Abstract
Identification of determinants of human tropism of porcine endogenous retrovirus (PERV) is critical to understanding the risk of transmission of PERV to recipients of porcine xenotransplantation products. Previously, we showed that a chimeric envelope cDNA encoding the 360 N-terminal residues of the human-tropic PERV envelope class A (PERV-A) SU and the 130 C-terminal residues of the pig-tropic PERV-C SU and all of TM (PERV-A/C) showed a 100-fold decrease in infectivity titer on human cells (M. Gemeniano, O. Mpanju, D. R. Salomon, M. V. Eiden, and C. A. Wilson, Virology 346:108-117, 2006). To identify residues important for human cell infection, we performed site-directed mutagenesis on each of the nine residues, singly or in combination, that distinguish the C-terminal region of PERV-C from PERV-A. Of the nine amino acids, two single-amino-acid substitutions, Q374R and I412V, restored the infectivity of human cells to the chimeric PERV-A/C to a titer equivalent to that of PERV-A. In contrast, PERV-A/C mutant envelope Q439P resulted in undetectable infection of human cells and an approximately 1,000-fold decrease in control pig cells. Mutation of K441R rescued mutants that carried Q439P, suggesting an incompatibility between the proline residue at this position and the presence of KK in the proteolytic cleavage signal. We confirmed this incompatibility with vectors carrying PERV-A envelope mutant R462K that were also rendered noninfectious. Finally, tropism of vectors carrying PERV-C envelope mutants with only four amino acid changes in the C terminus of PERV-C envelope, NHRQ436YNRP plus K441R, was shifted to one similar to that of PERV-A. Our results show an important and previously unrecognized role for infectivity and tropism for residues at the C terminus of SU.
Porcine to human xenotransplantation holds the long-term potential to address the serious shortage of human organs. Along with the challenge to overcome the antiporcine immune response that results in a particularly aggressive rejection of porcine organs comes the risk of transmission of infectious agents. In particular, porcine endogenous retroviruses (PERVs) have been an active area of concern and investigation. Although the transmission of PERV in vivo has not been reported in human clinical trials, the release of infectious particles from primary pig cells (6, 7, 26, 37, 41, 49, 52), the in vitro infection of human cells by PERV (19, 26, 44, 52), the generation of high-titer human-tropic PERV by recombination (16), the demonstration of transmission of PERV to mice after pig islet xenotransplantation (49), and a report on the productive in vivo PERV infection of HuPAR transgenic mice (27) all support the need for continued studies on the risk of porcine to human xenotransplantation.
PERVs are gammaretroviruses, integrated at multiple loci in the genomes of all pigs (29, 34, 35, 48), and can be classified into one of three envelope classes, envelope class A (PERV-A) or B (PERV-B) (23) or C (PERV-C) (1) based on a combination of sequence variation, receptor interference, and in vitro cell line specificity (46). Of the three classes of PERV, PERV-A and -B are capable of infecting both porcine and human cells in vitro, while PERV-C has a more restricted host range and is able to infect only cells of porcine origin (46).
It is assumed that the PERV envelope, like those of all retroviruses, is initially translated as a polyprotein that is then cleaved by cellular proteases into two subunits, the surface envelope glycoprotein (SU) and the transmembrane protein (TM). The SU and TM components are found in disulfide-linked heterodimers in trimeric configurations embedded within the lipid membranes of the viral particles that were derived from the host cell plasma membrane during the process of viral assembly and release (12, 42). For gammaretroviruses, the receptor binding domains are typically found in the N terminus of SU, with the fusion peptide found in the N terminus of TM (as reviewed in reference 33). Additional determinants that are important for viral entry have been found in both the amino and carboxyl regions of gammaretroviral SU (3, 5, 18, 21, 22, 32). Although the in vitro host range of PERV envelope classes has been studied (46, 53), more detailed mapping of determinants within the envelope that are critical to human cell infection by PERV has just begun (13, 16, 50).
During in vitro stimulation and culture of primary porcine peripheral blood mononuclear cells derived from NIH miniature swine, recombinant viruses with envelope and polymerase sequences comprised in part from PERV-A and PERV-C arise that are infectious for human cells (30, 53). Lack of detection of the PERV-A/C envelope recombinant in the pig genome has led to the hypothesis that these recombinant viruses are exogenous viruses derived from recombination events between the genomic sequences of PERV-A and PERV-C (43, 54). In addition, an isolate of human-tropic PERV with a recombinant A/C envelope, PERV 14/220, has been shown to have higher titers on human cells than the prototype PERV-A (16). Two regions of the SU, including I140V in variable region A of the SU and the proline-rich region (PRR), have been shown to be critical for efficient human cell infection (16).
Two related human cDNAs have been identified that function as receptors for PERV-A, designated HuPAR-1 and HuPAR-2 (10). The human receptor for PERV-B and the factors that limit PERV-C infection of human cells have not been identified. Recently, we have constructed truncated segments of PERV-A and PERV-C SU fused to immunoglobulin G (IgG) epitope tags along with corresponding chimeric envelope proteins between PERV-A and PERV-C to map the region of the PERV envelope that facilitates receptor binding and infectivity (13). These studies revealed the following. (i) Unlike murine leukemia virus (MLV) receptor binding domains (RBDs), the N-terminal 200 residues of PERV are not sufficient to bind receptor, rather the N-terminal 360 residues of PERV A comprising the PRR domain are required to allow binding to permissive cells. (ii) The N-terminal 360 amino acids of PERV-C can also bind human 293 cells, even though 293 cells do not allow infection by PERV-C. (iii) PERV-A binding to human cells is decreased when the C-terminal 130-amino-acid region of PERV-C replaces the corresponding region of PERV-A; conversely, binding of human cells increases when the C terminus of PERV-A replaces the corresponding region of PERV-C. In addition, using retroviral vectors bearing a subcloned chimeric PERV-A/C envelope resulted in a 100-fold decrease in titer on 293 human embryonic kidney cells relative to those bearing PERV-A envelopes (13). From these studies, we have concluded that the region required for receptor binding and host range specificity differs from most other gammaretroviruses, in that it extends beyond the conventional MLV RBD including residues in the PRR for efficient receptor binding interactions. Further, our previous results implicated a role for the C terminus of SU in binding and infection of human cells.
It is of note that the C-terminal regions of the PERV-A and PERV-C SU that affect the binding and infectivity properties of PERV-C differ by only nine residues. The purpose of this study is to use site-directed mutagenesis of these nine amino acids singly or in combination to identify the specific amino acids within the C-terminal region of the PERV-C SU glycoprotein that influence binding and infection of human cells. We use the PERV-A/C chimeric envelope as a starting point for our mutagenesis studies, reasoning that a mutation that restored the PERV-A-like entry activity could result in as much as a 100-fold increase in titer. Here we report the use of site-directed mutagenesis to identify residues within the C terminus of SU that are important for human cell infection and tropism. We also identified residues whose replacement results in noninfectious PERV.
MATERIALS AND METHODS
Cells.
