Abstract
The Escherichia coli dapB gene encodes one of the enzymes of the biosynthetic pathway leading to lysine and its immediate precursor, diaminopimelate. Expression of dapB is repressed by lysine, but no trans-acting regulator has been identified so far. Our analysis of the dapB regulatory region shows that sequences located in the −81/−118 interval upstream of the transcription start site are essential for full expression of dapB, as well as for lysine repression. Screening a genomic library for a gene that could alleviate lysine repression when present in multicopy led to the recovery of argP, a gene encoding an activating protein of the LysR-type family, known to use lysine as an effector. An argP null mutation strongly decreases dapB transcription that becomes insensitive to lysine. Purified His6-tagged ArgP protein binds with an apparent Kd of 35 nM to the dapB promoter in a gel retardation assay, provided that sequences up to −103 are present. In the presence of l-lysine and l-arginine, the binding of ArgP to dapB is partly relieved. These results fit with a model in which ArgP contributes to enhanced transcription of dapB when lysine becomes limiting.
Lysine biosynthesis in Escherichia coli involves nine successive enzymatic reactions, the last one being decarboxylation of diaminopimelate (DAP) by DAP decarboxylase, the lysA product (see Fig. S1 in the supplemental material) (reviewed in reference 19). DAP decarboxylase converts DAP, an essential molecule constituent of peptidoglycan, into l-lysine, another major cell component, and its synthesis is strictly dependent on the regulatory protein LysR, a transcriptional activator responding to the internal concentrations of DAP and lysine (24). The first eight steps are common to DAP and lysine biosynthesis. They are performed by enzymes encoded by eight genes scattered around the E. coli chromosome, all of which, except dapC, have been identified and studied in some detail. The last step is still obscure and might involve multiple nonspecific enzymes, among which is the argD product (15).
There is no single regulatory theme for these seven genes, and no trans-acting regulatory factor has been identified that could control expression of any of them in response to the lysine concentration. Two steps, performed by the dapE and dapF products, are insensitive to the lysine concentration (4, 19). Another step is controlled at the level of enzymatic activity by retroinhibition of the dapA product (19). The very first step, phosphorylation of aspartate, is performed by three isoenzymes, one of which, aspartokinase III, the lysC product, appears to be specific to the DAP-lysine pathway, whereas the other two belong to the threonine and the methionine biosynthesis pathways. Transcription of lysC is repressed through a riboswitch mechanism by direct binding of lysine to the 5′-terminal part of the lysC mRNA (25). The second step, under the control of the asd product, is common to the lysine, threonine, and methionine pathways, and expression of asd is subject to multivalent repression by these 3 amino acids. There is no attenuation-like sequence upstream of asd that could explain the complex regulation of its expression, which remains mysterious (13).
Finally, the steps performed by the dapB and dapD products are controlled by lysine through repression of the transcription of these two genes. Establishing the nucleotide sequences of dapB and dapD and identifying their transcription start site have ruled out the possible involvement of an attenuation mechanism (5, 21). The relatively high level of expression of dapB and dapD in the absence of lysine, as monitored from lacZ fusions, in contrast to their poor promoter signals, has led to the conclusion that involvement of a repressor, with lysine acting as a coeffector, was unlikely. Conversely, it was suggested that an activating molecule, antagonized by lysine, might be involved in dapB and dapD transcription (5, 21). Here, we report the identification of such an activator acting at the dapB promoter and target of the repressive lysine signals.
MATERIALS AND METHODS
Strains, cultures, and β-galactosidase assays.
The standard wild-type strain MG1655 (F− λ− rph-1) (1) and the recipient strain for the lacZ fusions, pop3125 [F− araD139 Δ(argF-lac)U169 deoC1 flbB5301 rpsL150 relA1 ptsF25 rbsR Φ(malP-lac)] (7) were from the laboratory collection. Growth of bacterial cells was performed aerobically at 37°C in rich (LB) or minimal (M63) medium (16). β-Galactosidase assays were performed, and activity was quantified as previously described (16).
