Abstract
Bordetella bronchiseptica is a gram-negative respiratory pathogen that infects a wide range of hosts and causes a diverse spectrum of disease. This diversity is likely affected by multiple factors, such as host immune status, polymicrobial infection, and strain diversity. In a murine model of infection, we found that the virulence of B. bronchiseptica strains, as measured by the mean lethal dose, varied widely. Strain 253 was less virulent than the typically studied strain, RB50. Transcriptome analysis showed that cyaA, the gene encoding adenylate cyclase toxin (CyaA), was the most downregulated transcript identified in strain 253 compared to that in strain RB50. Comparative genomic hybridization and genome sequencing of strain 253 revealed that the cya locus, which encodes, activates, and secretes CyaA, was replaced by an operon (ptp) predicted to encode peptide transport proteins. Other B. bronchiseptica strains from the same phylogenetic lineage as that of strain 253 also lacked the cya locus, contained the ptp genes, and were less virulent than strain RB50. Although the loss of CyaA would be expected to be counterselected since it is conserved among the classical bordetellae and believed to be important to their success, our data indicate that the loss of this toxin and the gain of the ptp genes occurred in an ancestral strain that then expanded into a lineage. This suggests that there may be ecological niches in which CyaA is not critical for the success of B. bronchiseptica.
Bordetella is a genus of Betaproteobacteria consisting of nine species, many of which cause respiratory disease. Two of the three classical bordetellae, Bordetella pertussis and Bordetella parapertussis, are highly infectious pathogens that cause the acute disease whooping cough in humans (5, 24, 37). The third, Bordetella bronchiseptica, infects a broad range of mammals resulting in a wide range of disease severities and chronically infects the upper respiratory tract (17, 37, 51). These three classical bordetellae are so closely related that it has been suggested that they be reclassified as subspecies (14, 39). B. pertussis and B. parapertussis appear to have independently emerged from a B. bronchiseptica-like progenitor through rearrangement and large-scale genome decay (12, 43). Chromosomal recombination appears to be infrequent among these bordetellae, and the population structure of each appears to be clonal (38, 39, 54).
Bordetellae express many virulence factors, such as adhesins, secretion systems, autotransporters, and toxins, that are regulated by a two-component signal transduction system, BvgAS (9, 52). B. bronchiseptica contains nearly all the genes encoding virulence factors in B. pertussis and B. parapertussis, but each subspecies expresses a different repertoire (10, 43). For example, promoter mutations in B. bronchiseptica have led to the lack of pertussis toxin (Ptx) expression, a B. pertussis-specific toxin (1). Similarly, gene loss in B. pertussis has resulted in the absence of O antigen, which is expressed by B. bronchiseptica and B. parapertussis (43). The difference in the virulence factor repertoire of each subspecies is thought to contribute to the variation in host range and severity of disease (10, 43).
Adenylate cyclase toxin (CyaA) is highly conserved, is the only secreted protein toxin known to be expressed by all three classical bordetellae, and is thought to be a main contributor to their success as respiratory pathogens (19, 27, 28, 37). CyaA is a 200-kDa, calmodulin-activated, bifunctional adenylate cyclase/hemolysin that belongs to the repeats-in-toxin family (16, 58). CyaA, activated by calmodulin present in host cells but not in bacteria, is the most active adenylate cyclase known, causing rapid supraphysiological cAMP accumulation in target cells, which inhibits invasivity, phagocytosis, and chemotaxis and causes apoptosis (3, 25, 28, 56). In a murine model of infection, CyaA contributes to pathology, lethality, and the colonization and persistence of B. bronchiseptica and B. pertussis in the lower respiratory tract (20, 23, 27, 57). The cya operon includes five genes that are responsible for the production and secretion of CyaA. cyaA encodes CyaA, cyaC encodes a protein that posttranslationally palmitoylates CyaA to form an enzymatically active toxin, and cyaB, -D, and -E genes are required for the secretion of the toxin (2, 16, 22). There is also a gene annotated as cyaX which is predicted to be a LysR family transcriptional regulator but has no known function associated with CyaA.
Despite the overall clonality of the classical bordetellae, phylogenetics and comparative genomic analyses indicate that B. bronchiseptica is more diverse than either B. pertussis or B. parapertussis (11, 12, 38, 39, 54). In fact, some strains of B. bronchiseptica are more distantly related to each other than they are to the two human-associated pathogens (12). The colonization of hosts by B. bronchiseptica can lead to anything from asymptomatic infection to lethal pneumonia (17, 37). The dose of B. bronchiseptica required to kill the host can differ up to 100,000-fold between isolates using a murine model of infection (18, 19). Strains of B. bronchiseptica can differ in virulence factor expression (4, 18, 34, 38, 47), and regulation of gene expression can correlate with phylogenetic lineage (15, 38). Considering observations like these, it has been hypothesized that different phylogenetic lineages of B. bronchiseptica may utilize distinct sets of virulence factors (11, 12, 37, 39). However, the relation of strain virulence and virulence factor expression to phylogenetic lineage has not been addressed.
Here, we test if B. bronchiseptica strains of different phylogenetic lineages use distinct sets of virulence factors that relate to virulence. To identify strains that are most likely to differ in virulence factor expression, we screened for strains that differed in virulence, as measured by the mean lethal dose (LD50) in a murine model of infection. Then, whole-genome and phylogenetic approaches were used to examine these strains to determine which virulence factors differed and whether these differences were linked phylogenetically. We found that CyaA was not expressed by a hypovirulent isolate of B. bronchiseptica, strain 253, due to a replacement of the cya operon with a novel set of genes (ptp) predicted to be involved in peptide transport. All strains from the same phylogenetic lineage as that of strain 253 also lacked cyaA, contained the ptp genes, and were less virulent than strain RB50. All strains analyzed from other lineages contained cyaA and lacked the ptp genes. These data support the conclusion that CyaA is not present in all classical bordetellae and is not essential to the success of B. bronchiseptica and that different lineages of this species contain distinct sets of virulence factors that correlate with their virulence. Furthermore, these data are consistent with the overarching idea that the diversity of B. bronchiseptica-related disease and broad host range may be due, in part, to the distinct sets of virulence factors used by strains of different phylogenetic lineages (10, 12, 15).
MATERIALS AND METHODS
Bacterial strains and growth.
B. bronchiseptica isolates, providers, locations, dates, and sites of isolation are listed in Table S3 in the supplemental material. A more detailed clinical report was not available for these isolates. All strains were maintained on Bordet-Gengou agar (Difco, Sparks, MD) containing 10% sheep's blood (Hema Resources, Aurora, OR) with 20 μg/ml streptomycin (Sigma, St. Louis, MO). For inoculation, bacteria were grown overnight at 37°C in Stainer-Scholte (SS) broth to the mid-logarithmic phase, bacterial density was measured by the optical density at 600 nm (OD600), and bacteria were diluted in sterile phosphate-buffered saline (Omnipur, Gibbstown, NJ) to the appropriate concentration. We confirmed inocula by plating dilutions on Bordet-Gengou agar and counting the resulting colonies after 2 days of incubation at 37°C (23, 29).
Animal experiments.