Three types of cells that differ in their molecular interaction with the three receptor classes of PERV were used (see Table 1). Human embryonic kidney (293) cells (ATCC, CRL-1573), 293T (obtained from Tom Dull, Cell Genesys, Foster City, CA), and swine testes ST-Iowa cells (obtained from R. Fister, Tufts University, Boston, MA) were cultured in Dulbecco's modified Eagle's medium (Cambrex BioScience, Walkersville, MD) supplemented as described below. Rabbit corneal fibroblasts (SIRC cells) expressing the human PERV-A receptor (10) (hereafter referred to as SIRC/PAR) (a kind gift from Clive Patience) were cultured in Eagle's minimum essential medium (EMEM) (Cambrex BioScience, Walkersville, MD) supplemented as described below. Both types of media were supplemented with 10% fetal bovine serum (HyClone, Logan, Utah), 1% l-glutamine, 1% penicillin/streptomycin, and 1% sodium pyruvate (Biofluids, Rockville, MD). 293 cells productively infected with PERV-A 14/220 (16) were a kind gift from Clive Patience. The ST-Iowa cells productively infected were described previously (45). All cells were cultured at 37°C with 5% CO2.
TABLE 1.
Infection and receptor binding properties of three receptor classes of PERV
Construction of mutant envelope clones.
The pC1neoPERVA/C plasmid, encoding residues 1 to 333 of PERV-A Env protein is fused in frame to the C-terminal 313 to 640 residues of the PERV-C envelope (13), served as a backbone to substitute either singly or in combination each of the nine amino acid residues of PERV-A Env for the analogous PERV-C Env amino acids in the C terminus of the PERV SU. The amino acid modifications R322G, K357M, Q374R, I412V, I428V, N436Y, H437N, Q439P, and K441R were introduced by site-directed mutagenesis according to the manufacturer's instructions (QuikChange mutagenesis kit; Stratagene, La Jolla, CA) and are shown schematically in Fig. 1. Individual amino acid changes were combined in subsequent order to give mutants with two, three, four, or five amino acid changes and accordingly are named Combo2, Combo3A, Combo4, and Combo5, respectively. Three of the four consecutive amino acids preceding the putative TM cleavage site were combined and mutated using a primer to form a mutant chimeric envelope termed Combo3B (NHRQ to YNRP at positions 436 to 439). The substitution mutation of the changes made in Combo3B were made on the backbone of Combo5 to make the retroviral mutant named Combo6. Then, Combo6 served as a backbone to introduce the substitution mutation of one of the dibasic amino acids from the putative protease cleavage site to form a mutant envelope termed Combo7. The sense primers, in combination with the corresponding antisense primers, used for the generation of the mutant envelopes corresponding to the primer names are as follows: PERV-C-R322G, 5′-GAAGGAATGGCTAGAGGAGGGAAATTCAATGTTG-3′; PERV-C-K357M, 5′-GGCACCTGCATAGGAATGGTTCCCCCATCCCACC-3′, PERV-C-Q374R, 5′-GAAGCCTTTAATAGAACCTCTGAGAGTC-3′, PERV-C-I412V, 5′-CTAAAGATTTTTGCGTTATGGTCCAAATTGT-3′, PERV-C-I428V, 5′-TATCCCGAAAAAGCAGTCCTTGATGAATATGAC-3′; PERV-C N436Y, 5′-TGACTACAGATATCATCGACAAAAGAAAGAACCC3′; PERV-C-H437N, 5′-TGACTACAGAAATAATCGACAAAAGAAAGAACC-3′; PERV-C-Q439P, 5′-TGACTACAGAAATCATCGACCAAAGAAAGAACCC-3′; PERV-C-NHQ436YNP, 5′-GACTACAGATATAATCGACCAAAGAAAGAACCC-3′; PERV-C-I468V, 5′-AGGAACAGCTGCCCTGATCAC GGGACCACAGCAGCT-3′; and PERV-CT476K, 5′-AACACAGCAGCTAGAAAAAGGACTTAGTAACCT-3′. Primers for substitution mutation at the cleavage site are PERV-C-K441R (5′-AATCATCGACAAAAAAGA GAGCCCATATCT-3′), PERV-A-R462K (5′-TATAGATATAATCGGCCAAAGAAAGAGCCCATA-3′), and PERV-A/C Combo6 K441R (5′-TATAATCGGCCAAAGAGAGAACCCATATCTCTG-3′). The sequences of the primers used are based on the cDNA sequence of the PERV-C envelope derived from cDNA of ST-Iowa pig cells exposed to NIH miniature pig plasma (13) (GenBank accession no. EU440732) with nucleotides in bold type indicating the changes made to alter the resulting residue to the PERV-NIH strain of PERV-A (53) (GenBank accession no. AF130444) All mutated cDNAs were sequenced using Big Dye chemistry on an ABI 310 (Applied Biosystems, Foster City, CA) to confirm that site-directed mutations were present and to verify that unscheduled mutations were absent. The putative protease cleavage site separating the SU glycoprotein gp70 and the p15E is as proposed previously (1, 23).
FIG. 1.
Schematic representation of a chimeric PERV envelope construct made from PERV-A and PERV-C envelopes (PERV-A/C) and alignment of the C-terminal regions of PERV-A and PERV-C SU highlighting the nine amino acids that distinguish these envelopes and the residues targeted for mutagenesis. Dots represent identical amino acids (not to scale), mutated residues are underlined, residues that differ are shown in bold type, and the residue number is based on PERV-C numbering (GenBank accession no. EU440732). Residues in italic type indicate the putative protease cleavage recognition sequences, and the arrow shows the predicted cleavage point following the dibasic amino acids(1, 23). Note that the PERV-A used in this study and in this alignment is the PERV-NIH strain (also termed PERV-1.15) previously described (53). (GenBank accession no. AF130444). The positions of PERV-A envelope SU (A) (amino acids 1 to 333) and PERV-C envelope (C) (amino acids 313 to 442 of SU plus TM) are shown. The PERV-C envelope is constructed from cDNA obtained by reverse transcription of RNA isolated from ST-Iowa cells exposed to NIH miniature pig plasma as previously described (13) (GenBank accession no. EU440732).
Generation of retroviral vector pseudotypes and infectivity assay.
As previously described, retroviral vector pseudotypes bearing each of the mutant or control envelopes were generated by calcium phosphate transient transfection (ProFection mammalian transfection system; Promega, Madison, WI) of 293T cells with three plasmids (obtained from Tom Dull, Cell Genesys, Foster City, CA) (47): (i) pC1neoPERV envelope expression plasmids expressing the mutant, chimeric, or wild-type envelope, (ii) pMLV-gagpol, expressing the core and enzymatic proteins derived from Moloney MLV, and (iii) pRT43.2Tnslβ-gal, the Moloney MLV-based retroviral vector genome encoding the β-galactosidase gene. Supernatant containing the pseudotyped viruses were collected 72 h posttransfection, filtered through an 0.45-μm filter, adjusted to a final concentration of 6 μg/ml Polybrene, and then used to expose target cells to the retroviral vectors. All target cells were seeded in 12-well plates 1 day prior to infection, and infectivity titers were determined after histochemical staining and microscopic enumeration of foci of cells expressing β-galactosidase 48 to 72 h postexposure to vector-containing supernatant as previously described (51). All infection assays included vectors pseudotyped with vesicular stomatitis virus G, generated in parallel to the PERV enveloped vectors, as a control for the transfection procedure (data not shown). Infectivity of each retroviral vector pseudotype was assayed in duplicate wells, and each assay was repeated three times.