Recombinant DNA techniques.
Isolation of plasmid DNA, PCR amplification using Taq DNA polymerase, digestion with restriction enzymes, ligation with T4 DNA ligase, and transformation of E. coli were carried out according to published protocols (22). The oligonucleotides used in this work are listed in Table 1.
TABLE 1.
Oligonucleotides used in this study
| Name | Sequence (5′→3′) |
|---|---|
| B1 | GCATAGCTATTCTCTTTTG |
| B2 | GCATGCAGTCATTCATCG |
| B3 | TCGACTCATGCCTTTCAC |
| B4 | ATCCCTCCCTGTTTATC |
| B5 | CATTAATTTCTAATTATCAGCG |
| B6 | TGGCGGCGTAGCGATGCGC |
| B7 | GGGGGAATTCAAAATGAGGGTATTTGCGCC |
| B8 | GGGGGAATTCAACACAACCAGGGCGCG |
| B9 | CCCGCCGACGGCTTCGGTATATGCAACCTGACACAAAATTGTGTCATAGTGCAGGGTGTAGGCTGGAGCTGCTTC |
| B10 | GAGGACAACGCCTGATATGTGCTATCAGGCATTATTTGATGGATTAATCCTGACGCATATGAATATCCTCCTTTAG |
| B11 | GGGGGAATTCATTTACCAGGAGCAGACAACAGC |
| B12 | GGGGAAGCTTTTTACGTGGGTTTGTCGCCAGC |
| argO1 | GAACTTGGTGCATCGTCG |
| argO2 | GCATTTTGTGGACCGAGCGG |
Construction of dapB-lacZ fusions.
The 154-bp SphI-NsiI DNA fragment carrying the dapB promoter region in plasmid pDB17 (5) was purified and cloned in vector pSB118 (3), yielding plasmid pDB68. The resulting 212-bp EcoRI DNA fragment was cloned in the promoter-probe plasmid pOM41 (26) and inserted by homologous recombination into the chromosome of strain pop3125, upstream of the Φ(malP-lacZ) transcriptional fusion, as described previously (11), leading to strain JCP75. Strain JCP76 was constructed similarly, except that a 120-bp EcoRV-NsiI DNA fragment carrying the dapB promoter was used. The constructions in strains JCP75 and JCP76 were checked by PCR amplification from chromosomal DNA using appropriate oligonucleotides.
Construction of argP-carrying plasmids.
Chromosomal DNA was extracted from strain MG1655 cells with the DNeasy tissue kit (Qiagen), according to the manufacturer's protocols, and partially digested with Sau3A. DNA fragments were ligated with BamHI-digested plasmid pCL57, a Bluescript derivative in which the bla gene has been replaced by a gene encoding spectinomycin resistance (6), and introduced by electrotransformation into strain JCP75. The transformed cells were plated on MacConkey-lactose agar in the presence of spectinomycin (100 μg·ml−1) and lysine (250 mM). The argP-carrying plasmid isolated in that experiment was then used as a template to amplify by PCR the argP reading frame with primers B11 and B12 (Table 1). After cleavage with EcoRI and HindIII, the argP fragment was cloned in pBAD18, downstream of the araB promoter (12).
Construction of argP strains.
Disruption of argP was performed as described previously (9). A DNA fragment carrying a chloramphenicol resistance gene was amplified using plasmid pKD3 as a template and primers B9 and B10 (Table 1), which created 55-bp extensions homologous to argP borders. Competent JCP75 cells, containing the lambda Red recombinase expression plasmid pKD46 (9), were electrotransformed with 100 ng of amplified DNA treated with DpnI and plated on LB agar in the presence of 30 μg·ml−1 chloramphenicol. After the transformants were streaked on LB agar plates without antibiotics to lose the helper plasmid, Amps Camr clones were purified, leading to strain JCP95, which was checked by PCR (with primers B7 and B8) for the argP deletion. The argP mutation was then introduced into JCP76 and MG1655 by P1 transduction and selection for chloramphenicol resistance, leading to strains JCP96 and JCP97, respectively.
