Abstract
In Aplysia californica, the serotonin-mediated translocation of protein kinase C (PKC) Apl II to neuronal membranes is important for synaptic plasticity. The orthologue of PKC Apl II, PKCɛ, has been reported to require phosphatidic acid (PA) in conjunction with diacylglycerol (DAG) for translocation. We find that PKC Apl II can be synergistically translocated to membranes by the combination of DAG and PA. We identify a mutation in the C1b domain (arginine 273 to histidine; PKC Apl II-R273H) that removes the effects of exogenous PA. In Aplysia neurons, the inhibition of endogenous PA production by 1-butanol inhibited the physiological translocation of PKC Apl II by serotonin in the cell body and at the synapse but not the translocation of PKC Apl II-R273H. The translocation of PKC Apl II-R273H in the absence of PA was explained by two additional effects of this mutation: (i) the mutation removed C2 domain-mediated inhibition, and (ii) the mutation decreased the concentration of DAG required for PKC Apl II translocation. We present a model in which, under physiological conditions, PA is important to activate the novel PKC Apl II both by synergizing with DAG and removing C2 domain-mediated inhibition.
Protein kinase Cs (PKCs) are a family of lipid-activated kinases that mediate a wide variety of cellular processes, including the regulation of synaptic strength in the nervous system (23, 42, 48). In Aplysia californica, behavioral sensitization is mediated in part by an increase in the strength of the connections between mechanoreceptor sensory neurons and motor neurons (19). This increase, called synaptic facilitation, is mediated by the neurotransmitter serotonin (5HT), which induces facilitation in isolated ganglia as well as in cocultures of sensory neurons and motor neurons (5, 6). Sensitizing stimulation causes the translocation of PKC to neuronal membranes (39, 53). In the Aplysia nervous system, there are two phorbol ester-regulated PKCs: PKC Apl I, which is homologous to the Ca2+-activated PKC family in vertebrates (α, β1, β2, and γ) that are called conventional or classical PKCs (cPKCs), and PKC Apl II, which is homologous to the Ca2+-independent epsilon family of PKC in vertebrates (ɛ and η) that are called novel PKCs (nPKCs) (21, 42, 43). PKC Apl I and PKC Apl II translocate under different conditions to mediate distinct types of synaptic facilitation (53). PKC Apl II, but not PKC Apl I, is translocated by the application of 5HT to sensory neurons and is required for 5HT-mediated facilitation at synapses that previously have been depressed (24). In contrast, combining 5HT and the firing of the sensory neuron leads to the additional translocation of PKC Apl I, and PKC Apl I is required for the intermediate-term facilitation induced by the combination of sensory neuron firing and 5HT (53).
Both cPKCs and nPKCs contain two C1 domains and one C2 domain. However, the C2 domain of nPKCs is located N terminal to the C1 domains and lacks critical aspartic acid residues involved in coordinating Ca2+ ions in cPKCs (29). The C2 domain of cPKCs mediates Ca2+-dependent binding to the membrane lipid phosphatidylserine (PS) (30, 31). This binding transiently recruits the enzyme to the membrane, where its physiological activator, diacylglycerol (DAG), resides. Then, in conjunction with the C1 domain interacting with DAG, the binding of the C2 domain to PS induces a conformational change that activates the enzyme (26, 32).
The function of the C2 domain of nPKCs is less clear. The C2 domain of PKCδ binds to phosphotyrosine, allowing for the regulation of the kinase (2, 41, 51). However, the structure of the C2 domain of the epsilon family of PKCs including PKC Apl II is considerably different (11, 15, 33, 35, 45), and these C2 domains do not bind phosphotyrosine (10, 42). It has been reported that the C2 domain of PKCɛ can bind to phospholipids, especially phosphatidic acid (PA), and that PA binding to the C2 domain of PKCɛ is required for translocation (11, 18). However, the amount of PA required to bind to the C2 domain is high compared to that for the Ca2+-dependent binding of the C2 domains of cPKCs to PS (11, 16, 25, 45). In PKC Apl II, the C2 domain binds poorly to lipids; however, the phosphorylation of the C2 domain increases binding (38). While lipid binding suggests a positive role for the C2 domain in translocation, the removal of the C2 domain of PKC Apl II allows for the activation of the enzyme at lower concentrations of PS, implicating the C2 domain as a negative regulator of kinase regulation (44). Moreover, the C2 domain of PKC Apl II lowers the affinity of phorbol esters to bind to the C1 domains, and this is removed by PA (37).
