent in murine models. Interestingly, we found here that encapsulated strains are killed more efficiently by α-defensins than are nonencapsulated strains in vitro. This difference in killing is not due to differences in binding of defensins to the bacteria. According to the widely accepted model, positively charged host AMPs bind to and insert into the microbial cell membrane, thereby killing the microbe (27). An increased net positive surface charge (mediated, for example, by d-alanylation of LTAs) repels cationic peptides. This model is supported by our data showing that nonencapsulated strains become sensitive to cationic α-defensins when net negative surface charge is increased by genetic inactivation of dltA. The sensitizing effect of the capsule brings in a novel aspect to this well-accepted model by putative mechanisms discussed below.
Apart from the removal of d-alanylation, the removal of the amidase LytA, a choline-binding protein, also sensitized nonencapsulated pneumococci against α-defensins. A less pronounced effect of protection against HNP1-3 could be observed for PgdA, a peptidoglycan deacetylase that confers protection from lysozyme by removing acetyl groups from N-acetylglucosamine (33). The resistance mechanism toward lysozyme was described to be due to altered substrate specificity. Here we found that this deacetylation, potentially via charge modification, also affects sensitivity toward charged AMPs but not to the same extent as d-alanylation.
The question remains as to why the above-mentioned mechanisms manifest only in the absence of the polysaccharide capsule. We found no reports addressing potential differences in d-alanylation, peptidoglycan deacetylation, or levels of choline binding proteins in encapsulated versus nonencapsulated strains that might explain why nonencapsulated strains are less sensitive to human defensins. Nevertheless, Swiatlo et al. showed that the removal of choline-binding proteins in a nonencapsulated background results in a significant alteration in the hydrophobic character of the pneumococcal cell surface (30). These findings do not exclude the possibility that the overall level of choline-binding proteins or other surface molecules is changed upon the inactivation of capsular expression.
Another explanation for the observed capsule-mediated effect might be a direct binding of AMPs to polysaccharides. Campos and coworkers described a mechanism of resistance to AMPs dependent on the Klebsiella pneumoniae capsule polysaccharide (4). Thus, a capsule polysaccharide mutant was shown to be more sensitive than the wild-type strain to human neutrophil defensin 1, β-defensin 1, lactoferrin, protamine sulfate, and polymyxin B. A higher susceptibility of encapsulated strains to defensins was also observed for Neisseria meningitidis (28). Possibly, the direct binding of AMPs to the polysaccharide moiety might lead to a higher local concentration, effectively killing encapsulated bacteria.
A third possibility is that the capsule masks the charge effects of d-alanylation, peptidoglycan deacetylation, and choline binding and thereby abrogates the protective effect of increased positive or decreased negative surface charge.
Thus, paradoxically, the pneumococcal capsule provides resistance against neutrophil phagocytosis and prevents capture in NETs (1, 34) but increases susceptibility to killing by α-defensins, which are abundant in neutrophil granules and the phagolysosome (16). We demonstrated previously that neither encapsulated nor nonencapsulated pneumococci are killed in AMP-rich NETs upon capture (1, 34) despite the presence of α-defensins (C. Urban and A. Zychlinsky, unpublished observations). It has previously been demonstrated, however, that α-defensins released from neutrophils are rapidly inactivated by serum proteins and by the ion concentration in the extracellular environment (9). The physiological concentration of α-defensins HNP1-3 within the neutrophil phagolysosome can be in the mg/ml range (7, 11), but extracellular concentrations due to degranulation are significantly lower (∼6 μg/ml) (10). The inactivating effect of salts and serum in vivo clearly reduces activity; however, the extent to which this occurs cannot be determined, and hence the in vitro concentrations (up to 15 μg/ml) used here need to be treated with caution when extrapolating into the in vivo setting. Our data, however, are consistent with the literature: in vitro individual defensins from humans kill bacteria, fungi, and enveloped viruses at peptide concentrations in the 1- to 50-μg/ml range and gram-positive bacteria at concentrations as low as 1 μg/ml (for a review, see reference 26).