Electrophoretic mobility shift experiments (EMSE).
ArgP protein bearing a C-terminal His6 tag (His6-ArgP) was produced from strain BL21(DE3) transformed with pHYD1705, a derivative of plasmid pET21b (17). Expression and purification of His6-ArgP were done as described previously (17). Strains BL21(DE3)/pHYD1705 grown in rich medium with 0.5 mM isopropyl-β-d-thiogalactopyranoside and MG1655/pBAD18-argP+ grown in rich medium with 1% arabinose overexpress His6-ArgP and intact ArgP, respectively. Crude extracts were prepared from these strains, as described previously (2). A 392-bp DNA fragment carrying the argO promoter region (17) or DNA fragments carrying various segments of the dapB promoter region were synthesized by PCR in the presence of 20 μCi of [α-32P]dATP, using appropriate pairs of oligonucleotide primers (O1 and O2 for argO) (Fig. 1; also see Fig. 3 for dapB) and chromosomal DNA or plasmid pDB17 (5) as templates. Then, 10-ng aliquots of the labeled DNA fragment were incubated for 15 min at room temperature with either various concentrations of purified His6-ArgP or 1 μg of crude extract in 20 μl of buffer B (2) supplemented with 50 mM NaCl and 1 μg of poly(dI-dC)/poly(dI-dC) competitor DNA (Pharmacia). The binding mixture was loaded on 5% polyacrylamide gels under 6-V/cm voltage and run at a constant voltage of 12 V/cm. After electrophoresis, the gels were dried and the DNA bands were revealed by autoradiography or phosphorimager analysis (Fuji FLA-3000 and Multi Gauge v3.0 software).
FIG. 1.
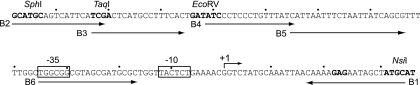
Sequence of the dapB regulatory region. The shortest arrow indicates the dapB transcription start site (+1), and the corresponding −10 and −35 sequences are boxed. Relevant restriction sites are shown in boldface type, as is the ribosome binding site. The longer arrows indicate the various oligonucleotides used to amplify that region (B1 to B6).
FIG. 3.
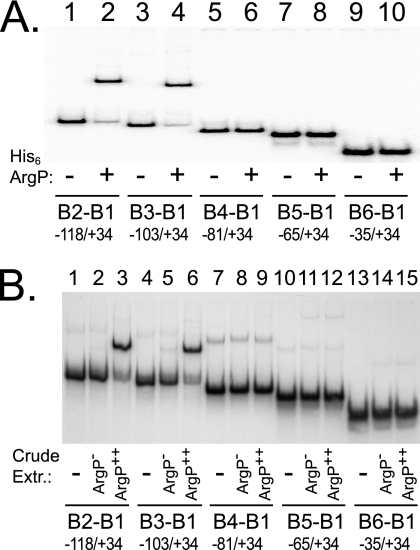
ArgP binds a site upstream of the dapB promoter. DNA fragments, extending from +34 to the upstream locations indicated at the bottom, were incubated in plain buffer (lanes −), with 1.5 μM of purified His6-ArgP (lanes + in panel A), or with crude extracts from the argP mutant JCP97 (lanes ArgP− in panel B) or from strain JCP97 harboring argP on a multicopy plasmid (lanes ArgP++ in panel B). Samples were loaded on a polyacrylamide gel, run, and analyzed by autoradiography.
RESULTS
Defining the dapB regulatory region.