For receptor interference assays, we used ST-Iowa cells productively infected with PERV-C or PERV-A, and 293 or SIRC cells stably expressing the human PERV-A receptor 2 cDNA (hPAR-2), productively infected with PERV-A 14/220 (16). One day prior to exposure, 5 × 104 to 1 × 105 matched uninfected and infected target cells were seeded in 12-well plates. On the day of superinfection, supernatants of the pseudotyped vectors from the 293T cells 72 h posttransfection were collected, filtered, adjusted to 6 μg/ml Polybrene, and applied in serial dilutions to the target cells. Infected cells were incubated at 37°C for 3 to 6 h and then rinsed, and the medium was replenished. Cells were further incubated for 48 h at 37°C before histochemical staining was performed as indicated above. To determine receptor interference, infectivity titer on uninfected and infected target cells is determined as described above. The ratio of the titer obtained on the infected cells to the uninfected cells is calculated to measure the extent of receptor interference.
Generation and characterization of PERV SU-IgG fusion proteins.
The PERV SU-IgG fusion proteins were comprised of a series of proteins derived by fusing in frame a portion or the entire reading frame for the PERV-SU with the rabbit immunoglobulin heavy chain, as described previously (13). The following SU-IgG fusion proteins used in this study as controls and were previously described: C-360, A-460, C-440, and A/C (13). Primers introducing mutations into the previously made PERV-A/C SU-IgG were used for constructing the plasmids encoding the PERV-A/C mutant SU-IgGs corresponding to the mutant PERV-A/C envelopes (A/C-Q374R, Combo3B, and A/C-I412V) (13). For PERV-A/C Combo4 SU-IgG, plasmid PERV-A/C SU-IgG was used as a template to acquire ApaI-NotI and ApaI-SpeI fragments bearing the rabbit IgG, and then two oligonucleotide primers were designed to introduce ApaI and SpeI sites at the 5′ and 3′ ends of the C terminus of PERV-C SU, respectively, and amplify a fragment, by PCR, from the PERV-A/C Combo4 plasmids. Conventional cloning methods were employed to ligate the PCR product of the mutants to the ApaI-NotI and ApaI-SpeI restriction fragments of the PERV-A/C SU-IgG plasmid. The plasmid constructs were then verified by restriction enzyme digestion and confirmed by DNA sequencing using Big-Dye chemistry on ABI 310 Prism sequence analyzer (Applied Biosystems, Foster City, CA).
The same procedures and methods described previously were used to generate and characterize the SU-IgG fusion proteins (13). Expression was confirmed by Western blot analysis and anti-rabbit IgG antibody enzyme-linked immunosorbent assay was used to determine the SU-IgG concentration as described previously (13). Detection of binding to various target cells by the SU-IgG proteins was determined according to the methods previously described (13).
RESULTS
Two residues present in the C terminus of PERV-C SU regulate infectivity of the PERV-A/C chimeric envelope on human cells.
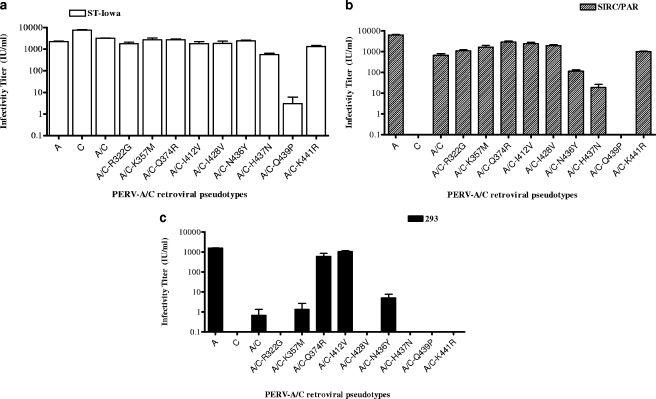
We showed that the infectivity and binding of PERV to human cells is modulated by the C terminus of SU, a region that differs by only nine residues between pig-tropic PERV-C and the analogous region of human-tropic PERV-A (13). This was shown by using a chimeric envelope that we constructed, PERV-A/C, that contains the PERV-A N-terminal 333 residues joined to the C-terminal approximately 130 residues of the PERV-C envelope. We showed that retroviral vector pseudotypes bearing this chimeric envelope have a 100-fold decrease in infectivity titer on human cells (13). To define which residues account for the reduced infectivity, we initially used site-directed mutagenesis to create PERV-A/C encoding cDNAs that express envelopes with single-amino-acid-residue substitution R322G, K357M, Q374R, I412V, I428V, N436Y, H437N, Q439P, or K441R (Fig. 1). It is of note that the last two amino acids are from the putative TM protease cleavage site (1, 13). To functionally assess the impact of each of the mutant envelopes on viral entry, we generated retroviral vectors bearing each mutant envelope and screened these vectors for infectivity on ST-Iowa cells, SIRC/PAR cells, and human 293 cells. The three cell lines chosen for analysis differ in their receptor expression and permissivity to PERV-A and PERV-C infection and binding of SU-IgG, as summarized in Table 1. As shown in Fig. 2, compared to the parental PERV-A/C chimeric envelope, most of the single-amino-acid mutations were able to support generation of infectious retroviral vectors as shown by comparable infectivity titers on ST-Iowa and SIRC/PAR cells. Similar to our prior report, vectors bearing our PERV-A/C chimeric envelope showed a 3-log-unit decrease in titer on human cells compared to those bearing PERV-A envelopes (the mean titer of PERV-A was 1,571 IU/ml; the mean titer of PERV-A/C was 0.7 IU/ml). Retroviral vectors bearing PERV-A/C with any of the following single point mutations R322G, K357M, I428V, N436Y, H437N, and K441R did not show an increased titer over the parental PERV-A/C titer and in some cases had undetectable infection on 293 cells. However, two of the single point mutations restored the titer of the PERV-A/C chimeric mutant to levels comparable to that of the wild-type envelope, PERV-A: Q374R (mean titer of 602 IU/ml) and I412V (mean titer of 1,045 IU/ml). Interestingly, the PERV-A/C mutant with the substitution mutation of Q439P demonstrates a 1,000-fold decrease or undetectable infectivity in all three types of cells tested.
FIG. 2.
Titers of retroviral vectors bearing control PERV-A (A), PERV-C (C), or PERV-A/C (A/C) envelopes with single-residue substitutions of PERV-A residues with the corresponding PERV-C residues. Shown are the average infectious units/ml of duplicate wells tested three times in each of three cell types (ST-Iowa [a], SIRC/PAR [b], and human 293 cells [c]). The error bars represent the standard errors of the means. Mutants are labeled by the amino acid position in PERV-C. See Fig. 1 for the specific amino acid change.
Receptor interference assays independently validate that the chimeric envelope mutants use the PERV-A receptor for entry.