We conclude that α-defensins are active against pneumococci in general; however, the capsule increases susceptibility toward α-defensins and interferes with the protective effect accomplished by d-alanylation. How this increased killing of encapsulated pneumococci translates into human infections remains to be elucidated.
Acknowledgments
We thank Björn Eilers for help with the hNGE preparation.
This research project was supported by two Marie Curie Early Stage Research Training Fellowships of the European Union's Sixth Framework Programme (called IMO-train and EIMID-EST), the Torsten and Ragnar Söderberg Foundation, the Swedish Royal Academy of Sciences, and the Swedish Research Council.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 12 May 2008.
REFERENCES
- 1.Beiter, K., F. Wartha, B. Albiger, S. Normark, A. Zychlinsky, and B. Henriques-Normark. 2006. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr. Biol. 16401-407. [DOI] [PubMed] [Google Scholar]
- 2.Bricker, A. L., and A. Camilli. 1999. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172131-135. [DOI] [PubMed] [Google Scholar]
- 3.Brinkmann, V., U. Reichard, C. Goosmann, B. Fauler, Y. Uhlemann, D. S. Weiss, Y. Weinrauch, and A. Zychlinsky. 2004. Neutrophil extracellular traps kill bacteria. Science 3031532-1535. [DOI] [PubMed] [Google Scholar]
- 4.Campos, M. A., M. A. Vargas, V. Regueiro, C. M. Llompart, S. Alberti, and J. A. Bengoechea. 2004. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect. Immun. 727107-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cross, A. S. 1990. The biologic significance of bacterial encapsulation. Curr. Top. Microbiol. Immunol. 15087-95. [DOI] [PubMed] [Google Scholar]
- 6.Dahlgren, C., and A. Karlsson. 1999. Respiratory burst in human neutrophils. J. Immunol. Methods 2323-14. [DOI] [PubMed] [Google Scholar]
- 7.Edgerton, M., S. E. Koshlukova, M. W. Araujo, R. C. Patel, J. Dong, and J. A. Bruenn. 2000. Salivary histatin 5 and human neutrophil defensin 1 kill Candida albicans via shared pathways. Antimicrob. Agents Chemother. 443310-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernebro, J., I. Andersson, J. Sublett, E. Morfeldt, R. Novak, E. Tuomanen, S. Normark, and B. H. Normark. 2004. Capsular expression in Streptococcus pneumoniae negatively affects spontaneous and antibiotic-induced lysis and contributes to antibiotic tolerance. J. Infect. Dis. 189328-338. [DOI] [PubMed] [Google Scholar]
- 9.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3710-720. [DOI] [PubMed] [Google Scholar]
- 10.Ganz, T. 1987. Extracellular release of antimicrobial defensins by human polymorphonuclear leukocytes. Infect. Immun. 55568-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganz, T., M. E. Selsted, and R. I. Lehrer. 1986. Antimicrobial activity of phagocyte granule proteins. Semin. Respir. Infect. 1107-117. [PubMed] [Google Scholar]
- 12.Haas, W., J. Sublett, D. Kaushal, and E. I. Tuomanen. 2004. Revising the role of the pneumococcal vex-vncRS locus in vancomycin tolerance. J. Bacteriol. 1868463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heaton, M. P., and F. C. Neuhaus. 1992. Biosynthesis of d-alanyl-lipoteichoic acid: cloning, nucleotide sequence, and expression of the Lactobacillus casei gene for the d-alanine-activating enzyme. J. Bacteriol. 1744707-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 1835709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenssen, H., P. Hamill, and R. E. Hancock. 2006. Peptide antimicrobial agents. Clin. Microbiol. Rev. 19491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kagan, B. L., T. Ganz, and R. I. Lehrer. 1994. Defensins: a family of antimicrobial and cytotoxic peptides. Toxicology 87131-149. [DOI] [PubMed] [Google Scholar]