Four- to six-week-old C57BL/6 mice were obtained from Jackson Laboratories (Bar Harbor, ME) and were bred in our specific pathogen- and Bordetella species-free rooms. For inoculation, mice were lightly sedated with 5% isoflurane (IsoFlo; Abbott Laboratories) in oxygen and the indicated number of CFU of B. bronchiseptica in 50 μl of phosphate-buffered saline (Omnipur, Gibbstown, NJ) was gently pipetted onto the external nares as previously described (23, 59). This method of inoculation efficiently distributes the bacteria throughout the respiratory tract (29). For survival curves, groups of three to four mice were inoculated with the indicated dose and the percent survival was monitored over a 28-day period. Mice with lethal bordetellosis, indicated by ruffled fur, labored breathing, and diminished responsiveness, were euthanized to alleviate unnecessary suffering (23, 36, 44). The statistical significance was calculated using a Fisher's exact test where groups of mice were compared in terms of survival or death at the same given dose of two different strains. Convalescent-phase sera were generated by inoculating mice with 105.7 CFU of either RB50, RB50ΔcyaA, or 253 and collecting sera 28 days postinoculation as previously described (29). To quantify the number of bacteria in the respiratory organs, groups of three to four mice were sacrificed at the indicated time point, and bacterial numbers in the lungs, tracheas, and nasal cavities were quantified as previously described (23, 29). The mean ± the standard error was determined for each treatment group. The statistical significance between strains in the bacterial load was calculated by using an analysis of covariance in Minitab (v13.30; Minitab Inc.). The explanatory variable used was the bacterial strain, and a covariate for day was fitted to control for the change in load over time. A significance level was set at P values of ≤0.05. All animal experiments were repeated at least twice with similar results. All mice were maintained in Pennsylvania State University approved housing facilities and were closely monitored in accordance with institutional guidelines and IACUC regulations.
RNA isolation and preparation of labeled cDNA.
For RNA isolation, two independent biological replicates of strains RB50 and 253 were grown in SS broth supplemented with 40 μg/ml streptomycin. Bacteria were then subcultured at a starting OD600 of 0.02 into 50 ml of SS broth and grown at 37°C for 24 h to the logarithmic phase (OD600, 1.0) while shaking. Bacteria were then harvested, and total RNA was extracted with Trizol (Invitrogen, Carlsbad, CA), treated with RNase-free DNase I (Invitrogen, Carlsbad, CA), and purified using RNeasy columns (Qiagen, Valencia, CA) according to the manufacturer's instructions. A two-color hybridization format was used, and dye swap experiments were performed analogously, in which the fluorescent labels were exchanged to ensure that uneven incorporation did not confound our results. For each reaction, fluorescently labeled cDNA copies of 5 μg from the total RNA pool were prepared as previously described (40).
Microarray hybridization and data analysis.
The two differentially labeled reactions to be compared were combined and hybridized to a long-oligonucleotide microarray specific to B. bronchiseptica strain RB50 as previously described (40). Slides were then scanned using a GenePix 4000B microarray scanner and analyzed with GenePix Pro software (Axon Instruments, Union City, CA). Spots were assessed visually to identify those of low quality, and arrays were normalized so that the median of the ratio across each array was equal to 1.0. Automatically and manually flagged spots, spots for which the sum of median (635/532) signal intensities was less than or equal to 100, and spots with signal intensities below the threshold (the sum of median intensities plus one standard deviation above the mean background) were filtered out prior to analysis. Ratio data from the two biological replicates were compiled and normalized based on the total Cy3 percent intensity and Cy5 percent intensity to eliminate slide-to-slide variation. Gene expression data were then normalized to 16S rRNA. The statistical significance of the gene expression changes observed was assessed by using the significant analysis of microarrays program (53) (see Table S1 in the supplemental material). A one-class unpaired significant analysis of microarrays using a false discovery rate of 0.60% (<0.1%) was performed (see Table S1 in the supplemental material).
Comparative genomic hybridization (CGH) analysis.
For DNA isolation, strains RB50 and 253 were grown in SS broth at 37°C with shaking overnight and genomic DNA was isolated from bacterial cultures using a DNA extraction kit (Qiagen, Valencia, CA) according to manufacturer's instructions and digested with DpnII. For each labeling reaction, 2 μg of digested genomic DNA was randomly primed using Cy5 and Cy3 dye-labeled nucleotides, with BioPrime DNA labeling kits (Invitrogen, Carlsbad, CA), and the two differentially labeled reactions to be compared were combined and hybridized to a long-oligonucleotide microarray specific to B. bronchiseptica strain RB50 (40). Dye swap experiments were also performed. Regions of difference (RDs), meaning genes that are either divergent or absent, were identified by combining the conventional ratio-based ranking along with a data rotation method (46). Briefly, all points belonging to genes with an M of >0 were removed from the MA plot, and the data were then rotated counterclockwise to enable higher specificity of the candidate RDs (see Fig. S4 in the supplemental material) (46). To further separate RDs from present genes, genes were then ranked by their M* value and assigned a ROTMIX score, which is an average of two posterior probabilities using plain and rotated data (50). M is the average log ratio of signal intensity of the sample strain over the signal intensity of the index strain, and M* is a rotated log ratio of M, as previously described (46). A is the average log intensity, as previously described (46). Each estimated posterior probability was derived using a two-component Gaussian model. Genes were then considered RDs by using a threshold of 0.6 to 1, with 1 representing the highest confidence of a gene being an RD.
qRT-PCR, RT-PCR, and PCR.
Quantitative real-time PCR (qRT-PCR) was completed as previously described (40), and RNA was extracted as described for the microarray experiment. RNA (1 μg) from each biological replicate was reverse transcribed using 300 ng of random oligonucleotide hexamers and SuperScript III RTase (Invitrogen, Carlsbad, CA). The resulting cDNA was diluted 1:1,000, and 1 μl was used in qRT-PCRs containing 300-nM primers designed with Primer Express software (Applied Biosystems, Foster City, CA) and 2× SYBR Green PCR master mix (Applied Biosystems, Foster City, CA). Primer sequences are listed in the supplemental material (Table S1). To confirm the lack of DNA contamination, reactions without reverse transcriptase were performed. Dissociation curve analysis was performed for verification of product homogeneity. Threshold fluorescence was established within the geometric phase of exponential amplification, and the cycle threshold (CT) was determined for each reaction. The CT from all biological replicates for each strain was compiled, and the 16S RNA amplicon was used as an internal control for data normalization. The fold change in transcript level was determined using the relative quantitative method (ΔΔCT) (49). For RT-PCR, the indicated bacterial strains were grown to the logarithmic phase (OD600, 0.3). The culture density was equalized between strains, and RNA was isolated using an RNAeasy kit (Qiagen, Valencia, CA). The RNA was reverse transcribed into cDNA according to the manufacturer's protocol (Promega, Madison, WI). cDNA from each strain was PCR amplified using either cyaA primers or adk primers (see Table S3 in the supplemental material) (12). For PCRs, all conditions and primers used are listed in the supplemental material (Table S3).
Western blotting.
B. bronchiseptica strains were grown to the logarithmic phase in SS broth. A total of 1 × 108 CFU of the indicated strain was treated with Laemmli sample buffer (32) and run on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels, and protein was transferred to a polyvinylidene diflouride membrane (Millipore, Bedford, MA). Membranes were probed with the monoclonal antibody 3D1 (anti-CyaA) (1:1,000 dilution) (35) and goat anti-mouse (immunoglobulin H+L) horseradish peroxidase-conjugated (1:10,000 dilution) (Southern Biotech, Birmingham, AL) antibody to detect CyaA. To detect anti-CyaA antibodies in mouse sera, 5 μl of 4 ng/ml of recombinant CyaA was run on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels, transferred to a polyvinylidene diflouride membrane (Millipore, Bedford, MA), and probed with serum from naïve, RB50-, RB50ΔcyaA-, or 253-inoculated mice (1:1,500 dilution) and goat anti-mouse (immunoglobulin H+L) horseradish peroxidase-conjugated (1:10,000) (Southern Biotech, Birmingham, AL) antibody. All membranes were visualized with ECL Western blotting detection reagents (Amersham Biosciences, Piscataway, NJ).