To clarify the role of PA and the C2 domain in the translocation of PKC Apl II, we examined the translocation of fluorescent protein-labeled PKC constructs for PKC Apl II, PKC Apl II ΔC2, and a PKC Apl II-C1b mutant that does not respond to PA. Our results demonstrate an important role for PA in the physiological translocation of PKC Apl II.
MATERIALS AND METHODS
Plasmid construction and mutagenesis.
The pNEX3 enhanced green fluorescent protein (eGFP)-PKC Apl I and eGFP-PKC Apl II, as well as the pNEX3 monocistronic red fluorescent protein (mRFP)-PKC Apl II, have been described previously (24, 53). eGFP-PKC Apl II ΔC2 was constructed by beginning with PKC Apl II ΔC2 in the baculovirus vector (44). eGFP-PKC Apl II ΔC2 was constructed in a manner similar to that for PKC Apl II, by first inserting a hemagglutinin tag at the N terminus and then excising the insert with KpnI and SacI and inserting it into the pNEX3 vector with Kpn and SacI (24). eGFP (N1) (Clontech) then was amplified using PCR and inserted at XhoI, followed by filling in the XhoI site (24). mRFP-PKC Apl II ΔC2 was constructed using overlap PCR similarly to what had been described previously for mRFP-PKC Apl II, except using KpnI and SphI to insert the modified mRFP-containing DNA such that the linker in eGFP-PKC Apl II ΔC2 was identical to that in mRFP-PKC Apl II ΔC2. The C1b constructs were generated using PCR to insert the domains into the pNEX3-eGFP construct for expression or into pGEX2.1 (Amersham Pharmacia Biotech) to generate the glutathione S-transferase (GST) fusion protein. Mutations in the C1b, PKC Apl II, and PKC Apl II ΔC2 constructs were accomplished using overlap PCR, as has been described already (38). All constructs were verified by sequencing.
Fusion protein synthesis and purification.
Maltose-binding protein (MBP)-C2 and GST fusion proteins GST-C1b and GST-C1b-R273H were expressed in DH5 Escherichia coli in Luria-Bertani medium containing 0.1 mM ampicillin. The MBP and GST fusion proteins were purified by affinity chromatography on amylose (New England Biolabs) or GST resins (Amersham Pharmacia Biotech), respectively, by following the recommendations of the manufacturer, with the exception that the MBP-C2 domain was purified in the presence of 0.3 mg/ml ovalbumin to prevent nonspecific binding to the column. The MBP-C2 domain was eluted from the beads using 10 mM maltose in ovalbumin-containing buffer, and the pure protein was kept at 4°C until use. To test for the binding of PKC Apl II-C1b and PKC Apl II-C1b-R273H domains to the PKC Apl II-C2 domain, GST-C1b and GST-C1b-R273H bound to glutathione-Sepharose beads were incubated with pure MBP-C2 protein in 10 mM HEPES, pH 7.4, and 150 mM NaCl for 2 h with shaking at room temperature. The beads were washed five times with 1 ml of phosphate-buffered saline, and the bound proteins were eluted with Laemmli sample buffer and used for Western blot analysis. The primary antibody directed against MBP clone MBP-17 was purchased from Sigma-Aldrich (Oakville, Ontario, Canada). Immunoblotting was performed by following the recommendation of the manufacturer.
Sf9 cell culture.
The Sf9 cells were purchased from Sigma-Aldrich. Sf9 cells were grown in Grace's medium (Invitrogen, Burlington, Ontario, Canada) supplemented with 10% fetal bovine serum (Cansera, Etobicoke, Ontario, Canada) as a monolayer at 27°C. For transfection, cells were plated on MatTek glass-bottom culture dishes (MatTek Corporation, Ashland, MA) with a glass surface of 14 mm and a coverslip thickness of 0.16 to 0.19 mm at a density of 0.11 × 106 cells/35-mm dish. Cells were transfected using the Cellfectin reagent (Invitrogen, Burlington, Ontario, Canada) by following the recommendation of the manufacturer. Live imaging on the confocal microscope was performed 48 to 72 h posttransfection. Cells were serum starved for 6 h before imaging sessions. 1,2-Dioctanoyl-sn-glycerol (DOG) and 1,2-dioctanoyl-sn-glycero-3-phosphate (DiC8-PA) were purchased from Avanti Polar Lipids (Alabaster, AL). DOG and DiC8-PA were dissolved in dimethyl sulfoxide and diluted to the final concentration with Grace's medium shortly before the experiment. During the experiment, the cells were not exposed to dimethyl sulfoxide concentrations of >1%. All of the experiments were performed in a temperature-controlled chamber at 27°C on at least two different occasions, and in each experiment recordings were obtained from two to six cells.