In order to determine whether the vectors bearing mutant envelopes that restored infection on 293 cells were entering using receptors utilized by PERV-A or PERV-C, we performed a receptor interference experiment. We compared the titers of each of these vectors on the infected cells to the corresponding uninfected cells using 293 or SIRC/PAR cells productively infected with PERV-A or ST-Iowa cells productively infected with PERV-C or PERV-A. In conventional receptor interference studies, if two viruses use the same receptor, the ratio should be <0.01 (40). As shown in Table 2, we observed receptor interference in 293, SIRC/PAR, and ST-Iowa cells infected with PERV-A to all the vectors bearing any of the PERV-A/C mutants. As expected, ST-Iowa cells infected with PERV-C were resistant to vectors carrying the PERV-C envelope, but not to any of the vectors carrying the PERV-A/C envelope or the mutant derivatives. These results demonstrate that vectors bearing envelope mutants of the PERV-A/C chimera enter 293 cells using the PERV-A receptor, but not with the PERV-C receptor.
TABLE 2.
Analysis of receptor interference to retroviral vectors bearing mutant PERV envelopes
| Envelope and/or mutationa | Ratio of titer on infected to uninfected cells
|
|||
|---|---|---|---|---|
| PERV-A-infected cells
|
PERV-C-infected ST-Iowa cells | |||
| 293 | SIRC/PAR | ST-Iowa | ||
| A | <0.001 | <0.001 | <0.001 | 0.98 ± 0.06 |
| C | NDb | ND | 0.447 ± 0.032 | <0.001 |
| A/C | ND | 0.026 | <0.001 | 0.327 ± 0.045 |
| A/C-R322G | ND | 0.003 | <0.001 | 0.358 ± 0.036 |
| A/C-K357M | ND | <0.001 | <0.001 | 0.315 ± 0.013 |
| A/C-Q374R | <0.001 | 0.009 | <0.001 | 0.859 ± 0.147 |
| A/C-I412V | <0.001 | 0.012 | <0.001 | 0.847 ± 0.213 |
| A/C-I428V | ND | 0.012 | <0.001 | 0.719 ± 0.292 |
| C-Combo3/K441R | ND | 0.019 | <0.001 | 0.374 ± 0.08 |
Numbers refer to amino acid position where mutagenesis was performed. Refer to Table 1 for specific amino acids changed. A, PERV-A; C, PERV-C; A/C, PERV-A/C.
ND, not detectable or ≤5 IU/ml on uninfected 293 human embryonic kidney cells.
Certain combinations of mutations of the C-terminal residues of SU in the PERV-A/C chimeric envelope result in a noninfectious phenotype.
PERV-A/C chimeric envelope carrying multiple amino acid substitutions were generated: Combo2, Combo3A and Combo3B, Combo4, Combo5, Combo6, and Combo7 (Fig. 1). As shown in Fig. 3, vectors carrying the Combo2 and Combo3A mutants were infectious for ST-Iowa and SIRC/PAR cells but had either very low or no detectable titers on 293 cells (Combo3 had a mean titer of 6 IU/ml), indicating that these sets of mutations were not sufficient to restore infectivity of the PERV-A/C chimera on 293 cells. In contrast, vectors carrying mutant envelopes Combo3B through Combo6 were all noninfectious or had very low titers in ST-Iowa, SIRC/PAR, and 293 cells (for example, Combo6 had a mean titer of 6 IU/ml on SIRC/PAR cells). All the noninfectious vectors shared the same set of altered residues from NHRQ to YNRP found in Combo3B (Fig. 1 and 3). For a control, we made Combo7, which contains all PERV-A residues in the C-terminal region. As expected, Combo7 had titers comparable to those of PERV-A on all cell lines tested.
FIG. 3.
Infectivity titers of retroviral vectors bearing PERV-A/C envelopes with multiple amino acid mutations. Shown are the average infectious units/ml (plus standard errors [error bars]) of triplicate wells tested three times. Mutants are labeled by the amino acid position in PERV-C. See Fig. 1 for the specific amino acid change. A, PERV-A; C, PERV-C; A/C, PERV-A/C.
SU-IgG binding to target cells correlates with infectivity.
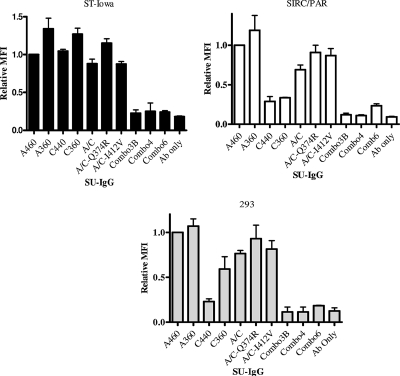
We derived SU-IgG fusion proteins corresponding to those PERV-A/C mutants that either restored or inhibited the infectivity of retroviral vector pseudotypes: PERV-A/C Q374R, PERV-A/C I412V, PERV-A/C Combo3B, Combo4, Combo6, and control PERV-A, PERV-C, and PERV-A/C. Since the A/C SU-IgG fusion protein was previously shown to have reduced binding on human cells relative to PERV-A SU-IgG, we sought to assess the influence of these mutations on cell binding properties relative to the parental A/C SU-IgG. As shown in Fig. 4, Western blot analysis was used to confirm expression and correct molecular weight of the SU-IgG fusion proteins, as described in Materials and Methods. As shown in Fig. 5, SU-IgG carrying the C-terminal mutations of Combo3B and its derivatives (Combo4 and Combo6) were unable to bind any of the cell types analyzed over that observed with the secondary antibody control only, suggesting that the lack of infectivity observed with these mutants may be due to the inability to bind. We further found that neither of the mutants that enhanced PERV-A/C infection on human cells (I412V and Q374R) increased binding relative to PERV-A/C (Fig. 5). In general, the ability of the various mutant PERV-A/C SU-Igs tested to bind all three cell lines, ST-Iowa, SIRC/PAR, or 293, correlated with the infectivity of the retroviral vectors carrying the corresponding mutated PERV-A/C envelopes, in that those that conferred an infectious phenotype in the form of a retroviral vector pseudotype showed comparable levels of binding to PERV-A-SU-Ig, while those that were noninfectious showed only background levels of binding.
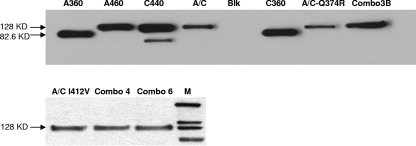
FIG. 4.
Western blot analysis of SU-IgG fusion proteins. Western blot analysis of representative fusion proteins of PERV-SU and mutants with rabbit IgG was performed. The samples tested are abbreviated as follows: A360, PERV-A 360SU-IgG; A460, PERV-A SU-IgG; C440, PERV-C SU-IgG; A/C, PERV-A/C SU-IgG; Blk, blank supernatant of nontransduced cells; C360, PERV-C 360 SU-IgG; A/C-Q374R, PERV-A/C Q374R SU-IgG; Combo3B, PERV-A/C NHQ436YNP SU-IgG (PERV-A/C Combo3BSU-IgG); A/C I412V, PERV-A/C I412V SU-IgG; Combo4, PERV-A/C Combo4 SU-IgG; Combo6, PERV-A/C Combo6 SU-IgG; M, MagicMark XP Western protein standard (Invitrogen, CA). The positions of molecular mass markers (in kilodaltons) are shown to the left of the blots.