MLST and phylogenetic tree construction.
Multilocus sequence typing (MLST) was performed as previously described (12). All alleles were double-strand sequenced from each strain at either The Pennsylvania State University sequencing center or Davis Sequencing (San Diego, CA). The sequences were trimmed, and alleles and sequence types (STs) were designated using the Bordetella MLST database (http://pubmlst.org/bordetella) (12, 26). All sequence typing data are available at the Bordetella MLST website. Using MEGA 3.0, the alleles were concatenated and aligned, and an unweighted-pair group method using average linkages (UPGMA) tree with 1,000 bootstraps using the K2 model was constructed (31).
Genomic sequencing and analysis.
The RB50 DNA sequence and detailed annotations are available through NCBI under the accession number NC_002927. The DNA from strain 253 was isolated using a DNA extraction kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The sequencing of strain 253 was performed as previously described (43), and the genomic sequence of strain 253 was obtained from the B. bronchiseptica complex sequencing project (http://www.sanger.ac.uk/Projects/B_pertussis). The comparative genomics of strains RB50, 253, and Rhodoferax ferrireducens T118 (NC_007908) were completed using the Artemis comparison tool (8). Detecting genomic island-related signatures to detect horizontal gene transfer was completed using Alien Hunter, an interpolated variable order motifs method (55).
Accession numbers.
All microarray data are available in the supplemental material and have been deposited in MIAMExpress under the accession numbers E-MEXP-1203 and E-MEXP-1205 (Tables S1 and S2). The preliminary annotation of the genomic sequence of strain 253 was performed using Artemis (48) as previously described (43) and has been submitted to NCBI under the accession number CU633843.
RESULTS
B. bronchiseptica strain 253 is less virulent than strain RB50.
To test our hypothesis that virulence and virulence factor expression relate to phylogenetic lineage, we first screened for B. bronchiseptica strains that differ in virulence, as measured by LD50 in a murine model of infection. In an initial survey, we identified strains that had higher or lower LD50s than those of the typically studied and sequenced B. bronchiseptica strain, RB50, but that did not differ in their in vitro growth rates (37, 43; data not shown). From just five B. bronchiseptica isolates, the LD50s varied up to 1,000-fold (data not shown). Consistent with what others have observed, there was no correlation between the host species from which the strain was isolated and the ability to efficiently infect lab animals or virulence in the murine model (18; data not shown). Here, we investigate the characteristics of a hypovirulent isolate of B. bronchiseptica, strain 253. When inoculated with 106.1, 106.3, or 106.6 CFU of strain RB50, 100%, 50%, and 0% of mice survived the infection, respectively (Fig. 1A). When inoculated with 106.5, 107.3, or 107.5 CFU of strain 253, 100%, 50%, and 33% of mice survived the infection, respectively (Fig. 1B), indicating that the LD50 of strain 253 is 10-fold higher, 17.9 million more CFU, than that of strain RB50. To determine if the increased LD50 of strain 253 correlated with the decreased bacterial load in the respiratory tract, groups of three mice were inoculated with 105.7 CFU of either strain RB50 or 253. In the lungs, strain RB50 was present at 104.9, 106.3, 104.3, and 103.5 CFU and was below the limit of detection (10 CFU) on days 3, 7, 14, 28, and 56 postinoculation, respectively (Fig. 1C). Strain 253 was present at 105.6, 105.7, 103.8, and 101.5 CFU and below the limit of detection on days 3, 7, 14, 28, and 56 postinoculation (Fig. 1C). Over the course of infection, the colonization of strain 253 in the lower respiratory tract was lower than that of strain RB50 (bacterial strain, F1, 29 = 5.0; P = 0.033) when controlling for the change in load over time (day, F1, 29 = 167.9; P < 0.001). In the trachea and nasal cavity, the bacterial load of strain 253 was not significantly different from that of strain RB50 over the course of infection (see Fig. S1 in the supplemental material). Combined, these data indicate that strain 253 is less virulent and grows in numbers less efficiently than strain RB50 in the lower respiratory tract.
FIG. 1.

LD50 and quantification of lung bacterial load of B. bronchiseptica strains RB50 and 253. Groups of three to four C57BL/6 mice were inoculated intranasally with the indicated doses of strains RB50 (A) or 253 (B). Survival curves were generated by inoculating mice with the indicated dose and determining the percent survival over a 28-day period. (C) Groups of three mice were inoculated intranasally with 105.7 CFU of strain RB50 (filled squares) or strain 253 (open squares). The bacterial load in the lungs was quantified 3, 7, 14, 28, and 56 days postinoculation. The dashed line indicates the lower limit of detection. Bacterial numbers are expressed as the log10 mean ± the standard error (error bars).
CyaA is not expressed by B. bronchiseptica strain 253.
Since these experiments were performed using genetically similar, inbred mice from an individual facility and were age and gender matched, the difference in virulence can be attributed to differences in the B. bronchiseptica strains rather than other factors such as host immune status. Therefore, we sought to identify candidate bacterial genes that correlated with the decreased virulence of strain 253. Whole-genome transcriptome analysis was performed to determine which genes were differentially expressed between strains 253 and RB50 (Fig. 2A). Of 5,013 genes, 205 genes were upregulated in strain 253 relative to those in strain RB50 (see Table S1 in the supplemental material). These included 21 predicted transcriptional regulators, 16 metabolism-associated transcripts, 12 transporters, 10 electron transporters, and 98 putative, hypothetical, or probable genes (see Table S1 in the supplemental material). Unexpectedly, ptx-associated genes were upregulated in strain 253 when examined by microarray and qRT-PCR (Fig. 2A; see Table S1 in the supplemental material). However, the Ptx protein was not detected in strain 253 by Western blot analysis (see Fig. S2 in the supplemental material). No other genes encoding known virulence factors were upregulated in strain 253 relative to those in RB50 (see Table S1 in the supplemental material).
FIG. 2.
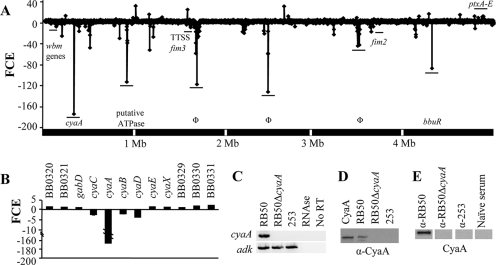
Whole-transcriptome analysis and adenylate cyclase toxin expression in B. bronchiseptica strains RB50 and 253. Comparison of whole-transcriptome analysis (A) and cya-related genes (B) between strains RB50 and 253. The x axis indicates the order of genes along the B. bronchiseptica strain RB50 5.3-megabase (Mb) chromosome. The y axis indicates the fold change in expression (FCE) of each gene. Negative-fold-change values indicate decreased expression of strain 253 genes compared to strain RB50 genes, while positive-fold-change values indicate increased gene expression. Genes of interest are labeled with corresponding underscores. Φ, phage-related gene; wbm, O-antigen-related genes; ptx, pertussis toxin gene; fim, fimbria-related genes; bbuR, urease regulator gene; cyaA, adenylate cyclase toxin gene. Error bars represent the mean ± the standard error. (C) RT-PCR of cyaA and adk in RB50, RB50ΔcyaA, and 253 with samples either including RNase treatment or lacking reverse transcriptase. (D) Western blot analysis for the presence of CyaA in bacterial lysates. Recombinant CyaA, RB50, RB50ΔcyaA, or 253 lysates were probed for CyaA using the monoclonal antibody 3D1. (E) Western blot analysis for the presence of anti-CyaA antibodies in sera from mice immunized with strains RB50, 253, and RB50ΔcyaA or naïve sera. Using these sera, recombinant CyaA was probed. α, anti.