A dapB-lacZ translational fusion carried by a multicopy plasmid and harboring 104 bp upstream of the dapB transcription start site is active and repressed by lysine (5). To further delineate the region of the dapB promoter involved in lysine-mediated repression, we constructed two dapB-lacZ transcriptional fusions that were integrated into the chromosome. Both fusions share the same downstream border that overlaps the dapB translational start codon, whereas they extend either 81 bp or 118 bp upstream of the dapB transcription start site (Fig. 1). The two strains harboring these fusions were grown in minimal glucose medium, with or without lysine or arginine, and β-galactosidase was assayed (Table 2). The larger fusion (strain JCP75) behaved as the fusion carried by a multicopy plasmid and showed a fourfold reduction in β-galactosidase synthesis in the presence of lysine (5). Only chemostat-induced lysine limitation can lead to a 10-fold derepression of the dapB gene (20). It also exhibited a twofold reduction in the presence of arginine. The shorter fusion (strain JCP76) was expressed at a much lower level and was insensitive to the presence of lysine or arginine. Therefore, some sequences located upstream of −81 enhance dapB transcription and are necessary for repression by lysine or, to a lesser extent, by arginine. These data suggest that this region contains a binding site for a protein that activates transcription at the dapB promoter and that is the target of lysine repression.
TABLE 2.
Regulation of dapB transcription by lysine, arginine, and ArgP
| Strain | Relevant genotypea | β-Galactosidase activity (Miller units) inb:
|
|||
|---|---|---|---|---|---|
| Minimal glucose | Minimal glucose + lysine (10 mM) | Minimal glucose + arginine (10 mM) | Minimal glucose + arginine (10 mM) + lysine (10 mM) | ||
| JCP75 | Φ(dapBp[−118/+35]-lac) | 99 | 23 | 57 | 24 |
| JCP95 | Φ(dapBp[−118/+35]-lac) ΔargP::Camr | 21 | 20 | 21 | 20 |
| JCP76 | Φ(dapBp[−81/+35]-lac) | 10 | 11 | 10 | 11 |
| JCP96 | Φ(dapBp[−81/+35]-lac) ΔargP::Camr | 11 | 11 | 10 | 11 |
The extent of the DNA fragment carrying dapBp is indicated between brackets.
Cells were grown aerobically at 37°C in M63 glucose. Overnight cultures were diluted 100-fold in the same medium and grown to an optical density at 600 nm of 0.8.
Identification of an activator of dapB transcription.
Repression by lysine of the larger dapB-lacZ fusion carried by strain JCP75 can be visualized on MacConkey lactose agar plates where colonies turn from red (Lac+) to white (Lac−) in the presence of a high concentration (250 mM) of lysine. We took advantage of this phenotype to screen for a gene encoding an activator of dapB transcription, assuming that its presence on a multicopy plasmid would reverse the negative effect of lysine. We built a genomic library of E. coli strain MG1655 in pCL57, a high-copy-number plasmid allowing selection for spectinomycin resistance (see Materials and Methods for details). The library was introduced by transformation in strain JCP75, and the transformants were plated on MacConkey lactose agar supplemented with spectinomycin and lysine. Among approximately 18,000 independent colonies, seven clones exhibited a reddish color. They were purified, and their plasmids were extracted and transformed back into JCP75. Only one of these plasmids was able to reconstitute the original derepressed phenotype on screening plates. It was named pDB102 and was shown to carry a 1.7-kb DNA fragment. Sequencing the borders of the insert indicated that this fragment contains only one complete reading frame, argP, which encodes a member of the LysR family of activating proteins (8, 23). The argP gene, including 183 bp of upstream regulatory sequence, was PCR amplified from the chromosome of strain MG1655, using oligonucleotides B7 and B8 (Table 1), and cloned in the unique EcoRI site of pCL57, yielding plasmid pDB105. Upon transformation into strain JCP75, this plasmid led to red colonies on MacConkey lactose agar plates supplemented with lysine, confirming that ArgP is able to enhance dapB transcription.