FIG. 5.
Binding data of SU-IgG of selected mutated PERV-A/C SUs compared to control PERV-A and PERV-C SU in ST-Iowa, SIRC/PAR, and 293 cells. The y axis shows the relative mean fluorescence intensity (MFI), representing results after normalization to A-360 binding as described previously (13). Results shown are from three experiments performed in duplicate. Antibody only (Ab only) refers to cells without SU-IgG treated with fluorescein isothiocyanate-conjugated anti-rabbit IgG antibody, representing the assay background. Samples were abbreviated as in the legend to Fig. 4.
Vectors carrying the PERV-C envelope with only four PERV-A-specific residue substitution mutations in the C terminus of SU were infectious on human cells.
Mutations were introduced into the PERV-C envelope in order to assess whether the residues that increased the infectivity titer on human cells of the PERV-A/C chimeric envelope would confer infectivity of human cells upon retroviral vectors bearing these mutated PERV-C envelopes. We made a series of mutant PERV-C envelopes wherein residues from PERV-A were substituted for the corresponding PERV-C residues: PERV-C-Q374R, PERV-C-I412V, C-Q374R+I412V, PERV-C-Q439P, PERV-C-K441R, PERV-C-Combo3B, and PERV-C-Combo3B/K441R. No infection of human cells was detected with vectors bearing the mutant PERV-C-Q374R, PERV-C-I412V, and PERV-C-K441R envelopes, although each of these retained infectivity for ST-Iowa cells with a titer comparable to those of vectors with the parental PERV-C envelope, showing that they are not impaired for infectivity generally (Table 3). In contrast, the single substitution mutation Q439P or the Combo3B mutant at the distal end of the carboxyl terminus of the PERV-C SU resulted in no detectable infection by retroviral vector pseudotypes bearing these mutant envelopes on either pig or human cells. Although a PERV-C mutant with a substitution mutation of the putative TM cleavage site, PERV-C-K441R, remains infectious to pig cells (Fig. 1 and Table 3), the analogous substitution mutation of the putative TM cleavage site of PERV-A envelope, PERV-A-R462K, resulted in lack of infection in all types of cells tested (Table 3). Unexpectedly, the infection of human cells was detected with vectors carrying PERV-C envelopes with mutations of four residues in the SU C-terminal region to the corresponding residues of PERV-A (PERV-C-Combo3B/K441R; Table 3). Analysis for interference further showed that PERV-A-infected ST, SIRC/PAR, and 293 cells prevented superinfection by vectors carrying PERV-C-Combo3B/K441R, while PERV-C-infected ST cells allowed infection by these vectors.
TABLE 3.
Titers of retroviral vectors bearing mutant PERV-C envelopes
| PERV envelopea | Titerb (IU/ml) of retroviral vector with mutant PERV-C envelope on the following cells:
|
|
|---|---|---|
| ST-Iowa | 293 | |
| A/C | 2.6 × 103 ± 192 | 1 × 101 ± 4 |
| C | 1.9 × 104 ± 337 | <0.001 |
| C-Q374R | 1.3 × 104 ± 390 | <0.001 |
| C-I412V | 9.4 × 103 ± 447 | <0.001 |
| C-Q374R + I412V | 1.09 × 104 ± 308 | <0.001 |
| C-Q439P | 0 | 0 |
| C-K441R | 3.93 × 104 ± 659 | 0 |
| C-Combo3B | <0.001 | <0.001 |
| C-Combo3B/K441R | 5.8 × 102 ± 266 | 1.6 × 103 ± 112 |
| A | 3.8 × 103 ± 239 | 1.57 × 103 ± 109 |
| A-R462K | 0 | 0 |
A/C, PERV-A/C; A, PERV-A; C, PERV-C.
Data represent the results from duplicate wells from each of three experiments, shown as the average infectious units per ml (IU/ml) ± standard error.
DISCUSSION
We mutated specific residues in the C terminus of SU that differ between PERV-A and PERV-C envelopes (Fig. 1) to gain insight into the function of the C terminus of PERV SU in viral entry. Viral envelope chimeras from groups of viruses have been used as a tool to map the viral determinants for entry of gammaretroviruses (14, 24, 25, 28, 31, 47). We previously showed that the C terminus of the PERV envelope carries determinants that influence infectivity and binding by using chimeric envelope constructs between PERV-A and PERV-C (13). In particular, a chimeric envelope we constructed carrying the C-terminal 130 amino acid residues of PERV-C in place of the corresponding residues of PERV-A was shown to reduce infectivity titers at least 100-fold on human 293 cells. In addition, the apparent RBD of PERV-C was shown to bind human cells, even though PERV-C is not able to infect human cells. The significance of this last observation was unknown, as the binding may have been to a molecule that is unable to support entry. However, the combination of results from our present study provides an opportunity to understand that result further.
Using the chimeric PERV-A/C envelope as a substrate for mutagenesis of this C-terminal region of SU, we found that retroviral vectors carrying the PERV-A/C envelope with either the Q374R or I412V mutation resulted in an increase of 2- to 3-log units of infectivity on human cells compared to our PERV-A/C chimeric envelope (Fig. 2). However, introduction of either or both of these same residue changes into PERV-C did not allow infection of human cells (Table 3). The finding that the Q374R and I412V residue changes were not sufficient to allow human cell infection when introduced into the PERV-C envelope but were sufficient to restore infectivity on human cells to PERV-A/C enveloped vectors indicates that additional residues are required for human cell infection. The PERV-A/C chimera results suggest an important role for the RBD in human cell tropism. However, our finding that mutation of as few as four residues at the C terminus of PERV-C (PERV-C-Combo3B-K441R) allows infection of human cells demonstrates that human cell infection does not require a PERV-A RBD. Even more remarkable is that in the absence of a PERV-A-RBD, the PERV-C-Combo3B-K441R envelope seems to infect human cells via the PERV-A receptor based on interference data. Combined with our prior results showing that the PERV-C RBD is capable of binding human cells, it appears that PERV-C binds cells through the human PERV-A receptor but that naturally occurring PERV-C envelope sequences identified thus far cannot functionally engage this molecule to trigger the postbinding events required to complete entry.
Our finding that the C-terminal sequences of SU can influence the entry function of the N terminus is supported by other studies that have shown that viruses acquired the receptor specificity and coupling of fusion after receptor recognition, when the N-terminal changes of the SU are complemented with C-terminal changes (8, 15, 21, 22, 31). For at least some gammaretroviruses, both the N- and C-terminal regions of SU harbor viral entry determinants (5, 11, 20). Regarding PERV, findings showing the extension of the receptor binding activity of the PERV SU to include the PRR (13), the presence of residues in the PRR that confer higher titer infectivity on human cells (16), and our finding that specific residues in the C terminus of SU enhance human cell infectivity when introduced into a chimeric PERV-A/C or PERV-C envelope demonstrate an interdependency of different domains of SU to facilitate efficient entry of PERV into human cells. It is also noteworthy that in some retroviruses, such as feline leukemia virus (8, 15) and MLV (4), there are variants in which a single residue change in the C terminus of SU is a key pathogenic determinant of the variant viruses; therefore, this region of envelope may influence more than one viral function.
