Abstract
Vaccinia virus was treated in a controlled manner with various combinations of nonionic detergents, reducing agents, and proteolytic enzymes, and successive products of the reactions were visualized using atomic force microscopy (AFM). Following removal of the outer lipid/protein membrane, a layer 20 to 40 nm in thickness was encountered that was composed of fibrous elements which, under reducing conditions, rapidly decomposed into individual monomers on the substrate. Beneath this layer was the virus core and its prominent lateral bodies, which could be dissociated or degraded with proteases. The core, in addition to the lateral bodies, was composed of a thick, multilayered shell of proteins of diverse sizes and shapes. The shell, which was readily etched with proteases, was thoroughly permeated with pores, or channels. Prolonged exposure to proteases and reductants produced disgorgement of the viral DNA from the remainders of the cores and also left residual, flattened, protease-resistant sacs on the imaging substrate. The DNA was readily visualized by AFM, which revealed some regions to be “soldered” by proteins, others to be heavily complexed with protein, and yet other parts to apparently exist as bundled, naked DNA. Prolonged exposure to proteases deproteinized the DNA, leaving masses of extended, free DNA. Estimates of the interior core volume suggest moderate but not extreme compaction of the genome.
Vaccinia virus, the best-characterized member of the Poxviridae family, is unusual among most double-stranded DNA viruses in that both transcription and replication occur in the cytoplasm of the host cell (5, 8, 24). The 191-kbp genome of the Copenhagen strain contains up to 266 open reading frames encoding proteins >65 amino acids in length (9, 15), with 63 to 80 vaccinia virus gene products being packaged in the virion (4, 53, 57, 60). Replication takes place within discrete cytoplasmic viral factories that are thought to be devoid of cellular organelles (3) but which contain ribosomes (16). Infectious virions can be isolated in several forms, depending on their degree of maturation, including mature virus (MV), wrapped virus, and enveloped virus (25).
Many general features of vaccinia virion architecture are readily apparent from a variety of studies (3), notably a genome/enzyme-containing proteinaceous core, proteinaceous lateral bodies (LBs), and surrounding surface tubular elements and lipid membrane. Nonetheless, despite ∼60 years of study by conventional electron microscopy (EM) (11, 14, 15, 45, 48, 54, 58), cryoelectron microscopy (1, 9, 54), and more recently cryoelectron tomography (4) freeze etch (31), deep etch (11), and atomic force (22) microscopies, specific controversies remain in issues such as the intrinsic rigidity of the core, the degree of order in genome packaging, the degree and affinity of the association of the genome with protein in the packaged state, the presence/absence of an “inner membrane” or “palisade” enwrapping the core, and the number of lipid bilayers that enwrap the intact virion. Structural details of the virion are relevant to several stages of the vaccinia life cycle, including mechanisms of fusion and entry, extrusion of early mRNA from the infecting virion, disassembly of the infecting virion, genome packaging into nascent virions during assembly, and virion maturation/membrane enwrapment.
A persistent drawback with EM images, irrespective of the technique applied, is that they generally provide two-dimensional projections of specimens onto a single plane. Three-dimensional information is usually absent, and approximations can be recovered only via reconstruction. Thus, ambiguities and uncertainties arise in the assignment of features to internal structures and surfaces. In addition, various artifacts may be introduced, particularly via the fixing and staining procedures generally employed to enhance contrast. Atomic force microscopy (AFM) complements EM in a number of ways. Although the resolution of AFM is theoretically lower than that of EM, it often reveals details of soft, biological specimens, including viruses (20), that cannot otherwise be obtained. The three-dimensionality of specimens can easily be preserved during imaging, and instead of the specimen being projected onto a plane, topographical features of a specimen's surface are revealed. This might appear to limit the utility of the technique for delineating the internal structures of specimens such as complex viruses, but these drawbacks can be largely overcome. Viruses, as we have now shown in several investigations (32, 35, 38-40, 42-44, 52), can be broken down or disassembled in controlled ways to reveal, sequentially, internal layers. By chemical and enzymatic dissection of virions accompanied by AFM visualization of the products, information regarding internal structures can be revealed. AFM is a single-particle analytical method that does not rely on a homogeneous population (as would, for example, cryoelectron microscopy reconstruction). It records not only the structures and states common within a population but also, with equivalent accuracy, features found in just a few individuals. AFM can be applied with equal ease to dried specimens or specimens fully hydrated in physiological media. AFM records topography irrespective of the electron density.
We have previously established, as a proof of principle, the amenability of vaccinia virus to AFM analysis (22). Here we report a more in-depth AFM analysis of vaccinia virus that we believe clarifies some significant structural questions and provides additional, general details. The images presented here are, we believe, a significant improvement over those shown in the previous study (22) in terms of resolution and population coverage within individual preparations. We have focused in particular upon the virion core and LBs, along with aspects of the packaged genome, such as its degree of order and association with protein and its appearance when released from virion cores. We focus exclusively on MV released from mammalian cells using standard protocols. We have imaged MV in standard buffers akin to those used for storage and infection and have also treated it with a number of vigorous reagents and reagent combinations.
MATERIALS AND METHODS
Sample preparation and AFM analysis.
The virus preparation described elsewhere (22) was used throughout the current study, at the titer described previously. Intact and treated viruses were imaged on glass coverslips or freshly cleaved mica coated with poly-l-lysine. Genomic DNA was imaged on freshly cleaved, uncoated mica. For virus imaging, 1.5 μl of 50-fold-diluted freshly resuspended MV in 10 mM Tris (pH 7.8) was deposited on the substrate and allowed to sediment for 30 min at room temperature. For imaging in air, the specimen, on the substrate, was rinsed multiple times with distilled water and then dried under a stream of N2 gas. Prior to imaging under liquid, specimens were treated with a solution of 5% glutaraldehyde in phosphate-buffered saline (PBS) for 15 min and then rinsed with distilled water. Fixation with glutaraldehyde has been shown in previous investigations to not perturb the surface structures of virus particles within the resolution of the AFM technique (32, 34, 35, 39, 40).
For enzyme digestion experiments, after deposition of the intact specimen onto the imaging substrate, the substrate was rinsed multiple times with PBS and excess PBS was wicked away with torn filter paper. After addition of 15 μl of enzyme solution, the substrate was sealed in a plastic cell (to minimize evaporation), followed by incubation either in an incubator at 37°C or at room temperature. For sequential treatments with different reagents, the specimen on the substrate was rinsed with PBS after removal of the first reagent before addition of the second.