Confocal microscopy in Sf9 cells.
Cells expressing eGFP-PKC and mRFP-PKC constructs for PKC Apl II were examined using a Zeiss laser-scanning microscope (Zeiss, Oberkochen, Germany) with an Axiovert 200 and a ×63 oil immersion objective. During imaging, DOG and/or DiC8-PA was added to the dish after 30 s, and a series of 20 confocal images was recorded for each experiment at time intervals of 30 s. Aggregates (or pigmentation) observed in Sf9 cells in the red channel were not due to mRFP, since those aggregates were visible in the red channel even in nontransfected cells (control cells).
Aplysia cell culture preparation.
Sensory neuron cultures and sensory-motor neuron cocultures were prepared by following published procedures with slight modifications (53). Adult Aplysia californica (76 to 100 g; University of Miami Aplysia Resource Facility, RSMAS, FL) organisms were anesthetized by an injection of 50 to 100 ml of 400 mM (isotonic) MgCl2. Abdominal and/or pleuropedal ganglia were removed and digested in L15 medium containing 1% protease type IX (Sigma). L15 medium was purchased from Sigma and supplemented with 0.2 M NaCl, 26 mM MgSO4·7H2O, 35 mM dextrose, 27 mM MgCl2·6H2O, 4.7 mM KCl, 2 mM NaHCO3, 9.7 mM CaCl2·2H2O, 15 mM HEPES, and the pH was adjusted to 7.4. Following digestion, siphon (LFS) motor neurons and/or tail sensory neurons were isolated and plated in L15 medium containing 50% Aplysia hemolymph on MatTek glass-bottom culture dishes (MatTek Corporation, Ashland, MA) with a glass surface of 14 mm and a coverslip thickness of 0.085 to 0.13 mm. The dishes were pretreated with poly-l-lysine (molecular weight, >300,000; Sigma). For sensory-motor neuron cocultures, sensory neurons were manually paired with motor neurons to allow for synapse formation.
Microinjection of plasmid vectors.
On day 1 after isolation, solutions of plasmids in distilled water containing 0.25% fast green were microinjected into neurons from back-filled glass micropipettes. The tip of the micropipette was inserted into the cell nucleus, and short pressure pulses (10 to 50 ms in duration; 20 lb/in2) were delivered until the nucleus became uniformly green. The cells were incubated for 4 to 5 h at room temperature and then kept at 4°C until use. Because the fast green was still visible in the red channel in live imaging studies, only eGFP-tagged vectors were used in Aplysia neurons.
Confocal microscopy of Aplysia neurons.
For live cell imaging, neurons expressing eGFP-PKC constructs for PKC Apl II were imaged on a Zeiss laser-scanning microscope (Zeiss, Oberkochen, Germany) with an Axiovert 200 and a ×40 oil immersion objective with a 25-mW argon laser with 25% laser output. The laser line was attenuated to 4% transmission output prior to live imaging. Confocal images were acquired before and after the addition of 5HT (10 μM) to the dish. 5HT then was washed away with artificial seawater (ASW; 10 mM HEPES, pH 7.5, 0.46 M NaCl, 10 mM KCl, 11.2 mM CaCl2·2H2O, 55 mM MgCl2·6H2O). All experiments were performed at room temperature (20 to 23°C).
Drug treatment.
1-Butanol, 2-butanol, U-73122, and U-73433 were purchased from Sigma-Aldrich. Propranolol hydrochloride and the diacylglycerol kinase inhibitor I R59022 were purchased from Calbiochem (San Diego, California). The drugs were used at the following concentrations: 1% 1-butanol, 1% 2- butanol, 10 μM U-73122, 10 μM U-73433, 100 μM propranolol, and 20 μM R59022. In neurons, 1-butanol and 2-butanol were present in the medium 10 min prior to the addition of 5HT (10 μM) to the dish. U-73122 and U-73433 were present in the medium 1 min prior to the addition of 5HT (10 μM) to the dish. In Sf9 cells, 1-butanol and R59022 were present in the medium 10 min prior to the addition of DOG to the dish, and propranolol was present in the medium 10 min prior to the addition of DiC8-PA to the dish. 1-Butanol can replace water in the reaction catalyzed by phospholipase D (PLD) to produce choline and phosphatidylbutan-1-ol instead of PA and choline; however, 2-butanol does not compete in this reaction and, thus, can be used as an appropriate control for the inhibition of the PA generated by PLD. Vehicle controls were always performed when drugs were used in Sf9 cells and in neurons.