Of 5,013 genes analyzed, 474 genes were downregulated in strain 253 relative to those in strain RB50 (see Table S1 in the supplemental material). Genes identified as being downregulated in strain 253 included 147 putative, hypothetical, or probable genes and 115 phage-related genes that are clustered along the chromosome (Fig. 2A; see Table S1 in the supplemental material). Additionally, 25 genes categorized as having roles in transcriptional regulation, 20 in metabolism, 24 in transport, and 23 in electron transport were downregulated in strain 253 (see Table S1 in the supplemental material). The following four known virulence factors were downregulated in strain 253: fimbrial serotypes 2 and 3 (fim2 and fim3, respectively), 2 of 17 O-antigen-related genes (wbmS and wbmO), eight type three secretion system (TTSS) genes, and genes in the cya operon (cyaA and cyaC) (Fig. 2A; see Table S1 in the supplemental material). While fim2, fim3, O-antigen-related, and TTSS-related genes were downregulated 2- to 12-fold in strain 253, genes required for CyaA production and activation were downregulated from 4- to 172-fold (Fig. 2B; see Table S1 in the supplemental material). Of all the downregulated transcripts identified in strain 253, the cyaA transcript had the largest fold decrease (Fig. 2B). The expression of genes directly upstream and downstream of these genes was not significantly different between strains RB50 and 253 (Fig. 2B).
To further analyze cyaA expression, RT-PCR was performed using strains RB50, 253, and RB50ΔcyaA (23). While the transcript of adenylate kinase (adk), a housekeeping gene, was detected in all of these strains, the transcript of cyaA was only detected in strain RB50 (Fig. 2C). These results were confirmed by qRT-PCR (see Table S1 in the supplemental material). Similarly, the CyaA protein could be detected by Western blot analysis in the lysate of strain RB50 but not in strains 253 or RB50ΔcyaA (Fig. 2D). Antibodies against CyaA were detected in sera from mice immunized with strain RB50, but these antibodies were not detected in sera from mice immunized with strains 253 or RB50ΔcyaA or from naïve mice (Fig. 2E). Together, these data suggest that CyaA is not expressed by strain 253 during growth in vitro or in vivo.
Genes encoding CyaA are absent in B. bronchiseptica strain 253.
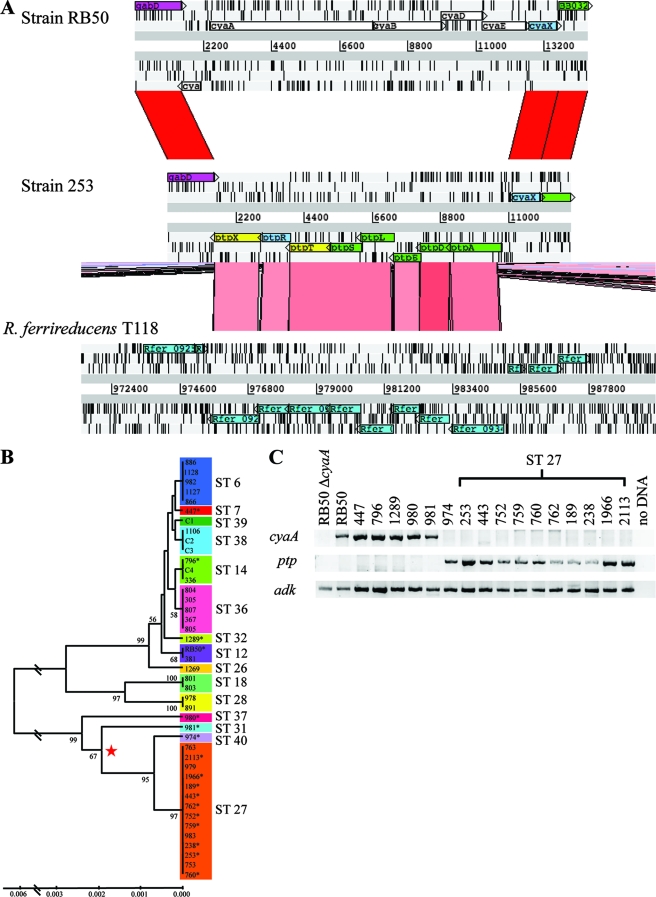
Two possibilities could lead to the lack of cyaA expression in strain 253. First, the cyaA gene may be present in strain 253 but not expressed. Second, the cyaA gene of strain 253 may be absent or so divergent that the cDNA inefficiently hybridizes to the microarray probe. These two possibilities were distinguished by CGH analysis which also allowed other candidate genes correlated with the decreased virulence of strain 253 to be identified. The absence of positive numbers indicated an absence of gene duplication events in strain 253 (Fig. 3A). Of the 5,013 genes analyzed, 252 genes were identified as RDs (absent or divergent genes) in strain 253 relative to those in the RB50 genome (see Table S2 in the supplemental material). Most of the RDs were clustered along the chromosome, indicating either a large-scale loss of genes in strain 253 or the insertion of genes in strain RB50 (Fig. 3A). Of the RDs identified, 139 are phage-related genes and comprised the majority of clustered RDs (Fig. 3A; see Table S2 in the supplemental material). Of the other RDs in strain 253, the following numbers of genes are annotated in the RB50 genome (43) as hypothetical, putative, or probable proteins (n = 57) and as involved in metabolism (n = 7), transcriptional regulation (n = 14), transport (n = 10), electron transport (n = 4), and two-component signal transduction (n = 1) (see Table S2 in the supplemental material). Interestingly, the only known virulence factors identified as RDs in strain 253 were fimbrial serotype 2 (fim2), 2 of 17 O-antigen-related genes, and 4 genes in the cya locus (cyaA, -B, -C, and -D) (Fig. 3A and 3B; see Table S2 in the supplemental material). The result that cyaB, -C, and -D were identified as RDs but were not identified as differentially expressed between strains RB50 and 253 is likely due to a low level of expression of these genes in strain RB50 (Fig. 2B and 3B). The gene upstream of the cya operon (gabD) and genes downstream of the cya operon (cyaX, BB0329, and BB0330) could be PCR amplified from both strains RB50 and 253 (Fig. 3B). While the genes in the cya operon (cyaA, -B, -C, -D, and -E) were amplified from strain RB50, these genes could not be amplified from strain 253 (Fig. 3B). The positions of these primer sets along the cya operon can be found in the supplemental material (see Table S3 in the supplemental material). Additionally, six other primer sets specific for regions across cyaA failed to produce a PCR product in strain 253 but were successful in producing a product in strain RB50 (data not shown). To further investigate the genetic basis for the apparent divergence or absence of cya-related genes, we searched the assembled contigs of the 253 genome (www.sanger.ac.uk/Projects/B_pertussis). In strain RB50, the distance between the stop codon of gabD and the start codon of cyaX is 11,223 bp (Fig. 4A). In strain 253, these genes are separated by a novel stretch of 9,630 bp that is not present in the genome of strain RB50 (Fig. 4A). While the genes upstream and downstream of the cya operon (gabD and cyaX) in strain 253 are >99% identical to strain RB50, the genes of the cya operon could not be identified in the 9,630-bp segment or in any assembled contig from the genome of strain 253 (Fig. 4A) (data not shown). Upon annotation of this novel DNA sequence, eight genes predicted to be an operon involved in peptide transport were identified (Fig. 4A). The putative function (and name) for each of these genes are putative ABC transporter, substrate binding protein (ptpA), putative peptide ABC transporter, permease protein (ptpD), putative peptide ABC transporter, permease protein (ptpE), putative oligopeptide/dipeptide ABC transporter, ATP-binding protein (ptpL), putative oligopeptide/dipeptide ABC transporter, ATP-binding protein (ptpS), putative deaminase (ptpT), transcriptional regulator, RpiR family (ptpR) and N-acyl-d-amino acid deacylase (ptpX) (Fig. 4A). No inverted repeats were found surrounding either the ptp operon in strain 253 or the cya operon in strain RB50 (data not shown). Additionally, there was no evidence to suggest that the ptp operon was recently horizontally transferred when the cya operon of strain RB50, the ptp operon of strain 253, and the surrounding regions of each operon were compared and analyzed using an interpolated variable order motifs method, which identifies differences in compositional biases at various levels (e.g., GC content, GC skew, codon, dinucleotide and amino acid bias, and structural constraints) (55; data not shown). Together, these suggest that both operons have existed in B. bronchiseptica for enough time that amelioration appears to have occurred (33). Further analysis of these sequences showed that the last 11 bp (two amino acids and a stop codon) of the cyaE gene remain intact at the 3′ end of the ptp operon in strain 253, suggesting that its ancestor may have contained the cya genes (Fig. 2E). While genes homologous to the ptp genes could not be found in any other sequenced Bordetella species, the most closely related operon was found in Rhodoferax ferrireducens strain T118, with 45 to 63% identity (Fig. 4A). The genes flanking this operon in R. ferrireducens do not have significant identity to that of strain 253 (Fig. 4A). Together, these data indicate that the cya operon was replaced by the ptp operon in strain 253.