Imaging procedures were essentially as previously described for other viruses (32, 34, 40, 42, 44) and RNA (33, 37), with use of a Nanoscope III multimode AFM instrument (Veeco Instruments, Santa Barbara, CA) calibrated to small lateral distances by imaging the 111 face of a thaumatin protein crystal as a standard, based on the known lattice spacings (19, 21). After the sample substrates were placed in a 75-μl fluid cell, images were acquired at 26°C in tapping mode (17, 18) using commercially available silicon tips (imaging under air) or oxide-sharpened silicon nitride tips (imaging under fluid) at a scan frequency of 1 Hz and an oscillation frequency of 9.2 kHz (under fluid) or 300 kHz (in air). In the AFM images presented here, increasing height above the substrate is indicated by an increasingly lighter color. Due to the distortion of lateral distances due to convolution of the cantilever tip shape and the surface features scanned, quantitative measures of size were based either on heights above the substrate or on center-to-center distances between particles.
RESULTS
The intact virion.
The shape, dimensions, and general surface characteristics of hydrated MV imaged by AFM (Fig. 1a and b) were as previously reported (22), with the virus assuming a familiar rectangular (“brick”) shape upon dehydration (Fig. 1c and d). Particles were pleomorphic, with an outer surface design varying from the more regular, short, haphazardly intersecting “railroad tracks” topography of naturally cell-extruded virus (see reference11 and references therein) to a more punctate topography, referred to in the early literature as the “M” or “mulberry” form of the virion (7, 25, 47, 58), which may be more diagnostic of intracellular virus released by cell rupture (10, 11). Parenthetically, the virion surface also closely resembled the external surfaces of various animal cells as imaged by AFM (34, 36, 39, 40), consistent with a comparable array of membrane proteins anchored in a lipid bilayer, perhaps with an underlying membrane skeleton.
FIG. 1.
Intact vaccinia virions, hydrated and dried. (a and b) AFM images of fully hydrated MV scanned in a fluid cell under PBS. Virion surfaces were very rough, and LBs were invisible or indistinct. (c and d) MV that had been dried on the AFM substrate and scanned in air. LBs in dried particles were prominent and readily visualized.
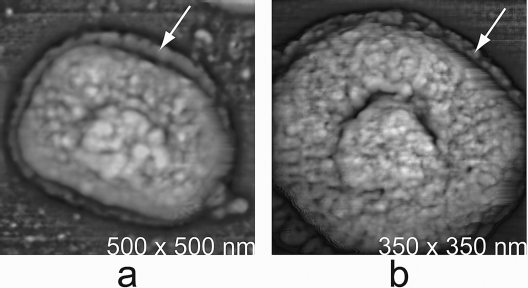
One LB (or very occasionally both) was discernible attached to hydrated specimens (Fig. 1 and 2). As observed previously (22), the profile of the LBs visibly associated with every particle became greatly accentuated upon virion dehydration (Fig. 1 and 2), and a deep trench formed around the visible LBs which clearly delineated them with respect to the background viral surface. LBs were about one-third the overall dimensions of dried intact MV (albeit the height could not be measured) but less rectangular in shape.
FIG. 2.
Two MV particles in panels a and b were dried in air on the substrate and imaged by AFM. Many dried particles showed a corona of membrane with embedded protein (indicated by arrows). This membrane appears to have withdrawn and disengaged from the particle. The prominence of LBs was also notable.
Virions in both hydrated and dehydrated preparations were frequently found to be aggregated with one another (a well-known property of vaccinia virus [17]), albeit isolated virions were also frequently observed. The isolated virions permitted observation of structural changes arising from chemical or enzymatic treatment (below). Insofar as individual particles were readily visualized against a very clean substrate background (Fig. 1), the MV preparation appeared almost completely devoid of large cellular assemblies or subviral materials (such as partially degraded particles, smaller viral fragments, or protein complexes).
In the dehydrated samples, a corona was frequently discerned surrounding the particle (Fig. 2). We interpreted this as a partial retraction of outer membrane(s) and associated proteins from the upper part of the virion surface toward the imaging substrate surface, leading to the exposure of a somewhat smoother underlying surface that may be diagnostic of a form previously referred to as the “C” form (6, 32). Brief treatment of virions with nonionic detergent-containing solution (e.g., NP-40 plus dithiothreitol [DTT]) dissolved the membrane quickly but left the ring of proteins at the periphery intact.
Interestingly, attempts to induce osmotic phenomena in hydrated particles by exposing them to pure water had no discernible effect on the integrity of virions, indicating a strong insulation from ion flux, consistent with the virions' tolerance to low-ionic-strength buffers, such as the standard storage buffer (10 mM Tris-HCl). Nonetheless, the severe change in volume upon dehydration/rehydration (Fig. 1) (22) suggests that water can be exchanged.
Chemical and enzymatic dissection of the vaccinia virus virion.
Before discussing the treatment of particles with various reagents, we should emphasize that the population of treated particles was an ensemble of individuals having a variety of structural strengths and weaknesses. Not all particles responded to a given treatment or became degraded in an identical or synchronous manner. Different particles, exposed to the same reagents, were observed in a variety of states of disassembly as a function of time. Since it seemed unlikely that the glutaraldehyde-fixed particles were not equivalently exposed to reagents, they were presumably structurally nonidentical (some being stronger/more resistant to degradation than others, which were more fragile/easily disrupted), presumably a result of the harvest of an asynchronous infection by cell rupture. Therefore, we have exercised caution in describing degradation pathways, particularly in terms of the exposure times necessary to induce structural changes. In general, we describe the response of the majority of particles.
Isolated STEs had a fibrous rather than tubular appearance.
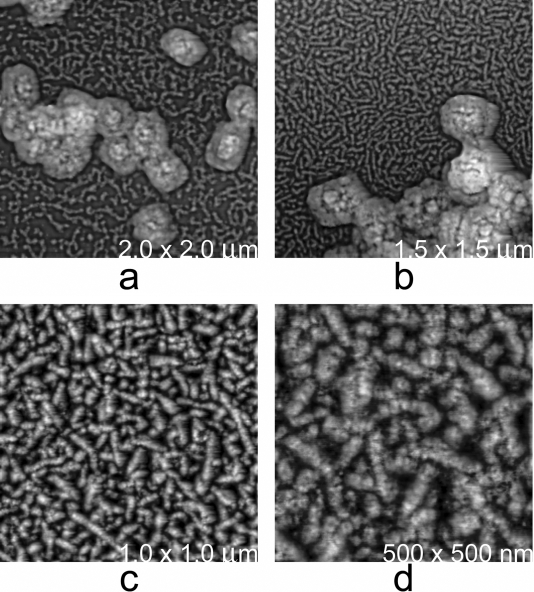
Surface tubular elements (STEs) have been previously observed (42, 45, 50, 51, 56, 58, 59) and are considered to be dissociated forms of the “short railroad tracks” or punctate spots previously observed associated with extracellular virions or “M”-form virions, respectively (see above) (11). In our hands, STEs dissociated from the virion in the presence of low concentrations of detergent and reducing agent (0.1% NP-40-10 mM DTT) (Fig. 3), revealing their structure quite clearly. Thus, although some minor proportion (perhaps 15%) was present as individual globular units, STEs appeared principally as thick fibers with a chain-like appearance (Fig. 3). By AFM, these fibers did not show evidence of a tubular (or otherwise hollow (33) morphology or of helicity. Rather, they appeared more as linear aggregates of one kind (or perhaps just a few kinds) of protein assembly. We will therefore henceforth refer to them as surface fibrous elements (SFEs). SFEs had a fairly uniform diameter of about 20 nm and lengths mostly in the range of 100 to 150 nm (though some appeared to be as long as 500 to 600 nm—twice the diameter of the virion). By comparison of SFE-positive and SFE-negative virions (Fig. 1c and d versus Fig. 3a and b), AFM results were consistent with the deep-etch EM (11, 14) observations that when attached to the virus, SFEs form a dense network or array oriented more or less parallel to the virion surface (“short railroad tracks”). However, by AFM, some individual SFEs, when packed around the virion core, appeared laterally twisted and/or bent and sometimes doubled back on themselves (Fig. 1c and d). The observed SFE diameter of 20 nm was consistent with the thickness of the virion “outer membrane” in virion thin sections (6, 9, 12, 21).