ArgP controls dapB transcription.
To assess the importance of ArgP in dapB expression, we inactivated the argP gene using the lambda Red recombinase procedure (9), as detailed in Materials and Methods. The argP strains carrying the longer or the shorter fusion were named JCP95 and JCP96, respectively. They were grown in minimal glucose medium, with or without lysine, and assayed for β-galactosidase activity (Table 2). Inactivating argP had no effect on the shorter fusion (JCP96), which remained at the same low level as its argP+ parent (JCP76) and was still insensitive to lysine. Conversely, the absence of ArgP strongly decreased expression of the larger fusion (compare JCP75 and JCP95) and eliminated repression by lysine.
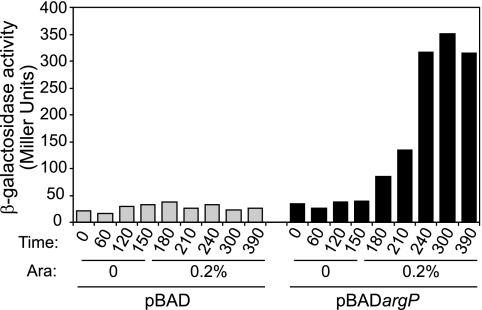
A reciprocal experiment was conducted in which ArgP was brought back to strain JCP95. The argP gene was put under the control of the arabinose-inducible promoter araBp in plasmid pBAD18 (12). Strain JCP95 was transformed with this plasmid (as well as with the empty parent vector), and expression of the dapB-lacZ fusion was monitored after addition of arabinose to the growth medium. As shown in Fig. 2, induction of ArgP synthesis was soon followed by a sharp stimulation of dapB-lacZ transcription. Altogether, these data indicate that ArgP is able to activate transcription from the dapB promoter, provided that some sequences upstream of −81 are present, and is necessary for lysine to repress dapB expression.
FIG. 2.
ArgP activates dapB transcription. Strain JCP95 transformed with either pBAD or pBADargP was grown in minimal glucose medium, and β-galactosidase activity was assayed at regular intervals. Arabinose (0.2% final concentration) was added 150 min after the start of the cultures.
ArgP binding to the dapB promoter.
To further investigate ArgP-mediated activation of dapB transcription, we purified His-tagged ArgP protein (17) and used it to perform EMSE. Radioactively labeled DNA fragments encompassing the dapB promoter region were synthesized by PCR, using oligonucleotides that created a fixed downstream border and a variable upstream one (Fig. 1). Incubation with His6-ArgP gave a single retarded band with DNA fragments extending up to −118 or −103 but none with the shorter fragments extending up to −81, −65, or −35 (Fig. 3A), demonstrating the existence of a binding site for ArgP located, at least in part, between positions −103 and −81.
Strain JCP97 (MG1655 ΔargP::Camr), expressing no ArgP, was transformed with the multicopy plasmid pDB105, expressing intact ArgP. The two strains were grown in rich medium until mid-exponential phase, and crude extracts were prepared and used to perform EMSE. Crude extracts enriched in ArgP gave a single retarded band of mobility similar to that observed with purified His6-ArgP protein, whereas crude extracts devoid of ArgP gave no retarded band (Fig. 3B; see also Fig. 5A), indicating that no protein other than ArgP is able to bind significantly to the dapB promoter region.
FIG. 5.