FIG. 3.
CGH and adenylate cyclase toxin locus in strains RB50 and 253. CGH analyses of whole genome (A) or cya-related and surrounding genes (B) between strains RB50 and 253. The x axis indicates the order of genes along the B. bronchiseptica 5.3-megabase (Mb) chromosome. The y axis indicates the fold change in hybridization (FCH) for each gene. Negative-fold-change values indicate the decreased hybridization of strain 253 genomic DNA compared to that of strain RB50, while positive-fold-change values indicate increased hybridization. Genes of interest have been labeled with corresponding underscores. HP, hypothetical protein; Φ, phage-related gene; wbm, O-antigen-related genes; cyaA, adenylate cyclase toxin gene. Error bars represent the mean ± the standard error. Marks under B designate if genes upstream, within, and downstream of the cya operon could (+) or could not (−) be PCR amplified from strains RB50 or 253.
FIG. 4.
Loss of cyaA is shared among a B. bronchiseptica lineage. (A) Artemis Comparison Tool snapshot of BLASTN comparison between strain RB50 (top) and 253 (middle) and TBLASTX comparison of strain 253 (middle) and R. ferrireducens (bottom) genomes. Areas of sequence similarity are indicated by red bands (99% identity) and pink bands (45 to 64% identity). Arrows indicate the direction of transcription. Each gene is labeled as annotated in the RB50, 253, and R. ferrireducens T118 genomes. (B) A UPGMA tree with 1,000 bootstraps based on the concatenated MLST gene sequence of 43 B. bronchiseptica isolates. The colors correspond to the indicated STs. The numbers on the tree branches indicate the branch strength. All branch strengths below 50 were removed. The identification number of each strain is listed. The asterisks next to the strain numbers in panel B indicate the isolates examined in panel C. The red star in indicates the emergence of a B. bronchiseptica lineage lacking cyaA and containing the ptp genes. (C) PCR amplification of cyaA, the novel sequence predicted to encode peptide transport proteins present in 253 and adk in six non-ST27 strains, one ST40 strain, 10 ST27 strains, and RB50ΔcyaA. A sample containing no DNA template was included to ensure the absence of DNA contamination.
Loss of cyaA and presence of ptp genes are shared among a B. bronchiseptica lineage.
Previous studies showed that CyaA is highly conserved among the classical bordetellae, and it enables the bacteria to grow to higher numbers in the lower respiratory tract (23, 43). Based on these findings, it would be predicted that a strain lacking CyaA should be less fit, and we therefore hypothesized that other strains closely related to strain 253 would contain cyaA. We completed MLST analyses on strain 253 and 29 more B. bronchiseptica isolates and constructed a UPGMA tree (Fig. 4B; see Table S3 in the supplemental material). As previously described, strain RB50 was found to be in ST12 (Fig. 4B) (12). Strain 253 was identified as an ST27 strain (Fig. 4B). Five new STs were identified (ST36 to ST40), with the newly assigned ST40 being the most closely related ST to ST27 (Fig. 4B).
To determine if other strains of the same ST as strain 253 contained a CyaA with hemolytic activity, we screened our ST27 strains and 13 strains previously identified as ST27, some of which were isolated from separate continents, for β-hemolysis, a CyaA-dependent function (Fig. 4B; see Table S3 in the supplemental material) (12, 16). Of all the B. bronchiseptica strains analyzed in this study, the only isolates that were not β-hemolytic were from ST27 and ST40, which suggests that these strains lost the hemolytic activity of CyaA (data not shown). Additionally, none of the ST27 strains analyzed produced CyaA protein when analyzed by Western blotting (see Fig. S2 in the supplemental material). To determine if strains of the same ST as strain 253 contained cyaA, we attempted to PCR amplify cyaA from the indicated B. bronchiseptica strains (Fig. 4B and 4C). cyaA could not be amplified from any ST27 strain, ST40 strain, or RB50ΔcyaA (Fig. 4C) (data not shown). In contrast, cyaA could be amplified from strains of all other STs examined (Fig. 4C). To ensure that a faulty genomic DNA isolation was not the cause of the lack of cyaA amplification, a housekeeping gene, adk, was amplified from all B. bronchiseptica strains (Fig. 4C). Together, these results indicate that all strains from the phylogenetic lineage including ST27 and ST40 lack cyaA, while all other lineages examined retain and express a functional CyaA. We were able to PCR amplify a region of the ptp locus, present between gabD and cyaX, from all ST27 and ST40 strains but were unable to amplify this DNA from all other B. bronchiseptica strains examined (Fig. 4C). These data suggest that all the strains examined from the B. bronchiseptica lineage including ST27 and ST40 contain the ptp operon, while strains from all other lineages lack the ptp genes. Together, these data suggest that the cya operon was replaced by the ptp operon in the last common ancestor to the B. bronchiseptica lineage encompassing ST27 and ST40 strains.
Decreased virulence is shared among ST27 strains.