FIG. 3.
Particles in the process of SFE shedding. (a) MV were treated for 5 min at 22°C with 0.1% NP-40 and 10 mM DTT, dried in air, and then imaged by AFM. (b) The particles underwent the same treatment, but for 15 min. The SFE layer lying immediately beneath the outer lipid membranes became exposed, with SFEs being rapidly shed onto the imaging substrate as linear aggregates (see the text). Note the substantial increase in density with time of the SFE on the substrate surrounding the particles. SFEs apparently occupy most if not all of the space between the outer membrane and the core. (c and d) Higher-magnification images of shed SFEs. After shedding, SFEs rapidly disassembled into individual globular units.
Upon exposure to higher concentrations of NP-40-DTT (0.5%-50 mM, respectively), SFEs appeared to depolymerize/disaggregate, prior to loss from virions (data not shown). Under these conditions, shedding and disaggregation into globular units was so rapid that it could scarcely be visualized. By comparison, after treatment in other ways, EM studies have shown partially disaggregated forms still attached to the virus (see references 14, 15, and 20 and references therein). After shedding, the individual globular units observed by AFM showed remarkable uniformity in size and shape, coating the substrate in the vicinity of the shedding particle (Fig. 3a and b). After 60 min of exposure, even at lower concentrations of NP-40-DTT (0.1%-10 mM, respectively), the fibrous layers of most particles were entirely stripped and monomerized and the underlying virion cores were exposed. SFEs appeared to be the principal material filling the space between the virion outer membrane(s) and the virion core and therefore presumably comprise some of the more abundant of the structural proteins in the virion, arguably virion protein p4b (33). Other possibilities might be actin or a residual fragment of the A-type inclusion body protein or inclusion body embedding factor VO.
MV cores and sacs.
Vaccinia virion cores could be reproducibly generated by 60 min of exposure of MV to 0.5% NP-40 plus 50 mM DTT at 37°C. Examples of vaccinia cores are shown in Fig. 4. Those in panels a and b were dried on the substrate, while those in panels c and d were imaged in their fully hydrated state. Dried cores appeared complex and pleomorphic in their structural detail. Most, however, shared some common features and exhibited similar behavior upon further exposure to reagents. The typical hydrated core, as revealed by AFM (Fig. 4c and d), was an oblong body of dimensions 220 nm by 200 nm and a height above the substrate of ∼120 nm. These dimensions shrank by approximately 15 to 20% when the cores were dried and then imaged in air. However, because of their apparent softness, the cores' surface structures were resolved in considerably greater detail when dry.
FIG. 4.
Longer-duration treatment produced fully stripped virion cores. (a and b) Two examples of dried virion cores prepared by 60-min exposure of MV to 0.5% NP-40-50 mM DTT at 22°C, followed by drying in air. SFEs have been lost, revealing the DNA-containing cores with their associated LBs. In these dried particles, the wall enclosing the nucleic acid appears to be heavily perforated. (c and d) Fully hydrated cores, treated in the same way. Dimensions of the hydrated cores were approximately 220 nm by 200 nm by 120 nm. In panel d, the proteins making up the LBs were particularly well defined.
The surfaces of dried cores appeared as dense networks of proteins, having a variety of shapes and sizes, and were extensively perforated with holes or channels. The outer surfaces of hydrated cores, by contrast, generally appeared quite smooth. Cores from most particles could be progressively etched by several hours of exposure to NP-40 and DTT (though a few individuals appeared to degrade much more rapidly). This process was noticeably accelerated if the incubation contained protease or was followed by protease treatment. In the presence of proteinase K, for example (after first washing thoroughly with water to remove NP-40), or in the presence of trypsin plus 0.5% NP-40, a progressive etching of the core outer surface could be observed, with the core surface becoming rougher in appearance with increased duration of proteolysis (data not shown).
Two interpretations of the “etching” observations were considered: (i) removal of a smoother outer protein layer revealing a rougher inner one or (ii) deepening of preexisting etch pits within a unitary wall. Our observations seemed consistent with the core wall being identical to the “palisade layer” (also referred to as the “core membrane”) identified in EM thin sections, since the deeply ridged or etched structure imaged by AFM could be expected to appear striated in section. Our observations are therefore consistent with the core wall having a structurally asymmetric unitary structure, without the need to invoke the greater structural complexity of a double wall or a membrane surrounding the core. It should be noted, however, that the particles had been treated with NP-40, which could disrupt any lipid membrane. A multiwalled architecture, however, remains a possibility.
Cores degraded in the presence of proteinase K finally lost or expelled their internal DNA. Although the remaining protein superstructure of the cores remained largely intact, the core superstructure collapsed on the substrate as flat, round “pancakes” of 12 to 15 nm in height (Fig. 5), as one would expect of deflated balloons or empty sacks (“ghosts” [14]). In Fig. 5d and e, the surfaces of the ghosts appeared to be very rough in texture, which was apparent whether or not they were hydrated. In the process of proteolytic etching of the outer layer of the cores, LBs were also degraded and lost. In some cases we observed what appeared to be intact LBs separated from cores, suggesting a particular susceptibility of the LB-core linkage. Most LBs, however, were progressively degraded until only remnants could be detected still associated with the cores.
FIG. 5.
(a to c) Virion cores prepared by treatment of MV with 0.5% NP-40 and 50 mM DTT followed by exposure to proteinase K in PBS for 16 h at 37°C, drying in air, and imaging by AFM. Virion DNA has been lost from the cores along with the LBs. Core walls have been substantially degraded and appear as flattened sacs on the imaging substrate. These sacs are 12 to 15 nm in height. Proteins and protein fragments can be seen on the substrate surrounding the sacs. (d and e) Higher-magnification images of the flattened sacs showing their rough and irregular surfaces, presumably maintained by residual protein-protein linkages. Note the absence of any LBs. (f) Residual sac of a core which had not been exposed to proteinase K at all but had nonetheless lost its DNA spontaneously. Its thickness is somewhat greater, 15 to 20 nm. Note the residual remnants of an LB (arrow).