In addition to identification of residues that enhance human cell infection, we also identified specific amino acid changes that rendered retroviral vectors bearing envelopes with those changes noninfectious. In particular, retroviral vectors with PERV envelopes carrying the Q439P mutation alone or combined with additional mutations in the C terminus of SU were noninfectious whether in the A/C chimeric envelope or full-length PERV-C (Fig. 2 and 3). SU-IgG bearing the Combo3B or Combo4 mutations (Fig. 1) were also unable to bind any of the cells tested, even though the SU-IgG epitope-tagged proteins were expressed as evidenced by Western blot analysis (Fig. 4 and 5), further underscoring the role of Q439P. The observed lack of infectivity of Combo6 suggested that additional residues that were different in Combo6 and PERV-A may have critical functions to mediate entry. PERV-A/C-Combo6 contains eight of the nine PERV-A-specific residues in the C-terminal region of SU, retaining only the KK from PERV-C SU and PERV-C TM. One explanation for the lack of infectivity and binding observed for Combo3B, Combo4, Combo5, or Combo6 is the possibility that there may be misfolding in the region required for binding. The finding that Combo7 was fully infectious on all cell lines tested underscores the importance of that last K441R mutation present in Combo7 but absent in Combo6. Supporting this observation was the demonstration that PERV-A/C Q439P showed no infection of human cells and an approximately 1,000-fold decrease in infection of control pig cells (Fig. 2). Moreover, a derivative of this mutant with three amino acid substitutions at the C terminus of PERV-A/C SU (PERV-A/C Combo3B) were unable to infect either human or pig cell lines (Fig. 3). Together, the results point to a critical interaction between the residue at position 439 with the particular dibasic residue in order to generate infectious virus. As shown in Table 4, a summary of key data clearly reveals a pattern that all mutations carrying Q439P in combination with the dibasic residue combination of KK were all noninfectious. In contrast, Q at position 439 is functional with either the KK or KR dibasic residue combination. We hypothesize that the residue at position 439 may play a role in the conformation so that the proline combined with the KK residues may prevent protease access, hence preventing maturation of the envelope due to proteolytic cleavage.
TABLE 4.
Summary of key mutations at the C terminus of SU impacting human cell tropism and infectivity
| Envelope mutanta | Envelope backbonea | C-terminal sequenceb | Dibasic residuesc | Infectiond of the following cells:
|
|
|---|---|---|---|---|---|
| ST-Iowa | 293 | ||||
| A | A | YNRP | KR | Yes | Yes |
| C | C | NHRQ | KK or KR | Yes | No |
| A/C-Q439P | A/C | NHRP (A) | KK (C) | No | No |
| A/C-Combo3B | A/C | YNRP (A) | KK (C) | No | No |
| A-R462K | A | YNRP (A) | KK (C) | No | No |
| C-Combo3B | C | YNRP (A) | KK (C) | No | No |
| C-K441R | C | NHRQ (C) | KR (A) | Yes | No |
| C-Combo3B-K441R | C | YNRP (A) | KR (A) | Yes | Yes |
A, PERV-A; C, PERV-C; A/C, PERV-A/C.
Amino acids 436 to 439 based on PERV-C numbering (53). Residues that were mutated from the indicated envelope backbone are shown in bold type; the envelope in which the mutated residues are naturally found is shown in parentheses following the sequence.
Dibasic residues in the putative proteolytic cleavage signal based on amino acids 440 to 441 of PERV-C and amino acids 461 to 462 with PERV-A numbering (53). Residues that were mutated from the indicated envelope backbone are shown in bold type; the envelope in which the mutated residues are naturally found is shown in parentheses following the sequence.
Infection as determined by retroviral vector pseudotypes bearing envelopes with the indicated sequence.
Several studies have shown that the proteolytic processing of the retrovirus envelope glycoprotein precursors by the host cell protease is a necessary step of the intracellular synthesis pathway to generate viral protein capable of incorporating into infectious virions (9, 36, 39, 55). It is also known that most gammaretroviruses require paired basic amino acid sequences (either Arg-Arg or Lys-Arg) for intracellular cleavage (9, 36). Interestingly, PERV-A and PERV-C SU either differ slightly or agree in their dibasic proteolytic cleavage signal sequence. Depending on the cDNA isolates of the PERV-C envelope from porcine cells, the PERV-C sequence at the C terminus of SU has been reported as either R-Q-KR (1, 38) or R-Q-KK (13, 17) (here and GenBank accession numbers DQ996276.1 and AF402663.1), while the corresponding residues reported in PERV-A have always been observed as R-P-KR (1, 23, 50, 53). The use of KK versus KR appears to be unique to PERV-C envelopes based on a GenBank database search of related exogenous gammaretroviruses (e.g., MLV and gibbon ape leukemia virus) and replication-competent endogenous retroviruses (e.g., baboon endogenous virus, Mus dunni endogenous virus, human endogenous retrovirus W, koala retrovirus, and RD114). While the presence of KK in PERV-C allows formation of entry-competent envelopes, the same combination in PERV-A prevents formation of entry-competent envelopes (PERV-A-R462K [Tables 3 and 4]).
In our experiments, we found that the retroviral vectors pseudotyped with the PERV-A/C envelope had higher infectious titers on SIRC/PAR than 293 cells. In addition, receptor interference (Table 2) showed that cells chronically infected with PERV-A, and not PERV-C, resisted superinfection with retroviral vectors pseudotyped with the PERV-A/C envelope and each of the derivative set of mutant envelopes that were human-tropic, thus demonstrating that the receptor specificity of the mutant viruses tested is the same as that of PERV-A. However, the observed reduced infection on 293 cells relative to SIRC/PAR cells could be due to either higher binding affinity of the virus envelope with its cognate receptor when exogenously introduced into nonpermissive rabbit cells, the requirement for auxiliary elements that enhance the entry process, or perhaps differences between the endogenous expression of HuPAR-1 and HuPAR-2 on 293 cells versus the expression of solely HuPAR-2 on SIRC/PAR cells. In support of this last possibility, we have observed that 293 cells express both HuPAR-1 and HuPAR-2 by reverse transcription-PCR and that HuPAR-1 is less efficient than HuPAR-2 in mediating PERV infection (D. Salomon, unpublished data).
In conclusion, it has been proposed that identification of pigs lacking genomic sequences encoding nonhuman tropic PERV-C may reduce the risk of generating recombinant human-tropic PERV-A/C virus, perhaps providing a means to identify a safer source for porcine to human xenotransplantation (2). Our findings underscore the need to understand the determinants that restrict infection of human cells by PERV-C and those that permit human cell tropism. The present study identified single residues in the C-terminal region of PERV SU that modulate infectivity of human cells as much as 1,000-fold (Q374R and I412V). Although these same residues do not confer human cell tropism to the PERV-C envelope, further analysis identified that as few as four amino acid changes in the PERV-C envelope are sufficient to convert this pig-tropic envelope into a human-tropic envelope (Table 3). Therefore, the risk of using pigs that carry genomic PERV-C loci may be twofold: the previously identified risk of providing a genomic reservoir for generating human-tropic PERV-A/C recombinants and the risk of naturally occurring mutants with as few as four amino acid changes, as exemplified in the present study, that may produce a human-tropic variant. It is hoped that the identification of residues that impact human cell tropism of PERV may allow development of antiviral approaches that target and inhibit PERV infection, potentially providing a means to reduce the risk of PERV transmission in recipients of porcine xenotransplantation products.