Since cya genes are absent in ST27 strains and strain 253 is less virulent than strain RB50, we sought to determine if decreased virulence is a common characteristic of ST27 strains. Groups of three to four mice were inoculated with 106.9 to 107.5 CFU of either B. bronchiseptica ST27 strain 443 or 189. The LD50 of strain 443 was 107.3 CFU, and mice did not succumb to 107.3 CFU of strain 189 (Fig. 5A). Strain 189 could be detected in the lungs, trachea, and nasal cavity 28 days postinoculation, which suggests that this strain is not an avirulent BvgAS mutant, which is cleared from the host by this time postinoculation (9; X. Zhang and E. T. Harvill, unpublished data). These data indicated that ST27 strains are less virulent than strain RB50. Groups of four mice were inoculated with 106.3 to 107.1 CFU of RB50ΔcyaA. The LD50 of RB50ΔcyaA was 106.7 CFU, a 2.5-fold increase compared to the LD50 of strain RB50 (Fig. 1A and 5B). These results demonstrate that CyaA contributes to the virulence of strain RB50. Since the LD50 of RB50ΔcyaA is more similar to the LD50 of RB50 than that of the ST27 strains, the decreased virulence of the ST27 strains is not likely to be solely due to the lack of CyaA expression.
FIG. 5.
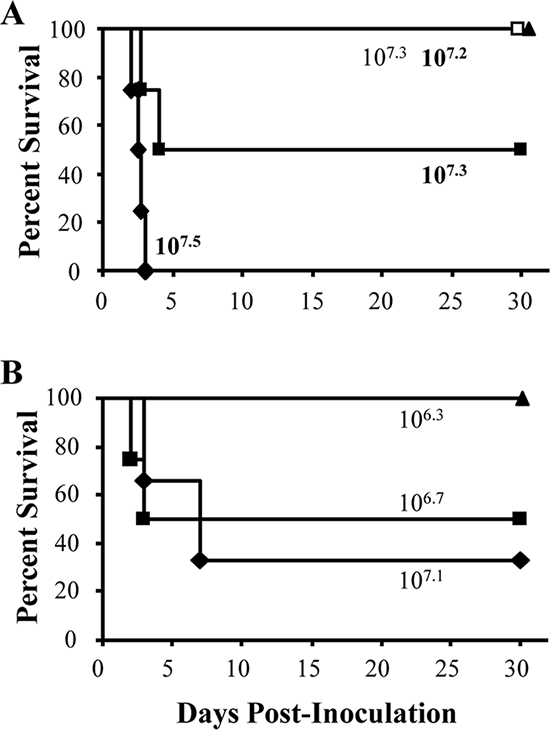
LD50 of B. bronchiseptica ST27 strains and RB50ΔcyaA. (A) Groups of three to four mice were inoculated intranasally with the indicated doses of strains 443 (filled) or 189 (open), which are both ST27 strains. (B) Groups of three to four C57BL/6 mice were inoculated intranasally with the indicated doses of RB50ΔcyaA. Survival curves were generated by inoculating mice and determining the percent survival over a 28-day period.
DISCUSSION
Although B. bronchiseptica strains are relatively closely related (12, 38, 39, 54), this species causes a wide variety of disease severities in a broad host range and individual strains differ in their expression of virulence factors (4, 17, 18, 34, 38, 47). Based on these and other observations, we hypothesized that strains in different phylogenetic lineages utilize distinct sets of virulence factors which relate to virulence. We tested this hypothesis by relating virulence, as measured by LD50 in a murine model of infection, to whole-genome analyses and phylogenetics. Transcriptome analysis revealed that cyaA was the most differentially expressed gene in the hypovirulent B. bronchiseptica strain 253 (Fig. 1 and 2). CGH analysis revealed and PCR and genomic sequencing confirmed that the cya operon was absent from strain 253 (Fig. 3). Genome sequencing and annotation were used to show that the cya operon was replaced by the ptp operon (Fig. 4A) (data not shown). Since CyaA is believed to be a critical toxin that is shared by all Bordetella species that infect mammals and that enables the bacteria to grow to higher numbers in the lower respiratory tract, a deletion of the cya operon would be expected to be strongly counterselected (23, 27, 57). Therefore, we were surprised to find an entire lineage, comprising at least two STs, that lacks the genes encoding this toxin and contains a novel set of genes in its place (Fig. 4).
While we focused on CyaA in this study, other genes were differentially expressed or identified as RDs between strains RB50 and 253 and may contribute to the difference in virulence between these two strains. Most of these were prophage-related genes present in RB50 but not in strain 253, which also caused them to be identified as differentially expressed between the two strains. Other studies comparing B. bronchiseptica isolates have also identified these prophage-related genes as RDs, indicating that these gene clusters represent insertion events into the RB50 genome rather than genes lost from strain 253 (see Tables S1 and S3 in the supplemental material) (11, 12). Another B. bronchiseptica strain lacking these prophage genes was more virulent than strain RB50, suggesting that it is unlikely that these genes are a main cause of the decreased virulence of strain 253 (A. M. Buboltz and E. T. Harvill, unpublished data). Virulence-associated genes that were downregulated and/or divergent or absent in strain 253 compared to those in strain RB50 included a few O-antigen- and TTSS-related genes, fim2, fim3, and the cya operon. The reason some O-antigen-related genes were identified as RDs and differentially expressed is because strain 253 contains the alternative O-antigen locus (43, 45) (Buboltz and Harvill, unpublished). The presence of either the classical or alternative O-antigen locus does not appear to correlate with the virulence of B. bronchiseptica strains in our murine model of infection. This suggests that these loci do not contribute to the difference in virulence between strains RB50 and 253 (Buboltz and Harvill, unpublished). Our initial attempt to determine how much the loss of CyaA contributed to the decreased virulence of strain 253 by transiently expressing the cya operon (approximately 11.1 kb) on a plasmid competent for replication in Bordetella isolates was unsuccessful due to a combination of insert size, low ligation efficiency, and retention of the plasmid during infection in vivo (30; data not shown). While it is tempting to speculate that CyaA is a main contributor to the decreased virulence of strain 253, our data showing that the LD50 of RB50ΔcyaA is closer to the LD50 of RB50 than it is to other ST27 strains indicates that CyaA could not be the sole contributor to the decreased virulence of ST27 strains (Fig. 5). The decreased expression of the other known virulence-associated genes mentioned or uncharacterized virulence-related genes may contribute to the decreased virulence of strain 253 compared to that of RB50. Additionally, other factors besides the loss of virulence factors in strain 253 may contribute to the decreased virulence of this strain. A few examples could be differences in gene expression between strains that change over the course of infection, which would not be observed during our in vitro analyses, or a gain of strain 253-specific genes that decrease the virulence of this isolate (13).
B. bronchiseptica strains lacking CyaA activity have been identified in previous studies (19, 38, 41). In a human case study, B. bronchiseptica expressed CyaA early in infection, but did not express CyaA when isolated later during the chronic infection of the same host (19). Since the isolates were otherwise indistinguishable, it was concluded that CyaA expression is important early in infection but is somehow turned off during chronic infection. Since the strains we examined all share the same replacement of the cya locus and phylogenetic lineage (Fig. 4), it appears highly unlikely that this genetic change in isolates collected from different hosts and separate continents (see Table S3 in the supplemental material) is occurring during individual infections by these strains. The widespread geographic distribution of ST27 strains demonstrates that the loss of cyaA has not resulted in a strong counterselection against this lineage and indicates that the last common ancestor that lost CyaA has since successfully spread geographically.
Previous studies have shown a correlation between the expression of particular virulence factors and the disease severity caused by certain strains of B. bronchiseptica (7, 41, 47). Since detailed clinical information was not available for the ST27 strains analyzed in this study, we cannot determine if this lineage of B. bronchiseptica causes disease. Although a clinical sample of B. bronchiseptica isolated from a human would be taken only if the patient sought medical attention, many human-related cases of B. bronchiseptica infection occur in immunocompromised patients, which can exacerbate disease (37). CyaA-deficient strains of the same multilocus enzyme electrophoretic type did not cause atrophic rhinitis in pigs, which suggested that an absence of CyaA activity correlates with a lack of disease in these animals (38, 41, 42). If these strains were found to be either ST27 or ST40 strains, it would suggest that this lineage is not associated with disease. If those strains were not of ST27 or ST40, then another cause leading to the loss of CyaA activity in B. bronchiseptica may exist (38).