- 17.Kovacs, M., A. Halfmann, I. Fedtke, M. Heintz, A. Peschel, W. Vollmer, R. Hakenbeck, and R. Bruckner. 2006. A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J. Bacteriol. 1885797-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristian, S. A., V. Datta, C. Weidenmaier, R. Kansal, I. Fedtke, A. Peschel, R. L. Gallo, and V. Nizet. 2005. d-Alanylation of teichoic acids promotes group A streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J. Bacteriol. 1876719-6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lacks, S., and R. D. Hotchkiss. 1960. A study of the genetic material determining an enzyme in Pneumococcus. Biochim. Biophys. Acta 39508-518. [DOI] [PubMed] [Google Scholar]
- 20.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49193-205. [DOI] [PubMed] [Google Scholar]
- 21.Masuda, K., R. Masuda, J. Nishi, K. Tokuda, M. Yoshinaga, and K. Miyata. 2002. Incidences of nasopharyngeal colonization of respiratory bacterial pathogens in Japanese children attending day-care centers. Pediatr. Int. 44376-380. [DOI] [PubMed] [Google Scholar]
- 22.Mayer-Scholl, A., P. Averhoff, and A. Zychlinsky. 2004. How do neutrophils and pathogens interact? Curr. Opin. Microbiol. 762-66. [DOI] [PubMed] [Google Scholar]
- 23.Mayer-Scholl, A., R. Hurwitz, V. Brinkmann, M. Schmid, P. Jungblut, Y. Weinrauch, and A. Zychlinsky. 2005. Human neutrophils kill Bacillus anthracis. PLoS Pathog. 1e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nathan, C. 2006. Neutrophils and immunity: challenges and opportunities. Nat. Rev. Immunol. 6173-182. [DOI] [PubMed] [Google Scholar]
- 25.Perego, M., P. Glaser, A. Minutello, M. A. Strauch, K. Leopold, and W. Fischer. 1995. Incorporation of d-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J. Biol. Chem. 27015598-15606. [DOI] [PubMed] [Google Scholar]
- 26.Selsted, M. E., and A. J. Ouellette. 1995. Defensins in granules of phagocytic and non-phagocytic cells. Trends Cell Biol. 5114-119. [DOI] [PubMed] [Google Scholar]
- 27.Sitaram, N., and R. Nagaraj. 1999. Interaction of antimicrobial peptides with biological and model membranes: structural and charge requirements for activity. Biochim. Biophys. Acta 146229-54. [DOI] [PubMed] [Google Scholar]
- 28.Spinosa, M. R., C. Progida, A. Tala, L. Cogli, P. Alifano, and C. Bucci. 2007. The Neisseria meningitidis capsule is important for intracellular survival in human cells. Infect. Immun. 753594-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sung, C. K., H. Li, J. P. Claverys, and D. A. Morrison. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 675190-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swiatlo, E., F. R. Champlin, S. C. Holman, W. W. Wilson, and J. M. Watt. 2002. Contribution of choline-binding proteins to cell surface properties of Streptococcus pneumoniae. Infect. Immun. 70412-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293498-506. [DOI] [PubMed] [Google Scholar]
- 32.Tomasz, A., and S. Waks. 1975. Mechanism of action of penicillin: triggering of the pneumococcal autolytic enzyme by inhibitors of cell wall synthesis. Proc. Natl. Acad. Sci. USA 724162-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vollmer, W., and A. Tomasz. 2002. Peptidoglycan N-acetylglucosamine deacetylase, a putative virulence factor in Streptococcus pneumoniae. Infect. Immun. 707176-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wartha, F., K. Beiter, B. Albiger, J. Fernebro, A. Zychlinsky, S. Normark, and B. Henriques-Normark. 2007. Capsule and d-alanylated lipoteichoic acids protect Streptococcus pneumoniae against neutrophil extracellular traps. Cell. Microbiol. 91162-1171. [DOI] [PubMed] [Google Scholar]
- 35.Wartha, F., K. Beiter, S. Normark, and B. Henriques-Normark. 2007. Neutrophil extracellular traps: casting the NET over pathogenesis. Curr. Opin. Microbiol. 1052-56. [DOI] [PubMed] [Google Scholar]