Acknowledgments
We thank Maribeth V. Eiden of NIMH/NIH for scientific discussions and critical reading of the manuscript. We are also indebted to Malou Gemeniano for technical advice and critical reading of the manuscript. We also appreciate the technical support provided by Elaina A. Berres during her Howard Hughes Medical Institute-funded teacher internship.
This research was supported by CBER/FDA and a CRADA between CBER/FDA and The Scripps Research Institute (NIH A152349-052A2 to D.R.S.).
Footnotes
Published ahead of print on 28 May 2008.
REFERENCES
- 1.Akiyoshi, D. E., M. Denaro, H. Zhu, J. L. Greenstein, P. Banerjee, and J. A. Fishman. 1998. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J. Virol. 724503-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartosch, B., D. Stefanidis, R. Myers, R. Weiss, C. Patience, and Y. Takeuchi. 2004. Evidence and consequence of porcine endogenous retrovirus recombination. J. Virol. 7813880-13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battini, J.-L., O. Danos, and J. M. Heard. 1995. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J. Virol. 69713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkhart, M. D., P. D'Agostino, S. C. Kayman, and A. Pinter. 2005. Involvement of the C-terminal disulfide-bonded loop of murine leukemia virus SU protein in a postbinding step critical for viral entry. J. Virol. 797868-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, H. H., M. M. Anderson, F. C. Hankenson, L. Johnston, C. V. Kotwaliwale, and J. Overbaugh. 2006. Envelope determinants for dual-receptor specificity in feline leukemia virus subgroup A and T variants. J. Virol. 801619-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham, D. A., G. J. Dos Santos Cruz, X. M. Fernandez-Suarez, A. J. Whittam, C. Herring, L. Copeman, A. Richards, and G. A. Langford. 2004. Activation of primary porcine endothelial cells induces release of porcine endogenous retroviruses. Transplantation 771071-1079. [DOI] [PubMed] [Google Scholar]
- 7.Deng, Y.-M., B. E. Tuch, and W. D. Rawlinson. 2000. Transmission of porcine endogenous retroviruses in severe combined immunodeficient mice xenotransplanted with fetal porcine pancreatic cells. Transplantation 701010-1016. [DOI] [PubMed] [Google Scholar]
- 8.Donahue, P. R., S. L. Quackenbush, M. V. Gallo, C. M. deNoronha, J. Overbaugh, E. A. Hoover, and J. I. Mullins. 1991. Viral genetic determinants of T-cell killing and immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J. Virol. 654461-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, J. Y., J. W. Dubay, L. G. Perez, and E. Hunter. 1992. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein define a requirement for dibasic residues for intracellular cleavage. J. Virol. 66865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ericsson, T. A., Y. Takeuchi, C. Templin, G. Quinn, S. F. Farhadian, J. C. Wood, B. A. Oldmixon, K. M. Suling, J. K. Ishii, Y. Kitagawa, T. Miyazawa, D. R. Salomon, R. A. Weiss, and C. Patience. 2003. Identification of receptors for pig endogenous retrovirus. Proc. Natl. Acad. Sci. USA 1006759-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faix, P. H., S. A. Feldman, J. Overbaugh, and M. V. Eiden. 2002. Host range and receptor binding properties of vectors bearing feline leukemia virus subgroup B envelopes can be modulated by envelope sequences outside of the receptor binding domain. J. Virol. 7612369-12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed, E. O., and R. Risser. 1987. The role of envelope glycoprotein processing in murine leukemia virus infection. J. Virol. 612852-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gemeniano, M., O. Mpanju, D. R. Salomon, M. V. Eiden, and C. A. Wilson. 2006. The infectivity and host range of the ecotropic porcine endogenous retrovirus, PERV-C, is modulated by residues in the C-terminal region of its surface envelope protein. Virology 346108-117. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, S., F. Zhou, C. E. Greer, H. Legg, T. Tang, P. Luciw, J. zur Megede, S. W. Barnett, J. J. Donnelly, D. T. O'Hagan, J. M. Polo, and M. Vajdy. 2006. Antibody responses against HIV in rhesus macaques following combinations of mucosal and systemic immunizations with chimeric alphavirus-based replicon particles. AIDS Res. Hum. Retrovir. 22993-997. [DOI] [PubMed] [Google Scholar]
- 15.Gwynn, S. R., F. C. Hankenson, A. S. Lauring, J. L. Rohn, and J. Overbaugh. 2000. Feline leukemia virus envelope sequences that affect T-cell tropism and syncytium formation are not part of known receptor-binding domains. J. Virol. 745754-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison, I., Y. Takeuchi, B. Bartosch, and J. P. Stoye. 2004. Determinants of high titer in recombinant porcine endogenous retroviruses. J. Virol. 7813871-13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hector, R. D., S. Meikle, L. Grant, R. A. Wilkinson, J. A. Fishman, and L. Scobie. 2007. Pre-screening of miniature swine may reduce the risk of transmitting human tropic recombinant porcine endogenous retroviruses. Xenotransplantation 14222-226. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda, H., K. Kato, T. Suzuki, H. Kitani, Y. Matsubara, S. Takase-Yoden, R. Watanabe, M. Kitagawa, and S. Aizawa. 2000. Properties of the naturally occurring soluble surface glycoprotein of ecotropic murine leukemia virus: binding specificity and possible conformational change after binding to receptor. J. Virol. 741815-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krach, U., N. Fischer, F. Czauderna, and R. R. Tonjes. 2001. Comparison of replication-competent molecular clones of porcine endogenous retrovirus class A and class B derived from pig and human cells. J. Virol. 755465-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauring, A. S., M. M. Anderson, and J. Overbaugh. 2001. Specificity in receptor usage by T-cell-tropic feline leukemia viruses: implications for the in vivo tropisms of immunodeficiency-inducing variants. J. Virol. 758888-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavillette, D., B. Boson, S. J. Russell, and F. L. Cosset. 2001. Activation of membrane fusion by murine leukemia viruses is controlled in cis or in trans by interactions between the receptor-binding domain and a conserved disulfide loop of the carboxy terminus of the surface glycoprotein. J. Virol. 753685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavillette, D., A. Ruggieri, B. Boson, M. Maurice, and F. L. Cosset. 2002. Relationship between SU subdomains that regulate the receptor-mediated transition from the native (fusion-inhibited) to the fusion-active conformation of the murine leukemia virus glycoprotein. J. Virol. 769673-9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Tissier, P., J. P. Stoye, Y. Takeuchi, C. Patience, and R. A. Weiss. 1997. Two sets of human-tropic pig retroviruses. Nature 389681-682. [DOI] [PubMed] [Google Scholar]
- 24.Lu, C. W., and M. J. Roth. 2001. Functional characterization of the N termini of murine leukemia virus envelope proteins. J. Virol. 754357-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier, C. C., S. Delagrave, Z. X. Zhang, N. Brown, T. P. Monath, K. V. Pugachev, and F. Guirakhoo. 2007. A single M protein mutation affects the acid inactivation threshold and growth kinetics of a chimeric flavivirus. Virology 362468-474. [DOI] [PubMed] [Google Scholar]