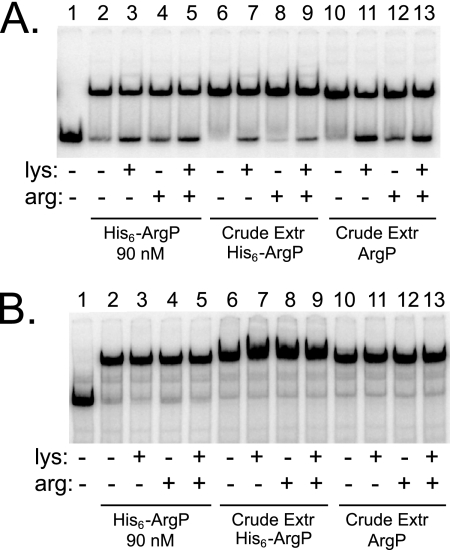
Effects of lysine and arginine on binding of ArgP at dapB and argO. DNA fragments carrying the dapB (A) and argO (B) binding sites for ArgP were incubated alone (lanes 1), with 90 nM of purified His6-ArgP (lanes 2 to 5), or with crude extracts (1 μg total protein) containing overexpressed His6-ArgP (lanes 6 to 9) or overexpressed intact ArgP (lanes 10 to 13). Lysine or arginine (10 mM) was added as indicated. Samples were loaded on a polyacrylamide gel, run, and analyzed by autoradiography.
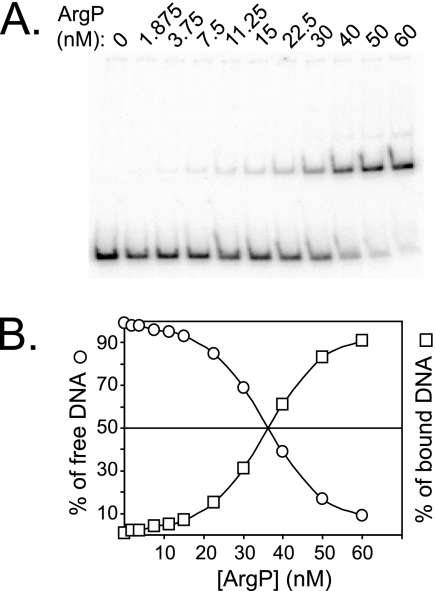
EMSE performed with various concentrations of His6-ArgP demonstrated an apparent Kd of 35 nM for the binding of ArgP to the dapB regulatory region (Fig. 4). As a control, we also performed EMSE with a mixture of two DNA fragments, the −118/+35 dapB fragment, and a −293/+109 argO fragment shown to harbor a binding site for ArgP with an apparent Kd of 6.5 nM (17). In agreement with these data and those from Fig. 4, we observed that binding of His6-ArgP to argO occurred at protein concentrations much lower than binding to dapB (data not shown).
FIG. 4.
Apparent Kd for ArgP binding near the dapB promoter. (A) The −118/+34 dapB DNA fragment was incubated with the indicated concentrations of purified His6-ArgP, loaded on a polyacrylamide gel, run, and analyzed on a phosphorimager. (B) The percentages of free and retarded DNA were quantified and plotted against the concentration of His6-ArgP.
Finally, we also investigated the effects of lysine and arginine on the binding of ArgP. As expected from earlier work (17), neither lysine nor arginine affected the binding of ArgP (or His6-ArgP) to argO (Fig. 5B). In contrast, lysine and arginine could partially displace ArgP from retarded complexes with dapB (Fig. 5A). This effect was observed with both intact ArgP or His6-ArgP in crude extracts and purified His6-ArgP. Similar experiments showed that the retarded complex was insensitive to the presence of d-lysine, ornithine, or DAP (data not shown). Therefore, binding of ArgP is specifically relieved by l-lysine and l-arginine.
Altogether, these in vitro results are in good agreement with those obtained in vivo, indicating that dapB-lacZ fusions harboring a fragment extending up to −118 or −103 are transcribed at a higher level and are subject to lysine repression, whereas a fusion extending up to −81 is not.
DISCUSSION
A DNA region located between 81 and 118 bp upstream of the dapB transcription start site is required for full expression of dapB, as well as for its repression by lysine. Gel retardation experiments using both purified His-tagged ArgP and crude cell extracts with or without ArgP showed that the same region is necessary for binding ArgP in a lysine-dependent manner. Altogether, these results indicate that dapB transcription is controlled by the activating protein ArgP, whose binding upstream of −81 is antagonized by lysine. Previous analysis of the dapB regulatory region had led us to propose such a regulatory mechanism, i.e., an apparent repression resulting from the loss of activation (5).