LBs.
As illustrated in Fig. 1 and 2, LBs were sometimes apparent even in fully hydrated, intact MV but became more distinct upon drying. The profound trench developing around the LBs upon drying showed them to be distinct entities (“organelles”) of the virion. As expected, the LBs were associated with opposing sides of the virion core (Fig. 6a and b). In many images, only one lateral surface of a particle was visible to the AFM tip, and therefore, only one core-associated LB could be verifiably ascertained. In virtually all of those cases in which the orientation of a particle was such that both LBs would be visible to the AFM tip, two symmetrically placed LBs were in fact observed. Thus, we feel that most particles did indeed possess two LBs. In Fig. 4d, the features of a hydrated LB attached to a virion core were particularly well resolved. The LB was significantly rougher and more irregular in texture than the remainder of the core and appeared to contain aggregates or assemblies of proteins having a range of sizes and shapes (as also indicated in Fig. 4c).
FIG. 6.
Additional virion cores. (a) After exposure of MV to 0.5% NP-40 and 50 mM DTT for 60 min at 22°C and imaging under water. Associated with the core were two LBs disposed on opposite sides. Dissociated proteins cluttered the substrate surface. (b) Again, a pair of LBs can be seen associated with some core particles. (c and d) Low- and high-magnification images of a native, untreated MV particle that had been air dried and imaged by AFM. An LB has been lost from the particle (the cause is unknown), and a deep pit remains in the otherwise still-intact virion.
The LBs were firmly attached to (and apparently covalently continuous by other than disulfide bonds with) the core, as indicated by their resistance to prolonged treatment with NP-40 plus DTT (though for some cores, an LB appeared to have become completely sheared away even in the absence of added protease, consistent with some heterogeneity in the preparation of intracellular vaccinia virus). LBs could, however, be consistently degraded by treatment with proteinase K (Fig. 7). As shown in Fig. 6c and d, LBs could occasionally be lost even from intact MV, and their removal left a deep pit or indentation in the host particle. It was difficult to approximate the depth of the pit for various reasons, including the radius of curvature of the AFM tip. However, insofar as the genome did not spill out and DNA (see below) was not imaged within the pit, we do not believe the pit to represent a perforation in the core wall. The pit was even more clearly revealed in additional images of cores immediately following LB removal by proteolysis with trypsin (Fig. 7a and b) (11). While many cores exhibited a profound central depression resulting from loss of LBs (as shown in Fig. 7a and b), occasionally trypsin treated cores did not. Interestingly, those that did not appeared to increase in number with time of exposure to protease. It is possible that cores lacking pits were simply the occasional products of a somewhat arbitrary proteolytic process. On the other hand, we suspected the progressive assumption of a “relaxed” shape from the initially pitted cores. That is, following loss of the LBs, the core wall may be prone to relax or “pop out” into a fuller, more loaf-like shape (as exhibited by the particles in Fig. 7c and d).
FIG. 7.
Cores produced by treatment of MV with a mixture of 0.5% NP-40 plus 50 mM DTT plus 0.1 mg/ml trypsin at 37°C for 60 min and then imaged by AFM in a fluid cell (thus, the particles represent fully hydrated cores). (a and b) LB loss left a deep, central depression. (c and d) Similarly treated cores, though these had more of a loaf shape and failed to exhibit the pits left by degraded LBs. In panel d, one of the three cores (indicated by an arrow) had lost its DNA and collapsed to a flattened sac, while its two nucleic acid-containing neighbors retained their full shape.
Nucleic acid bundles within the cores.
As described above, once the DNA had exited the core, the vacant protein sac collapsed like a plastic bag or a deflated ball onto the imaging substrate. The genomic DNA, visualized by AFM at various times both within and after leaving the cores and in different states of unraveling, displayed a variety of interesting features. In some cases, after NP-40-DTT treatment, the DNA within the virion cores was relatively unperturbed even when the surrounding sac was damaged. This is shown in Fig. 8a and b, where the sac was “peeled back” without disturbing its contents. In other instances after prolonged NP-40/DTT treatment, the entire sac was separated from the nucleic acid, leaving the DNA as a condensed packet or bundle with some unraveling at its edges (Fig. 8c). This bundle was reminiscent of the virion nucleoids observed by EM after sodium dodecyl sulfate treatment of particles (13). The dried nucleic acid bundle had a loaf-like shape, ∼100 to 120 nm in length, reaching up to ∼80 nm in height. The observation of this “nucleoid” as an almost entirely intact entity in the absence of the sac implied an absence of strong internal pressure within the “relaxed” core (after a loss of LB indentation, albeit there could have been slightly greater pressure with LBs in place). Although the nucleoid showed no inclination to spontaneously burst or spread, it was able to unravel, starting to assume the more recognizable features of DNA strands, mostly decorated with proteins (Fig. 8C and 9).
FIG. 8.
Core contents after treatment with 0.5% NP-40 and 50 mM DTT for 20 min at 22°C. (a) The wall/sac of a core has been partially sheared away (arrow), revealing an inner packet of nucleic acid, some of which was apparently associated with protein. (b) A higher-resolution image of the sheared sac and the core contents. Note that the nucleic acid and its associated protein had not burst from the opened core but remained intact. (c) Another unusual observation: the internal contents of a virion core, which has been entirely shorn of its sac, are shown on the AFM substrate. At the top, the nucleic acid with associated protein can be seen initially unraveling and spreading on the substrate around it. The packets of DNA inside the core were present as unique entities that in general unraveled only slowly into recognizable strands of nucleic acid.
FIG. 9.
Nucleoprotein loss from sac. (a) Cores were produced by treatment of MV with 0.5% NP-40 and 50 mM DTT at 22°C, washed with PBS, and subsequently treated with 0.1 mg/ml proteinase K for 30 min at 37°C. The core has begun disgorging its contents via a rupture in the core sac/wall. Spilled nucleic acid appears to be almost entirely associated with protein. (b) Nucleoprotein complex imaged at higher magnification. (c and d) the same nucleoprotein after a 60-min exposure; strands of naked DNA are beginning to appear (indicated by arrows). Note, however, that most of the DNA is still protein coated.
Vaccinia DNA.