Image analysis.
The same quantification technique was used for both Sf9 cells and neurons. The time series was analyzed using NIH Image J software as previously described (53). An individual analysis of protein translocation for each cell was performed by tracing three rectangles at random locations at the plasma membrane and three rectangles at random locations in the cytosol. For synapses, the entire membrane and cytoplasm of varicosities that adjoined the motor neuron were traced using NIH Image, and the intensity was measured. The translocation ratio was measured as the average intensity (membrane)/average intensity (cytosol) (Im/Ic) normalized to the degree of translocation before the addition of pharmacological agents (Post/Pre). The Student's t test or one-way analysis of variance was used to compare translocation ratios in Sf9 cells and in neurons. When two fluorescent proteins were coexpressed within a single cell in Sf9 cells, a paired Student's t test was used. Since there was an a priori expectation that the C2 domain would inhibit translocation, one-tailed tests were used for these comparisons. All data are presented as means ± standard errors of the means.
RESULTS
PA translocates PKC Apl II through its C1b domain.
To study the mechanism by which PA contributes to the plasma membrane localization of PKC Apl II, Sf9 cells were transfected with either eGFP-PKC Apl II or eGFP-PKC Apl II ΔC2 and treated with a cell-permeable phosphatidic acid called DiC8-PA, which can enter the cell but then partitions into membranes (14). The translocation ratio was measured as Im/Ic and was normalized to the degree of translocation before the addition of pharmacological agents. No translocation for either eGFP-PKC Apl II or eGFP-PKC Apl II ΔC2 was seen when the cells were treated with a concentration of 5 μg/ml DiC8-PA (Fig. 1C). However, when this concentration was increased to 10, 25, or 50 μg/ml DiC8-PA, translocation takes place for both proteins (Fig. 1A). Translocation induced by DiC8-PA is slow, with maximal translocation ratios reached only after 300 s of imaging (data not shown). There were no differences at any time point (data not shown) or any concentration of DiC8-PA (Fig. 1C) between the translocation of PKC Apl II and that of PKC Apl II ΔC2. Thus, the translocation of PKC Apl II by DiC8-PA alone neither requires the C2 domain nor is inhibited by the C2 domain. However, translocation to DiC8-PA is specific, in that DiC8-PA could not induce PKC Apl I translocation (Fig. 1A and B). Furthermore, translocation to DiC8-PA did not require PA to be converted to DAG, because the addition of DiC8-PA in the presence of propranolol, which inhibits DAG formation from PA by inhibiting phosphatidate phosphohydrolase (20, 22, 28), did not affect translocation (data not shown).
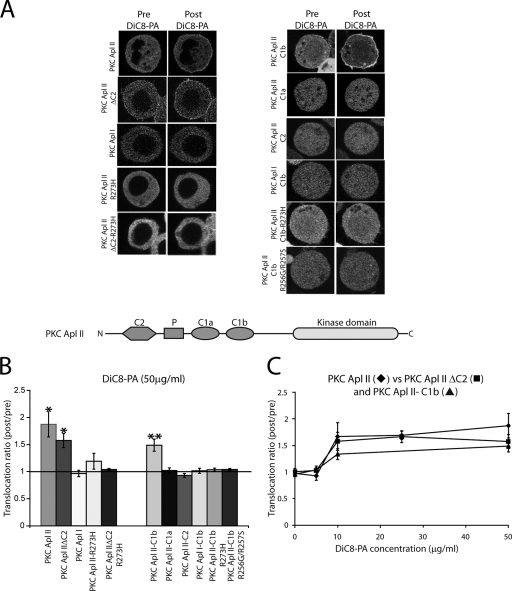
FIG. 1.