There is a strict correlation between the presence of ArgP and a high level of lysine-repressed dapB transcription, provided that the dapB regulatory region contains the necessary cis-acting upstream sequences. Interestingly, lysine is known to virtually abolish transcription activation of argO, a bona fide ArgP target. ArgP belongs to the family of LysR-type regulatory proteins, most of which use a small effector molecule as a coactivator (23), whereas lysine appears to inhibit ArgP function.
ArgP is essential for argO transcription (17), as LysR itself is essential for lysA transcription (24). However, ArgP plays only an auxiliary role in dapB transcription, enhancing its level about fourfold under our experimental conditions. An argP mutation does not lead to a DAP− phenotype, but the associated osmosensitivity observed in some genetic backgrounds (18) might reflect a reduced level of DAP. Evidently, a full dependence on ArgP would abolish dapB transcription in the presence of lysine, whereas synthesis of DAP needs to be sustained. Recruiting a lysine-sensitive activating protein to boost dapB transcription and synthesis of a lysine precursor under growth conditions where lysine becomes limiting is an efficient and economical regulatory strategy that allows the cell to maintain a basal level of DAP synthesis independently of the lysine concentration.
The molecular mechanisms of lysine repression of ArgP-dependent transcription appear to differ betweeen argO and dapB. Lysine traps RNA polymerase at the argO promoter, where it is recruited by ArgP (14), whereas lysine prevents binding of ArgP to the dapB promoter region. These differences might reflect a different topology of interactions between ArgP and RNA polymerase at the argO and dapB promoters. A putative primary ArgP binding site, ATTAGTTTTTCTGAT, where the conserved T-N11-A motif of the LysR-type protein binding site is underlined, is centered at −63 in the argO promoter (14). Our results show that ArgP binding requires sequences located upstream of −81 in the dapB promoter. Comparison of the genomes of various E. coli strains indicates a 100% conservation of the dapB regulatory region up to −98, where it suddenly diverges (data not shown). Only one potential ArgP binding site can be found in the −81/−98 interval, ATGCCTTTCACTGAT, which is centered at −89 and is conserved in the dapB region of closely related enterobacteria of the genera Shigella and Salmonella (data not shown). If functional, this ArgP binding site at dapB would be on the opposite side of the double helix compared to argO, which would significantly alter ArgP interaction with RNA polymerase. While this paper was under revision, very similar results were reported for lysine-sensitive activation of the Klebsiella pneumoniae gdhA gene by ArgP, and similar conclusions were drawn (10).
The mechanisms of lysine repression of two other genes involved in DAP biosynthesis, asd and dapD, remain obscure. The same rationale could be at work, namely, a basal level of transcription ensuring DAP synthesis whatever the environmental conditions and its lysine-modulated enhancement by an activator. Our preliminary experiments suggest that ArgP could also play this role.
Supplementary Material
Acknowledgments
We thank J. Gowrishankar for kindly providing plasmid pHYD1705 and P. Polard for helpful discussions.
Part of this work was supported by a grant from the Génopôle of Toulouse to C.G.