After protease treatment, the virion DNA was not visualized as a sac-free packet, as shown in Fig. 8c, but rather as nucleoprotein flowing out of a perforated or protease-damaged core onto the substrate surface in the immediate vicinity of the disintegrating core (Fig. 9). During the time course of protease digestion (which is relatively short), DNA seemed to be most heavily studded with protein at the earliest times after its emergence from the cores (e.g., see Fig. 9), with some strands being completely coated with protein (particularly evident in Fig. 9c and d). However, some protein-free strands could also be seen even at early times. Later in the proteolytic time course, the DNA appeared more as shown in Fig. 10, with significantly smaller amounts of associated protein. Most of the protein units associated with the DNA appeared fairly uniform in size, as though there was a predominant nucleoprotein unit (consistent with the four-protein “subnucleoid” structure described in reference 12), albeit DNA-free protein aggregates of other sizes and shapes were also observed (Fig. 9 and 10). Presumably a variety of proteins (e.g., enzymes) were included in the mass of protein and DNA. After prolonged exposure to proteinase K (for several hours or overnight), protein was almost entirely stripped from the DNA, and the DNA appeared ultimately as complex networks of folded, tangled, and looped, mostly naked, strands (Fig. 11). Often the most stubbornly remaining proteins appeared to be “soldering” together various strands and loops in the expelled DNA.
FIG. 10.
Nucleoprotein. (a) A cluster of vaccinia cores at the edge of the image (indicated by arrow) had emptied its nucleic acid entirely, which is seen expanded about it on the AFM substrate. The remains of the sacs are still present. (b to d) Higher-magnification images of various regions of disgorged DNA from the cores in panel a and others (of which there were many). DNA was also frequently exuded from cores after exposure to a mixture of 0.5% NP-40 plus 50 mM DTT plus 0.1 mg/ml proteinase K for 30 min.
FIG. 11.
Naked genomic DNA. (a to d) Images of vaccinia DNA which has emerged from ruptured core particles after exposure to proteinase K for 12 h. The protein was almost entirely degraded, leaving only naked DNA on the AFM substrate.
When condensed in packets, as in the intact virion core, the strands did not appear to be arranged or packed in an ordered manner. It seemed as though the DNA or nucleoprotein was simply stuffed inside the sac-like core in a disordered manner, as one might stuff a down parka or sleeping bag inside a nylon sac. Furthermore, when the nucleic acid unraveled at the edges of the DNA packets, its arrangement appeared arbitrary with no obvious systematic mode of packing. It appeared like a bag full of wet yarn. We saw no evidence for an ordered, tube-like nucleoprotein structure (27).
Radically ruptured MV.
The chemical and enzymatic dissection of vaccinia virions described above was a result of slow, reasonably controlled digestion with specific reagents and may bear some resemblance to what occurs during the early stages of vaccinia virus infection. We also identified reagents leading to a more radical rupture kinetic. “Radical rupture” led to the display of nucleic acid on the substrate directly from intact, native virions, even in the absence of any detergent or protease. For example, exposure to high concentrations of DTT alone (0.25 M) (Fig. 12a and b) or brief exposure to a high pH (pH 12) (Fig. 12c and d) led some particles to spontaneously rupture, followed by an apparently sudden release of nucleic acid to the exterior. When nucleic acid was spilled by a native particle undergoing chemical or physical “radical rupture,” a massive egress of what appeared to be almost entirely naked DNA, with little or no accompanying protein, was apparent. Our impression was that only the naked portions of the DNA exited the particle, with the more protein-rich portions perhaps remaining attached to the interior surface of the protein sac. This can be seen, to some extent, in Fig. 12b and c.
FIG. 12.
(a and b) MV treated with 0.25 M DTT only at 37°C for about 15 min and imaged in air by AFM. (c and d) MV exposed briefly to pH 12. Under either condition, a few MV ruptured precipitously and DNA poured from the damaged virions onto the substrate.
mRNA synthesis.
We made extensive efforts to treat virion cores with buffer/reagent mixtures known to support mRNA synthesis in the hope of visualizing newly made mRNA on the substrate or on the surfaces of particles. In this way we might have characterized the production of early mRNA from the encapsidated DNA. However, no evidence was ever detected of mRNA emerging from the particles, as might have been observable by AFM (33, 37). It must be emphasized, however, that because of protein loss induced by the transcription buffer (which also contained NP-40 and DTT), the substrate around the particles became very rough and cluttered with shed and damaged proteins. Similarly, the surfaces of the virion cores themselves became very uneven because of their associated protein. This surface irregularity may have precluded observation of mRNA by AFM, since a clean, flat surface would normally be required.
Anomalous particles.
We made a final, curious observation. In one or two images, we observed what appeared to be a particle that was not recognizable as vaccinia virus. An example can be seen in Fig. 13. It was nonenveloped, had a diameter of about 160 nm, and exhibited what appeared to be an ordered or partially ordered arrangement of protein clusters, much like capsomers, on its surface, as one might expect of a conventional icosahedral virus. However, we were not able to unambiguously verify the symmetry of the particles from AFM images. Presumably this was a contaminating virus of minor abundance or some aberrant vaccinia virus.
FIG. 13.
Particle X: occasionally, particles like this were observed among untreated MV. These particles were larger in diameter than MV, almost completely spherical (even after drying), and appeared to have an ordered arrangement of protein clusters (or capsomers) on their surfaces.
DISCUSSION
There have been a significant number of structural studies of vaccinia over the decades, principally via various forms of EM (see the introduction) but also by AFM (22). This body of work is extensive and has recently been reviewed (3). From the current AFM work, we have been able to confirm and lend weight to specific interpretations of EM observations, which, along with some additional findings, suggest a consensus model of vaccinia virion structure. In particular, we have been able to address SFEs, LBs, the nature of the core wall, and the packaged genome.
The fibers observed surrounding the vaccinia core (likely consistent with the “coats” observed in a previous study [22]), which were named SFEs, appeared to be composed predominantly of a multimerized protein complex, which reverted efficiently to a monomer form upon treatment with NP-40 and DTT. The fibers appeared to be oriented parallel to the virion surface (Fig. 1 and 2), an observation consistent with both the “short railroad tracks” surface topography described by Heuser (20) and the very notable ropelike helical surface fibers of parapoxvirus (46, 48, 49, 55). Although somewhat cytoskeletal in appearance, the mode of depolymerization of these SFEs, i.e., primarily under reducing conditions, would seem atypical for common cellular cytoskeletal proteins. Overall, our observations would support a view that the inner portion of the virion double outer layer, visible in some EM thin sections, may comprise these fibers in sections covered with a virion outer lipid bilayer, itself studded with proteins.