CyaA allows B. bronchiseptica and B. pertussis to grow to higher numbers, persist longer in the respiratory tract, and cause more severe pathology (20, 23, 57). For the hypovirulent lineage described herein to be successful, it may be that the loss of CyaA was compensated for by some other beneficial effect. For example, an exciting possibility is that the acquisition of the ptp genes may facilitate the success of these strains despite the loss of CyaA. The predicted function of the ptp genes suggests they are involved in oligopeptide/dipeptide binding and transport which may provide a growth advantage in a particular host niche. Our laboratory is currently investigating the function and regulation of these genes, with particular focus on conditions under which the ptp genes enhance this strain's growth and the role of these genes in vivo. Another possibility is that strains lacking CyaA may also be selected for by a strong host immune response against CyaA that was conferred by prior B. bronchiseptica infection (21). Since CyaA is antigenic (Fig. 2E) (18), B. bronchiseptica strains lacking this antigen would evade anti-CyaA antibody responses, potentially giving this strain a selective advantage in host populations where CyaA expressing strains are endemic. Another possibility is that strains lacking CyaA may not be as lethal and may allow more hosts to survive and transmit the bacteria (6).
Importantly, no strain should be considered representative of the species in view of the strain and phylogenetically linked differences observed among B. bronchiseptica isolates. In a lineage of B. bronchiseptica that is closely related to B. pertussis, most strains lacked the dermonecrotic toxin gene and Ptx-related genes and had polymorphic lipopolysaccharide profiles (12). Regulation of alcaligin synthesis also appears to be associated with the phylogenetic lineage (15). Strains of the same electrophoretic type lacked CyaA activity, but the cause for this loss was not examined (38). Here, we describe a lineage of B. bronchiseptica that is hypovirulent and lacks the cya operon due to replacement by a novel ptp operon. The recovery of isolates of this lineage from different continents indicates that CyaA is not required for the success of B. bronchiseptica. The data presented herein suggest that different phylogenetic lineages utilize distinct sets of virulence factors. Therefore, it appears that even a species historically characterized by little genetic diversity is flexible enough in its metagenome to cause an overall change in virulence, which likely contributes to the wide variety of disease severities observed and the ability to persist in a broad range of host populations.
Supplementary Material
Acknowledgments
Many thanks go to the Sanger Institute for access to the unpublished genomic sequence of strain 253, Erik Hewlett and Gina Donato for reagents and critical discussion of this work, and Stephan Schüster for help in obtaining the preliminary automated annotation of the genomic sequence of strain 253. We thank Frits Mooi at the Laboratory for Vaccine-Preventable Diseases, the National Institute of Public Health and the Environment, Bilthoven, The Netherlands; Gary Sanden at the Center for Disease Prevention and Control; and Bob Livingston at the University of Missouri Research Animal Diagnostic Laboratory for B. bronchiseptica isolates. Thanks to all members of the Harvill lab, especially Daniel Wolfe, for discussion and critical review of the manuscript.
We have no conflicting financial interests. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
This work was supported by NIH grants AI 053075, AI 065507, and GM083113 (E.T.H.).
Footnotes
Published ahead of print on 13 June 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aricò, B., and R. Rappuoli. 1987. Bordetella parapertussis and Bordetella bronchiseptica contain transcriptionally silent pertussis toxin genes. J. Bacteriol. 1692847-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry, E. M., A. A. Weiss, I. E. Ehrmann, M. C. Gray, E. L. Hewlett, and M. S. Goodwin. 1991. Bordetella pertussis adenylate cyclase toxin and hemolytic activities require a second gene, cyaC, for activation. J. Bacteriol. 173720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassinet, L., P. Gueirard, B. Maitre, B. Housset, P. Gounon, and N. Guiso. 2000. Role of adhesins and toxins in invasion of human tracheal epithelial cells by Bordetella pertussis. Infect. Immun. 681934-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bemis, D. A., H. A. Greisen, and M. J. Appel. 1977. Bacteriological variation among Bordetella bronchiseptica isolates from dogs and other species. J. Clin. Microbiol. 5471-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjornstad, O. N., and E. T. Harvill. 2005. Evolution and emergence of Bordetella in humans. Trends Microbiol. 13355-359. [DOI] [PubMed] [Google Scholar]
- 6.Blaser, M. J., and D. Kirschner. 2007. The equilibria that allow bacterial persistence in human hosts. Nature 449843-849. [DOI] [PubMed] [Google Scholar]
- 7.Brockmeier, S. L., K. B. Register, T. Magyar, A. J. Lax, G. D. Pullinger, and R. A. Kunkle. 2002. Role of the dermonecrotic toxin of Bordetella bronchiseptica in the pathogenesis of respiratory disease in swine. Infect. Immun. 70481-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carver, T. J., K. M. Rutherford, M. Berriman, M. A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 213422-3423. [DOI] [PubMed] [Google Scholar]
- 9.Cotter, P. A., and J. F. Miller. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 623381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cummings, C. A., H. J. Bootsma, D. A. Relman, and J. F. Miller. 2006. Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J. Bacteriol. 1881775-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cummings, C. A., M. M. Brinig, P. W. Lepp, S. van de Pas, and D. A. Relman. 2004. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J. Bacteriol. 1861484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diavatopoulos, D. A., C. A. Cummings, L. M. Schouls, M. M. Brinig, D. A. Relman, and F. R. Mooi. 2005. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog. 1e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foreman-Wykert, A. K., and J. F. Miller. 2003. Hypervirulence and pathogen fitness. Trends Microbiol. 11105-108. [DOI] [PubMed] [Google Scholar]
- 14.Gerlach, G., F. von Wintzingerode, B. Middendorf, and R. Gross. 2001. Evolutionary trends in the genus Bordetella. Microbes Infect. 361-72. [DOI] [PubMed] [Google Scholar]
- 15.Giardina, P. C., L. A. Foster, J. M. Musser, B. J. Akerley, J. F. Miller, and D. W. Dyer. 1995. bvg repression of alcaligin synthesis in Bordetella bronchiseptica is associated with phylogenetic lineage. J. Bacteriol. 1776058-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glaser, P., H. Sakamoto, J. Bellalou, A. Ullmann, and A. Danchin. 1988. Secretion of cyclolysin, the calmodulin-sensitive adenylate cyclase-haemolysin bifunctional protein of Bordetella pertussis. EMBO J. 73997-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodnow, R. A. 1980. Biology of Bordetella bronchiseptica. Microbiol. Rev. 44722-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gueirard, P., and N. Guiso. 1993. Virulence of Bordetella bronchiseptica: role of adenylate cyclase-hemolysin. Infect. Immun. 614072-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gueirard, P., C. Weber, A. Le Coustumier, and N. Guiso. 1995. Human Bordetella bronchiseptica infection related to contact with infected animals: persistence of bacteria in host. J. Clin. Microbiol. 332002-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guiso, N., M. Rocancourt, M. Szatanik, and J. M. Alonso. 1989. Bordetella adenylate cyclase is a virulence associated factor and an immunoprotective antigen. Microb. Pathog. 7373-380. [DOI] [PubMed] [Google Scholar]