- 26.Martin, U., V. Kiessig, J. Blusch, A. Haverich, K. von der Helm, T. Herden, and G. Steinhoff. 1998. Expression of pig endogenous retrovirus by primary porcine endothelial cells and infection of human cells. Lancet 352692-694. [DOI] [PubMed] [Google Scholar]
- 27.Martina, Y., K. T. Marcucci, S. Cherqui, A. Szabo, T. Drysdale, U. Srinivisan, C. A. Wilson, C. Patience, and D. R. Salomon. 2006. Mice transgenic for a human porcine endogenous retrovirus receptor are susceptible to productive viral infection. J. Virol. 803135-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan, R. A., O. Nussbaum, D. D. Muenchau, L. Shu, L. Couture, and W. F. Anderson. 1993. Analysis of the functional and host range-determining regions of the murine ecotropic and amphotropic retrovirus envelope proteins. J. Virol. 674712-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niebert, M., and R. R. Tönjes. 2005. Evolutionary spread and recombination of porcine endogenous retroviruses in the Suiformes. J. Virol. 79649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oldmixon, B. A., J. C. Wood, T. A. Ericsson, C. A. Wilson, M. E. White-Scharf, G. Andersson, J. L. Greenstein, H.-J. Schuurman, and C. Patience. 2002. Porcine endogenous retrovirus transmission characteristics of an inbred herd of miniature swine. J. Virol. 763045-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Reilly, L., and M. J. Roth. 2000. Second-site changes affect viability of amphotropic/ecotropic chimeric enveloped murine leukemia viruses. J. Virol. 74899-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ott, D., and A. Rein. 1992. Basis for receptor specificity of nonecotropic murine leukemia virus surface glycoprotein gp70SU. J. Virol. 664632-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Overbaugh, J., A. D. Miller, and M. V. Eiden. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 65371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patience, C., W. M. Switzer, Y. Takeuchi, D. J. Griffiths, M. E. Goward, W. Heneine, J. P. Stoye, and R. A. Weiss. 2001. Multiple groups of novel retroviral genomes in pigs and related species. J. Virol. 752771-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patience, C., Y. Takeuchi, and R. A. Weiss. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 3282-286. [DOI] [PubMed] [Google Scholar]
- 36.Perez, L. G., and E. Hunter. 1987. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein that block processing to gp85 and gp37. J. Virol. 611609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popp, S. K., D. A. Mann, P. J. Milburn, A. J. Gibbs, P. J. McCullagh, J. D. Wilson, R. R. Tonjes, and C. J. Simeonovic. 2007. Transient transmission of porcine endogenous retrovirus to fetal lambs after pig islet tissue xenotransplantation. Immunol. Cell Biol. 85238-248. [DOI] [PubMed] [Google Scholar]
- 38.Preuss, T., N. Fischer, K. Boller, and R. R. Tonjes. 2006. Isolation and characterization of an infectious replication-competent molecular clone of ecotropic porcine endogenous retrovirus class C. J. Virol. 8010258-10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ragheb, J. A., and W. F. Anderson. 1994. Uncoupled expression of Moloney murine leukemia virus envelope polypeptides SU and TM: a functional analysis of the role of TM domains in viral entry. J. Virol. 683207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rein, A. 1982. Interference grouping of murine leukemia viruses: a distinct receptor for the MCF-recombinant viruses in mouse cells. Virology 120251-257. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt, P., A. Forsman, G. Andersson, J. Blomberg, and O. Korsgren. 2005. Pig islet xenotransplantation: activation of porcine endogenous retrovirus in the immediate post-transplantation period. Xenotransplantation 12450-456. [DOI] [PubMed] [Google Scholar]
- 42.Schultz, A., and A. Rein. 1985. Maturation of murine leukemia virus env proteins in the absence of other viral proteins. Virology 145335-339. [DOI] [PubMed] [Google Scholar]
- 43.Scobie, L., S. Taylor, J. C. Wood, K. M. Suling, G. Quinn, S. Meikle, C. Patience, H. J. Schuurman, and D. E. Onions. 2004. Absence of replication-competent human-tropic porcine endogenous retroviruses in the germ line DNA of inbred miniature swine. J. Virol. 782502-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Specke, V., S. Rubant, and J. Denner. 2001. Productive infection of human primary cells and cell lines with porcine endogenous retroviruses. Virology 285177-180. [DOI] [PubMed] [Google Scholar]
- 45.Takefman, D. M., S. Wong, T. Maudru, K. Peden, and C. A. Wilson. 2001. Detection and characterization of porcine endogenous retrovirus in porcine plasma and porcine factor VIII. J. Virol. 754551-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takeuchi, Y., C. Patience, S. Magre, R. A. Weiss, P. T. Banerjee, P. L. Tissier, and J. P. Stoye. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 729986-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ting, Y.-T., C. A. Wilson, K. B. Farrell, G. J. Chaudry, and M. V. Eiden. 1998. Simian sarcoma-associated virus fails to infect Chinese hamster cells despite the presence of functional gibbon ape leukemia virus receptors. J. Virol. 729453-9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tönjes, R. R., and M. Niebert. 2003. Relative age of proviral porcine endogenous retrovirus sequences in Sus scrofa based on the molecular clock hypothesis. J. Virol. 7712363-12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Laan, L. J. W., C. Lockey, B. C. Griffeth, F. S. Frasier, C. A. Wilson, D. E. Onions, B. J. Hering, Z. Long, E. Otto, B. E. Torbett, and D. R. Salomon. 2000. Infection by porcine endogenous retrovirus after islet xenotransplantation in SCID mice. Nature 40790-94. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe, R., T. Miyazawa, and Y. Matsuura. 2005. Cell-binding properties of the envelope proteins of porcine endogenous retroviruses. Microbes Infect. 7658-665. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, C., and M. Eiden. 1991. Viral and cellular factors governing hamster cell infection by murine and gibbon ape leukemia viruses. J. Virol. 655975-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, C., S. Wong, J. Muller, C. Davidson, T. Rose, and P. Burd. 1998. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J. Virol. 723082-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson, C. A., S. Wong, M. V. Brocklin, and M. J. Federspiel. 2000. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J. Virol. 7449-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood, J. C., G. Quinn, K. M. Suling, B. A. Oldmixon, B. A. Van Tine, R. Cina, S. Arn, C. A. Huang, L. Scobie, D. E. Onions, D. H. Sachs, H. J. Schuurman, J. A. Fishman, and C. Patience. 2004. Identification of exogenous forms of human-tropic porcine endogenous retrovirus in miniature swine. J. Virol. 782494-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zavorotinskaya, T., and L. M. Albritton. 1999. Failure to cleave murine leukemia virus envelope protein does not preclude its incorporation in virions and productive virus-receptor interaction. J. Virol. 735621-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]