LBs were clearly imaged by AFM. Other than to say that the LBs surely do exist, we can only speculate as to their composition and function. The sizes and gross appearances of the two LBs associated with a core were equivalent (or roughly so). The LBs appeared similar to large protein complexes found in cells and may have comparable functions, such as the anchoring of DNA telomeres, orientation or anchoring of membrane proteins at the virion surface, or compression of the internal volume of the core (clamping the core wall down over the genome by taking up the slack in the core wall). The attachment sites for the LBs appeared symmetrically placed, indicating that they do not adhere arbitrarily to the core but instead have specific attachment or binding sites. This evokes two possible models for LB/virion morphogenesis. In one, the protomers of the core sac are constructed prior to LB attachment, and yet instead of being randomly distributed, some (at least) occupy discrete locations in the shell to provide the LB anchoring sites. In an alternative view, the LBs might appear early in the virion morphogenic pathway, acting as nucleation points for core wall growth so that once the LBs are saturated with core wall proteins, the latter would continue to assemble by interacting only with one another. Indeed, the collapsibility of the empty sac comprising the core wall (Fig. 5) (14) might be consistent with such a model. The collapsibility of the core wall would be consistent with the presence of an apparent external bracing during virion assembly (provided by the vaccinia virus D13 protein's clathrin-like networks) (11, 31). The core wall may be built within such a template, or mold, within the crescents that are characteristic of the early stages of virion morphogenesis.
With regard to genome packaging, we should first point out that in a previous AFM study (22), images of degraded vaccinia virus particles were presented that contained, or so it was believed, segmented tubules of about 16 nm in diameter which appeared on the exterior to have a helical character. These were visualized in association with thin fibers thought to be extended strands of DNA. The suggestion was advanced that the DNA might be enclosed in the cores within the tubules and might be superhelically stressed. These observations and conclusions were almost certainly incorrect. We have subsequently observed segmented 16-nm-diameter tubules, virtually identical to those previously shown, in media contaminated with bacteria. We are now convinced that these tubules were in fact of bacterial origin and were in no way a part of the vaccinia virus structure. We have taken some additional care in the experiments reported here to avoid any repetition of their appearance. Moreover, we have found no evidence in the current study to suggest that the packaged vaccinia virus genome is superhelically stressed, at least not in the quiescent state of the virion.
By contrast, our AFM images of DNA “nucleoid” structures indicated that a large portion of the packaged genome may be moderately tightly packed and perhaps not extensively complexed with protein. With regard to intravirion pressure, if one calculates the volume occupied by the DNA in the condensed packets seen in Fig. 8 or calculates an approximate volume for the interiors of the hydrated cores, a value of about 4 × 106 nm3 is obtained. From this, along with a genome size of ∼191 kbp, a DNA density of about 0.05 bp/nm3 may be computed. Similar calculations for bacteriophages lambda (28) and P22 (2), which are fairly representative of double-stranded DNA phages, give a value of around 0.60 bp/nm3. In the cases of lambda and P22, it is known that the encapsidated DNA is not accompanied by any protein and that it is packed with about as high a density as is believed to be theoretically possible (28) (about equivalent to that of DNA in a crystal [1]). Comparison of the packing density of the DNA in the vaccinia virus cores with corresponding densities for the bacteriophages (29, 41) clearly shows there to be at least 10 times more space in the vaccinia virus cores than in the heads of bacteriophages in proportion to the amount of nucleic acid.
The absence of extreme packaging pressures notwithstanding, our AFM images do suggest that the interior bulk of the vaccinia virus DNA is likely to consist primarily of naked DNA strands that are relatively closely packed. This is supported by low-angle X-ray scattering evidence, with ∼25-Å spacing being observed for both vaccinia virus (23) and the bacteriophages (7). A relatively low packing density for vaccinia virus DNA would be consistent with the observation that when the core is ruptured by proteases, the DNA does not burst out of the sac but pours out like a thick fluid and unravels gradually. It is interesting to speculate that stable interstrand connections may have constrained the DNA from radical ejection in terms of flexibility/degrees of freedom. The apparent protein-based “soldering” of DNA strands and its apparent stability during virion degradation (Fig. 11) may be consistent with this and with the stability of DNA “nucleoids” observed previously under extractive conditions (12, 13) and may serve to mitigate the internal pressure arising during moderate genome compaction.
In light of the evidence presented here that the packing of the DNA chiefly determines the core shape, which in turn is reflected in the virion shape, then ultimately the distribution or packing of the nucleic acid determines virion shape characteristics. In such a case, how is the genome packed that it imposes a rectangular shape on the core? One possibility, consistent with all of the observations, is that the DNA is simply a bundle of long loops, sometimes referred to as a “climber's rope,” equivalent to a coil collapsed about an axis in its plane. Such a folded bundle of rope, soldered predominantly at positions toward its equator, would likely appear relatively disordered when imaged from one end (“loop-on”) or in a partially disassembled state. An attraction of such an arrangement is that it has two distinct directions, one parallel and one perpendicular to the direction of the strands in the loops, yielding roughly rectilinear and biconcave morphologies, respectively. Moreover, it would provide consistency with prior observations of an internal tube-like structure (27). It has biological virtue too, in that it may be possible to bundle and unbundle in a relatively straightforward and uncomplicated way when its genetic content is required.
The sac containing the DNA we know to be flexible and able to assume multiple shapes (rectangular and dumbbell shaped in intact cores, loaf shaped in cores lacking LBs, or round and flat following a loss of DNA). If, as we suspect, there is a change in the overall shape of partially degraded cores which still contain their DNA, upon loss of LBs, from particles having profound central depressions to more loaf-like shapes (Fig. 7), then there would almost certainly have to be some rearrangement of the nucleic acid and its associated proteins within the core that either produces or accompanies the change in core shape. Conversely, the DNA may be more compressed with the LBs present.
In numerous cases (e.g., see Fig. 9 and 10), the DNA observed pouring from ruptured cores appeared to be partially associated with protein in addition to the “soldering sites,” with some strands appearing to be even more fully protein coated. One possibility (though not unequivocally demonstrated) is that while much of the nucleic acid may be packaged as bundles of naked or “soldered” DNA, portions of the DNA near the inside surface of the core wall (i.e., the outer surface of the DNA bundle) may be more extensively protein associated. If so, then a possibly related observation might be the action of the vaccinia virion as a transcriptosome early during infection, prior to virion disassembly. A key criterion might be the rate at which mRNA could (directionally) exit the bundle given the speed, after the initiation of transcription, at which mRNA starts to be extruded from the virion cores into the external milieu (30). As a result of this efficient extrusion in the context of a moderately compact genome, mRNA may not be synthesized throughout the bulk of the DNA bundle within the sac but only near the DNA bundle surface. Channels in the shell may then be more immediately available for mRNA escape from the core.
This is speculative, we concede, though it would have a number of implications for the packaging of the DNA and the internal structure/protein composition of the core. We will make no effort to address such a model here, other than to point to one obvious implication. For the purposes of intravirion transcription of early genes, the packaging of the DNA could not be an entirely random stuffing of pliable DNA into the sac as some of the AFM images might suggest. The genome would need to follow a path in which the early genes interfaced more closely with the interior of the sac. In this regard, early transcription complexes may be poised at early promoters during the late gene expression and/or the packaging phase. Clearly the question of genome packaging requires further study before clear conclusions can be drawn. Moreover, not having imaged DNA within the core under transcription conditions, i.e., in the context of changes known to occur under conditions that would support transcription (18, 26, 29), it is unclear how DNA morphology might be affected.