PA binding domain in PKC Apl II is located in the C1b domain. (A) Confocal fluorescence images of Sf9 cells expressing eGFP-PKC Apl II, eGFP-PKC Apl II ΔC2, eGFP-PKC Apl I, eGFP-PKC Apl II-R273H, eGFP-PKC Apl II ΔC2-R273H, the eGFP-PKC Apl II-C1b domain, the eGFP-PKC Apl II-C1a domain, the eGFP-PKC Apl II-C2 domain, the eGFP-PKC Apl I-C1b domain, eGFP-PKC Apl II-C1b-R273H, and eGFP-PKC Apl II-C1b-R256G/R257S at different points of the time-lapse experiment (Pre DiC8-PA was 0 s and Post DiC8-PA was 300 s, when translocation to DiC8-PA was maximal). For all of the constructs shown in this figure, except for those of eGFP-PKC Apl II and eGFP-PKC Apl II ΔC2, DiC8-PA (50 μg/ml) was added to the dish after 30 s of recording. For eGFP-PKC Apl II and eGFP-PKC Apl II ΔC2, DiC8-PA (25 μg/ml) was added to the dish after 30 s of recording. P, pseudosubstrate sequence. (B) The translocation ratios at 300 s of the constructs cited in panel A are shown in the presence of DiC8-PA (50 μg/ml). PKC Apl II, PKC Apl II ΔC2, and the PKC Apl II-C1b domain show significant translocation in the presence of DiC8-PA (50 μg/ml). **, P < 0.001; *, P ≤ 0.01; both were determined by two-tailed paired Student's t tests. n > 8 for each construct. (C) Dose-response of PKC Apl II (diamonds), PKC Apl II ΔC2 (squares), and PKC APl II-C1b domain (triangles) translocation at 300 s at different concentrations of PA. The translocation ratio for PKC Apl II was similar to that of PKC Apl II ΔC2 and to that of the PKC Apl II-C1b domain at all concentrations of DiC8-PA examined (5, 10, 25, and 50 μg/ml, respectively). n > 5 for each concentration of DiC8-PA.
To identify the domain of PKC Apl II that allowed DiC8-PA-mediated translocation, we generated fusion proteins of eGFP coupled to the PKC Apl II C2 domain alone, the C1a domain alone, or the C1b domain alone. These constructs were individually expressed in Sf9 cells, and their ability to translocate from the cytoplasm to the plasma membrane in response to DiC8-PA (50 μg/ml) was examined (Fig. 1A and B). Note that unlike the full-length enzymes, the smaller eGFP fusion proteins partitioned into the nucleus as well. eGFP-PKC Apl II-C1b was the only construct to exhibit translocation in response to DiC8-PA (50 μg/ml), suggesting that the PA interaction site was located in the C1b domain of PKC Apl II (Fig. 1B). The amount of PA required for the translocation of the isolated C1b domain was similar to that of the full-length enzyme (Fig. 1C). Translocation to DiC8-PA was specific, in that the eGFP-PKC Apl I-C1b domain did not translocate in response to DiC8-PA (Fig. 1A and B). These results are consistent with results from in vitro experiments that showed that the C1 domain, but not the C2 domain, of PKC Apl II could bind PA (36).
Identification of residues required for translocation to DiC8-PA.
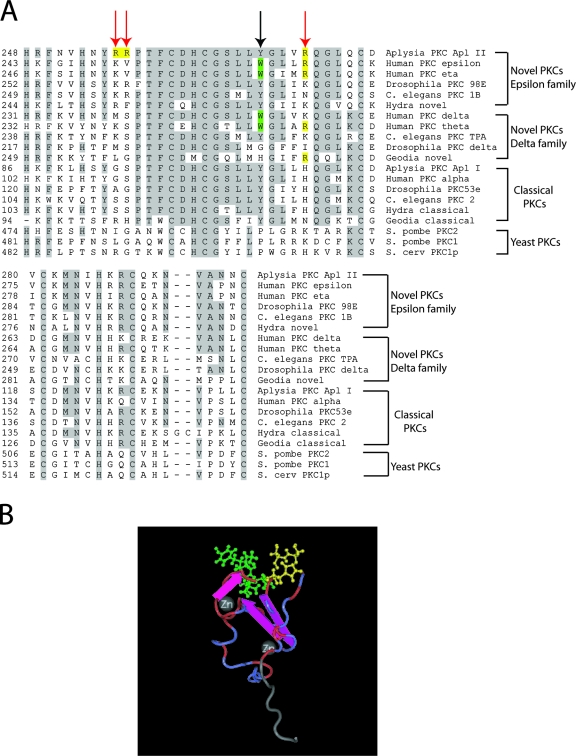
To identify the residues involved in PA binding to the C1b domain, we compared the C1b domain among different PKCs (Fig. 2A). We chose residues to mutate by using the following criteria: (i) the residues should have the ability to interact with the membrane adjacent to the DAG/phorbolester (DAG/PE) binding site based on the structures of the C1b domain of PKCδ and PKCγ (50, 52); (ii) the residues should be positively charged and, thus, be more likely to interact with PA; and (iii) the residues should be related in the nPKCs similarly to PKC Apl II but distinct from the residues in the C1b domain of cPKCs like PKC Apl I that do not appear to interact with PA. Based on these criteria, Arg 256, Arg 257, and Arg 273 were the most