Footnotes
Published ahead of print on 23 May 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtis III, C. A. Gross, J. L. Ingraham, E. C. C. Lin, K. B. Low, Jr., B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 2.Bordes, P., J. Bouvier, A. Conter, A. Kolb, and C. Gutierrez. 2002. Transient repressor effect of Fis on the growth phase-regulated osmE promoter of Escherichia coli K12. Mol. Genet. Genomics 268206-213. [DOI] [PubMed] [Google Scholar]
- 3.Bouvier, J., A. P. Pugsley, and P. Stragier. 1991. A gene for a new lipoprotein in the dapA-purC interval of the Escherichia coli chromosome. J. Bacteriol. 1735523-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouvier, J., C. Richaud, W. Higgins, O. Bogler, and P. Stragier. 1992. Cloning, characterization, and expression of the dapE gene of Escherichia coli. J. Bacteriol. 1745265-5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvier, J., C. Richaud, F. Richaud, J. C. Patte, and P. Stragier. 1984. Nucleotide sequence and expression of the Escherichia coli dapB gene. J. Biol. Chem. 25914829-14834. [PubMed] [Google Scholar]
- 6.Campo, N., M.-L. Daveran-Mingot, K. Leenhouts, P. Ritzenthaler, and P. Le Bourgeois. 2002. Cre-loxP recombination system for large genome rearrangements in Lactococcus lactis. Appl. Environ. Microbiol. 682359-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104541-555. [DOI] [PubMed] [Google Scholar]
- 8.Celis, R. T. 1999. Repression and activation of arginine transport genes in Escherichia coli K 12 by the ArgP protein. J. Mol. Biol. 2941087-1095. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goss, T. J. 18 April 2008. The ArgP protein stimulates the Klebsiella pneumoniae gdhA promoter in a lysine-sensitive manner. J. Bacteriol. doi: 10.1128/JB.00295-08. [DOI] [PMC free article] [PubMed]
- 11.Gutierrez, C., and J. C. Devedjian. 1991. Osmotic induction of gene osmC expression in Escherichia coli K12. J. Mol. Biol. 220959-973. [DOI] [PubMed] [Google Scholar]
- 12.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haziza, C., P. Stragier, and J. C. Patte. 1982. Nucleotide sequence of the asd gene of Escherichia coli: absence of a typical attenuation signal. EMBO J. 1379-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laishram, R. S., and J. Gowrishankar. 2007. Environmental regulation operating at the promoter clearance step of bacterial transcription. Genes Dev. 211258-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ledwidge, R., and J. S. Blanchard. 1999. The dual biosynthetic capability of N-acetylornithine aminotransferase in arginine and lysine biosynthesis. Biochemistry 383019-3024. [DOI] [PubMed] [Google Scholar]
- 16.Miller, J. H. 1992. A short course in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Nandineni, M. R., and J. Gowrishankar. 2004. Evidence for an arginine exporter encoded by yggA (argO) that is regulated by the LysR-type transcriptional regulator ArgP in Escherichia coli. J. Bacteriol. 1863539-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nandineni, M. R., R. S. Laishram, and J. Gowrishankar. 2004. Osmosensitivity associated with insertions in argP (iciA) or glnE in glutamate synthase-deficient mutants of Escherichia coli. J. Bacteriol. 1866391-6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patte, J. C. 1996. Biosynthesis of threonine and lysine, p. 528-541. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 20.Patte, J. C., P. Morand, E. Boy, C. Richaud, and F. Borne. 1980. The relA locus and the regulation of lysine biosynthesis in Escherichia coli. Mol. Gen. Genet. 179319-325. [DOI] [PubMed] [Google Scholar]
- 21.Richaud, C., F. Richaud, C. Martin, C. Haziza, and J. C. Patte. 1984. Regulation of expression and nucleotide sequence of the Escherichia coli dapD gene. J. Biol. Chem. 25914824-14828. [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47597-626. [DOI] [PubMed] [Google Scholar]
- 24.Stragier, P., F. Richaud, F. Borne, and J. C. Patte. 1983. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. I. Identification of a lysR gene encoding an activator of the lysA gene. J. Mol. Biol. 168307-320. [DOI] [PubMed] [Google Scholar]
- 25.Sudarsan, N., J. K. Wickiser, S. Nakamura, M. S. Ebert, and R. R. Breaker. 2003. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 172688-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vidal-Ingigliardi, D., and O. Raibaud. 1985. A convenient technique to compare the efficiency of promoters in Escherichia coli. Nucleic Acids Res. 135919-5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.