Acknowledgments
This research was supported by a grant from the NIH (GM58868-02).
We thank Aaron Greenwood for assistance in preparation of figures.
Footnotes
Published ahead of print on 28 May 2008.
REFERENCES
- 1.Baker, T. S., N. H. Olson, and S. D. Fuller. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbol. Mol. Biol. Rev. 63862-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casjens, S., L. Sampson, S. Randall, K. Eppler, H. Wu, J. B. Petri, and H. Schmieger. 1992. Molecular genetic analysis of bacteriophage P22 gene 3 product, a protein involved in the initiation of headful DNA packaging. J. Mol. Biol. 2271086-1099. [DOI] [PubMed] [Google Scholar]
- 3.Casjens, S., and P. Weigele. 2005. Headfull DNA packaging by bacteriophage P 22, p. 80-88. In C. Calalano (ed.), Viral genome packaging machines. Genetics, structure and mechanisms. Landis Publishing, Georgetown, TX.
- 4.Chung, C. S., C. H. Chen, M. Y. Ho, C. Y. Huang, C. L. Liao, and W. Chang. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 802127-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condit, R. C., N. Moussatche, and P. Traktman. 2006. In a nutshell: structure and assembly of the vaccinia virion. Adv. Virus Res. 6631-124. [DOI] [PubMed] [Google Scholar]
- 6.Cyrklaff, M., C. Risco, J. J. Fernández, M. V. Jiménez, M. Estéba, W. Baumeister, and J. L. Carrascosa. 2005. Cryo-electron tomography of vaccinia virus. Proc. Natl. Acad. Sci. USA 1022772-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dales, S. 1962. An electron microscope study of the early association between two mammalian viruses and their hosts. J. Cell Biol. 13303-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dales, S., and B. G. T. Pogo (ed.). 1981. Biology of poxviruses. Springer-Verlag, Vienna, Austria.
- 9.Dubochet, J., M. Adrian, K. Richter, J. Garces, and R. Wittek. 1994. Structure of intracellular mature vaccinia virus observed by cryoelectron microscopy. J. Virol. 681935-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earnshaw, W. C., and S. C. Harrison. 1977. DNA arrangement in isometric phage heads. Nature 268598-602. [DOI] [PubMed] [Google Scholar]
- 11.Easterbrook, K. B. 1966. Controlled degradation of vaccinia virions in vitro: an electron microscopic study. J. Ultrastruct. Res. 14484-496. [DOI] [PubMed] [Google Scholar]
- 12.Fenner, F., R. Wittek, and K. R. Dumbell. 1989. The orthopoxviruses. Academic Press, Inc., New York, NY.
- 13.Goebel, S. J., G. P. Johnson, M. E. Perkus, S. W. Davis, J. P. Winslow, and E. Paoletti. 1990. The complete DNA sequence of vaccinia virus. Virology 179247-266. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths, G., N. Roos, S. Schleich, and J. Krijne-Locker. 2001. Structure and assembly of intracellular mature vaccinia virus: thin-section analysis. J. Virol. 7511056-11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths, G., R. Wepf, R. Wendt, J. Krijnse-Locker, M. Cyrklaff, and N. Roos. 2001. Structure and assembly of intracellular mature vaccinia virus: isolated-particle analysis. J. Virol. 7511034-11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grubisha, O., and P. Traktman. 2003. Genetic analysis of the vaccinia virus I6 telomere-binding protein uncovers a key role in genome encapsidation. J. Virol. 7710929-10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansma, H. G., and J. H. Hoh. 1994. Biomolecular imaging with the atomic force microscope. Annu. Rev. Biophys. Biomol. Struct. 23115-139. [DOI] [PubMed] [Google Scholar]
- 18.Hansma, H. G., and L. Pietrasanta. 1998. Atomic force microscopy and other scanning probe microscopies. Curr. Opin. Chem. Biol. 2579-584. [DOI] [PubMed] [Google Scholar]
- 19.Harris, W. J., and J. C. Westwood. 1964. Phosphotungstate staining of vaccinia virus. J. Gen. Microbiol. 34491-495. [DOI] [PubMed] [Google Scholar]
- 20.Heuser, J. 2005. Deep-etch EM reveals that the early poxvirus envelope is a single membrane bilayer stabilized by a geodetic “honeycomb” surface coat. J. Cell Biol. 169269-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollinshead, M., A. Vanderplassen, G. L. Smith, and D. J. Vaux. 1999. Vaccinia virus intracellular virions contain only one lipid membrane. J. Virol. 731503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holowczak, J. A. 1982. Poxvirus DNA. Curr. Top. Microbiol. Immunol. 9727-79. [DOI] [PubMed] [Google Scholar]
- 23.Holowczak, J. A., V. L. Thomas, and L. Flores. 1975. Isolation and characterization of vaccinia virus “nucleoids.” Virology 67506-519. [DOI] [PubMed] [Google Scholar]
- 24.Ichihashi, Y., M. Oie, and T. Tsuruhara. 1984. Location of protein-binding proteins and disulfide-linked proteins in vaccinia virus structural elements. J. Virol. 50929-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikoma, K., Y. Hiramatsu, F. Uno, M. Yoshida, and S. Nii. 1992. Ultra-high-resolution scanning electron microscopy of vaccinia virus and its recombinant carrying the gag gene of human immunodeficiency virus type 1. J. Electron Microsc. (Tokyo) 41167-173. [PubMed] [Google Scholar]
- 26.Johnson, G. P., S. J. Goebel, and E. Paoletti. 1993. An update on the vaccinia virus genome. Virology 196381-401. [DOI] [PubMed] [Google Scholar]
- 27.Katsafanas, G. C., and B. Moss. 2007. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2221-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, K. S., and D. G. Sharp. 1966. Electron microscopic observations on the nature of vaccinia virus particle aggregation. J. Immunol. 97197-202. [PubMed] [Google Scholar]
- 29.Kindt, J., S. Tzlil, A. Ben-Shaul, and W. M. Gelbart. 2001. DNA packaging and ejection forces in bacteriophage. Proc. Natl. Acad. Sci. USA 9813671-13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleiman, J. H., and B. Moss. 1975. Characterization of a protein kinase and two phosphate acceptor proteins from vaccinia virions. J. Biol. Chem. 2502430-2437. [PubMed] [Google Scholar]
- 31.Ko, T. P., J. Day, A. Greenwood, and A. McPherson. 1994. Structures of three crystal forms of the sweet protein thaumatin. Acta Crystallogr. D Biol. Crystallogr. 50813-825. [DOI] [PubMed] [Google Scholar]