- 21.Gupta, S., M. C. Maiden, I. M. Feavers, S. Nee, R. M. May, and R. M. Anderson. 1996. The maintenance of strain structure in populations of recombining infectious agents. Nat. Med. 2437-442. [DOI] [PubMed] [Google Scholar]
- 22.Hackett, M., L. Guo, J. Shabanowitz, D. F. Hunt, and E. L. Hewlett. 1994. Internal lysine palmitoylation in adenylate cyclase toxin from Bordetella pertussis. Science 266433-435. [DOI] [PubMed] [Google Scholar]
- 23.Harvill, E. T., P. A. Cotter, M. H. Yuk, and J. F. Miller. 1999. Probing the function of Bordetella bronchiseptica adenylate cyclase toxin by manipulating host immunity. Infect. Immun. 671493-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heininger, U., K. Stehr, S. Schmitt-Grohe, C. Lorenz, R. Rost, P. D. Christenson, M. Uberall, and J. D. Cherry. 1994. Clinical characteristics of illness caused by Bordetella parapertussis compared with illness caused by Bordetella pertussis. Pediatr. Infect. Dis. J. 13306-309. [DOI] [PubMed] [Google Scholar]
- 25.Hewlett, E. L., G. M. Donato, and M. C. Gray. 2006. Macrophage cytotoxicity produced by adenylate cyclase toxin from Bordetella pertussis: more than just making cyclic AMP! Mol. Microbiol. 59447-459. [DOI] [PubMed] [Google Scholar]
- 26.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet - distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khelef, N., H. Sakamoto, and N. Guiso. 1992. Both adenylate cyclase and hemolytic activities are required by Bordetella pertussis to initiate infection. Microb. Pathog. 12227-235. [DOI] [PubMed] [Google Scholar]
- 28.Khelef, N., A. Zychlinsky, and N. Guiso. 1993. Bordetella pertussis induces apoptosis in macrophages: role of adenylate cyclase-hemolysin. Infect. Immun. 614064-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirimanjeswara, G. S., P. B. Mann, and E. T. Harvill. 2003. Role of antibodies in immunity to Bordetella infections. Infect. Immun. 711719-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166175-176. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 33.Lawrence, J. G., and H. Ochman. 1997. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44383-397. [DOI] [PubMed] [Google Scholar]
- 34.Le Blay, K., P. Gueirard, N. Guiso, and R. Chaby. 1997. Antigenic polymorphism of the lipopolysaccharides from human and animal isolates of Bordetella bronchiseptica. Microbiology 1431433-1441. [DOI] [PubMed] [Google Scholar]
- 35.Lee, S. J., M. C. Gray, L. Guo, P. Sebo, and E. L. Hewlett. 1999. Epitope mapping of monoclonal antibodies against Bordetella pertussis adenylate cyclase toxin. Infect. Immun. 672090-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mann, P. B., M. J. Kennett, and E. T. Harvill. 2004. Toll-like receptor 4 is critical to innate host defense in a murine model of bordetellosis. J. Infect. Dis. 189833-836. [DOI] [PubMed] [Google Scholar]
- 37.Mattoo, S., and J. D. Cherry. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin. Microbiol. Rev. 18326-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musser, J. M., D. A. Bemis, H. Ishikawa, and R. K. Selander. 1987. Clonal diversity and host distribution in Bordetella bronchiseptica. J. Bacteriol. 1692793-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Musser, J. M., E. L. Hewlett, M. S. Peppler, and R. K. Selander. 1986. Genetic diversity and relationships in populations of Bordetella spp. J. Bacteriol. 166230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicholson, T. L. 2007. Construction and validation of a first-generation Bordetella bronchiseptica long-oligonucleotide microarray by transcriptional profiling the Bvg regulon. BMC Genomics 8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novotny, P., A. P. Chubb, K. Cownley, and J. A. Montaraz. 1985. Adenylate cyclase activity of a 68,000-molecular-weight protein isolated from the outer membrane of Bordetella bronchiseptica. Infect. Immun. 50199-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novotny, P., A. P. Chubb, K. Cownley, J. A. Montaraz, and J. E. Beesley. 1985. Bordetella adenylate cyclase: a genus specific protective antigen and virulence factor. Dev. Biol. Stand. 6127-41. [PubMed] [Google Scholar]
- 43.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 3532-40. [DOI] [PubMed] [Google Scholar]
- 44.Pilione, M. R., L. M. Agosto, M. J. Kennett, and E. T. Harvill. 2006. CD11b is required for the resolution of inflammation induced by Bordetella bronchiseptica respiratory infection. Cell. Microbiol. 8758-768. [DOI] [PubMed] [Google Scholar]
- 45.Preston, A., A. G. Allen, J. Cadisch, R. Thomas, K. Stevens, C. M. Churcher, K. L. Badcock, J. Parkhill, B. Barrell, and D. J. Maskell. 1999. Genetic basis for lipopolysaccharide O-antigen biosynthesis in bordetellae. Infect. Immun. 673763-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Repsilber, D., A. Mira, H. Lindroos, S. Andersson, and A. Ziegler. 2005. Data rotation improves genomotyping efficiency. Biom. J. 47585-598. [DOI] [PubMed] [Google Scholar]
- 47.Roop, R. M., II, H. P. Veit, R. J. Sinsky, S. P. Veit, E. L. Hewlett, and E. T. Kornegay. 1987. Virulence factors of Bordetella bronchiseptica associated with the production of infectious atrophic rhinitis and pneumonia in experimentally infected neonatal swine. Infect. Immun. 55217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16944-945. [DOI] [PubMed] [Google Scholar]
- 49.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34374-378. [DOI] [PubMed] [Google Scholar]
- 50.Snipen, L., D. Repsilber, L. Nyquist, A. Ziegler, A. Aakra, and A. Aastveit. 2006. Detection of divergent genes in microbial aCGH experiments. BMC Bioinformatics 7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staveley, C. M., K. B. Register, M. A. Miller, S. L. Brockmeier, D. A. Jessup, and S. Jang. 2003. Molecular and antigenic characterization of Bordetella bronchiseptica isolated from a wild southern sea otter (Enhydra lutris nereis) with severe suppurative bronchopneumonia. J. Vet. Diagn. Invest. 15570-574. [DOI] [PubMed] [Google Scholar]
- 52.Stibitz, S., and M.-S. Yang. 1991. Subcellular localization and immunological detection of proteins encoded by the vir locus of Bordetella pertussis. J. Bacteriol. 1734288-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Zee, A., F. Mooi, J. Van Embden, and J. Musser. 1997. Molecular evolution and host adaptation of Bordetella spp.: phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J. Bacteriol. 1796609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vernikos, G. S., and J. Parkhill. 2006. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 222196-2203. [DOI] [PubMed] [Google Scholar]
- 56.Weingart, C. L., and A. A. Weiss. 2000. Bordetella pertussis virulence factors affect phagocytosis by human neutrophils. Infect. Immun. 681735-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiss, A. A., and M. S. Goodwin. 1989. Lethal infection by Bordetella pertussis mutants in the infant mouse model. Infect. Immun. 573757-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss, A. A., E. L. Hewlett, G. A. Myers, and S. Falkow. 1983. Tn5-induced mutations affecting virulence factors of Bordetella pertussis. Infect. Immun. 4233-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolfe, D. N., G. S. Kirimanjeswara, and E. T. Harvill. 2005. Clearance of Bordetella parapertussis from the lower respiratory tract requires humoral and cellular immunity. Infect. Immun. 736508-6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.