- 32.Kuznetsov, Y., A. Malkin, R. Lucas, M. Plomp, and A. McPherson. 2001. Imaging of viruses by atomic force microscopy. J. Gen. Virol. 822025-2034. [DOI] [PubMed] [Google Scholar]
- 33.Kuznetsov, Y. G., S. Daijogo, J. Zhou, B. L. Semler, and A. McPherson. 2005. Atomic force microscopy analysis of icosahedral virus RNA. J. Mol. Biol. 34741-52. [DOI] [PubMed] [Google Scholar]
- 34.Kuznetsov, Y. G., S. Datta, N. H. Kothari, A. Greenwood, H. Fan, and A. McPherson. 2002. Atomic force microscopy investigation of fibroblasts infected with wild-type and mutant murine leukemia virus (MuLV). Biophys. J. 833665-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuznetsov, Y. G., J. R. Gurnon, J. L. Van Etten, and A. McPherson. 2005. Atomic force microscopy investigation of a chlorella virus, PBCV-1. J. Struct. Biol. 149256-263. [DOI] [PubMed] [Google Scholar]
- 36.Kuznetsov, Y. G., A. J. Malkin, and A. McPherson. 1997. Atomic force microscopy studies of living cells: visualization of motility, division, aggregation, transformation, and apoptosis. J. Struct. Biol. 120180-191. [DOI] [PubMed] [Google Scholar]
- 37.Kuznetsov, Y. G., and A. McPherson. 2006. Atomic force microscopy investigation of Turnip Yellow Mosaic Virus capsid disruption and RNA extrusion. Virology 352329-337. [DOI] [PubMed] [Google Scholar]
- 38.Kuznetsov, Y. G., P. Ulbrich, S. Haubova, T. Ruml, and A. McPherson. 2007. Atomic force microscopy investigation of Mason-Pfizer monkey virus and human immunodeficiency virus type 1 reassembled particles. Virology 360434-446. [DOI] [PubMed] [Google Scholar]
- 39.Kuznetsov, Y. G., J. G. Victoria, A. Low, W. E. Robinson, Jr., H. Fan, and A. McPherson. 2004. Atomic force microscopy imaging of retroviruses: human immunodeficiency virus and murine leukemia virus. Scanning 26209-216. [DOI] [PubMed] [Google Scholar]
- 40.Kuznetsov, Y. G., J. G. Victoria, W. E. J. Robinson, and A. McPherson. 2003. Atomic force microscopy investigation of HIV and HIV-infected lymphocytes. J. Virol. 7711896-11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaMarque, J., and S. Harvey. 2004. Packaging double helical DNA into viral capsids. Biopolymers 73348-360. [DOI] [PubMed] [Google Scholar]
- 42.Malkin, A. J., A. McPherson, and P. D. Gershon. 2003. Structure of intracellular mature vaccinia virus visualized by in situ atomic force microscopy. J. Virol. 776332-6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malkin, A. J., M. Plomp, T. J. Leighton, A. McPherson, and K. E. Wheeler. 2006. Unraveling the architecture and structural dynamics of pathogens by high resolution in vitro atomic force microscopy, p. 32-85. In Microscopy and microanalysis, vol. 11. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 44.Malkin, A. J., M. Plomp, and A. McPherson. 2004. Unraveling the architecture of viruses by high-resolution atomic force microscopy, p. 85-108. In P. M. Lieberman (ed.), Virus structure and imaging, DNA viruses, methods and protocols. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 45.Medzon, E. L., and H. Bauer. 1970. Structural features of vaccinia virus revealed by negative staining, sectioning, and freeze-etching. Virology 40860-867. [DOI] [PubMed] [Google Scholar]
- 46.Mitchiner, M. B. 1969. The envelope of vaccinia and orf viruses: an electron-cytochemical investigation. J. Gen. Virol. 5211-220. [DOI] [PubMed] [Google Scholar]
- 47.Muller, G., and J. D. Williamson. 1987. Poxviridae, p. 421-433. In M. V. Nermut and A. C. Steven (ed.), Animal virus structure. Elsevier, New York, NY.
- 48.Nagington, J., and R. W. Horne. 1962. Morphological studies of orf and vaccinia viruses. Virology 16248-260. [DOI] [PubMed] [Google Scholar]
- 49.Nagington, J., A. A. Newton, and R. W. Horne. 1964. The structure of Orf virus. Virology 23461-472. [DOI] [PubMed] [Google Scholar]
- 50.Noyes, W. F. 1962. Further studies on the structure of vaccinia virus. Virology 18511-516. [DOI] [PubMed] [Google Scholar]
- 51.Noyes, W. F. 1962. The surface fine structure of vaccinia virus. Virology 17282-287. [DOI] [PubMed] [Google Scholar]
- 52.Plomp, M., M. K. Rice, E. K. Wagner, A. McPherson, and A. J. Malkin. 2002. Rapid visualization at high resolution of pathogens by atomic force microscopy: structural studies of herpes simplex virus-1. Am. J. Pathol. 1601959-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Resch, W., K. K. Hixson, R. J. Moore, M. S. Lipton, and B. Moss. 2007. Protein composition of the vaccinia virus mature virion. Virology 358233-247. [DOI] [PubMed] [Google Scholar]
- 54.Roos, N., M. Cyrklaff, S. Cudmore, R. Blasco, J. Krijnse-Locker, and G. Griffiths. 1996. A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J. 152345-2355. [PMC free article] [PubMed] [Google Scholar]
- 55.Spehner, D., S. De Carlo, R. Drillien, F. Weiland, K. Mildner, D. Hanau, and H. J. Rziha. 2004. Appearance of the bona fide spiral tubule of ORF virus is dependent on an intact 10-kilodalton viral protein. J. Virol. 788085-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stern, W., and S. Dales. 1976. Biogenesis of vaccinia: isolation and characterization of a surface component that elicits antibody suppressing infectivity and cell-cell fusion. Virology 75232-241. [DOI] [PubMed] [Google Scholar]
- 57.Takahashi, T., M. Oie, and Y. Ichihashi. 1994. N-terminal amino acid sequences of vaccinia virus structural proteins. Virology 202844-852. [DOI] [PubMed] [Google Scholar]
- 58.Westwood, J. C. M., W. J. Harris, H. T. Zwartouw, D. H. J. Titmuss, and G. Appleyard. 1964. Studies on the structure of vaccinia virus. J. Gen. Microbiol. 3467-78. [DOI] [PubMed] [Google Scholar]
- 59.Wilton, S., A. R. Mohandas, and S. Dales. 1995. Organization of vaccinia envelope and relationship to the structure of intracellular mature virions. Virology 214503-511. [DOI] [PubMed] [Google Scholar]
- 60.Yoder, J. D., T. S. Chen, C. R. Gagnier, S. Vemulapalli, C. S. Maier, and D. E. Hruby. 2006. Pox proteomics: mass spectrometry analysis and identification of vaccinia virion proteins. Virol. J. 310. [DOI] [PMC free article] [PubMed] [Google Scholar]