promising. Indeed, an examination of the three-dimensional structure of the C1b domain based on PKCγ (50) suggests that these residues (Fig. 2B) are positioned close to the DAG/PE binding site (Fig. 2B) and face the membrane. Since the C1b domain of PKC Apl I did not translocate in response to DiC8-PA, the arginines in the PKC Apl II-C1b domain were mutated to the residues present in PKC Apl I, as these should minimize structural alterations to the C1 domain. As shown in Fig. 1A and B, eGFP-PKC Apl II-C1b-R256G/R257S and eGFP-PKC Apl II-C1b-R273H mutants did not translocate in response to DiC8-PA (50 μg/ml). Similar results were obtained when full-length eGFP-PKC Apl II-R273H or truncated eGFP-PKC Apl II ΔC2-R273H or eGFP-PKC Apl II-R256G/R257S translocation was examined in response to DiC8-PA (50 μg/ml); (Fig. 1A and B and data not shown). Taking the above results together, the PA interaction site in PKC Apl II is located in the C1b domain and Arg 256, Arg 257, and Arg 273 are required for PA-mediated translocation.
FIG. 2.
Sequence alignment of the C1b domains and a representation of the overall structure of the C1b domain in nPKCs. (A) Comparison of the C1b domains of the nPKC epsilon family, the nPKC delta family, the cPKCs, and yeast PKCs. Gray shaded residues are conserved in at least 10 isoforms. Red arrows point to arginine residues that were mutated in the C1b domain of PKC Apl II (residues shaded in yellow). Arginines in positions 256 and 257 were mutated to a glycine and a serine, respectively, and arginine in position 273 was mutated to a histidine. The black arrow points to a tyrosine residue in position 269 that was shown to tune the affinity of the C1b domain for DAG-containing membranes (13). C. elegans, Caenorhabditis elegans; S. pombe, Saccharomyces pombe; S. cerv, Saccharomyces cerevisiae. (B) Three-dimensional representation of the structure of the C1b domain based on the C1b domain of PKCγ (50). The ribbon diagram shows conservation between the C1b domain of PKCγ and the C1b domain of PKC Apl II (red ribbons are conserved, blue ribbons are not). Residues colored in green (S111, T113, F114, L122, Y123, and G124 in PKCγ) are involved in phorbol ester binding. Residues colored in yellow were mutated in the C1b domain of PKC Apl II. These residues are R256, R257, and R273 in PKC Apl II, and the corresponding residues in PKCγ (shown in the diagram) are S109, S110, and H126. The structure was generated using CN3D 4.1 (produced by the National Center for Biotechnology Information; http://www.ncbi.nlm.nih.org).
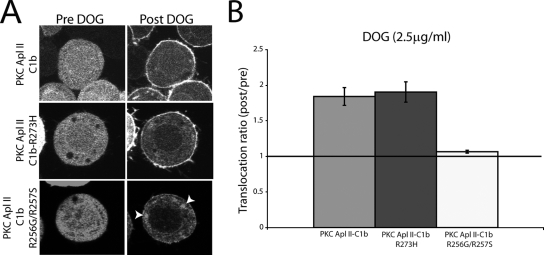
To rule out the possibility that the lack of translocation is due to the misfolding of the proteins or an inability to interact with other cofactors, such as DAG and PS, we next examined the effect of the mutations on the translocation of the C1b domain in response to a cell-permeable analog of DAG, termed DOG, that can enter the cell but then partitions into membranes. Sf9 cells were transfected with eGFP-PKC Apl II-C1b, eGFP-PKC Apl II-C1b-R273H, or eGFP-PKC Apl II-C1b-R256G/R257S, and translocation from the cytoplasm to the membrane was examined following the treatment of the cells with DOG. Figure 3A shows that while eGFP-PKC Apl II-C1b and eGFP-PKC Apl II-C1b-R273H translocated to the plasma membrane to a similar extent after DOG addition, eGFP-PKC Apl II-C1b-R256G/R257S was not translocated to the plasma membrane by DOG, although this protein did translocate to internal membranes (Fig. 3A). Specific translocation to the plasma membrane may indicate specificity for PS (27, 45, 46), and thus this mutation may affect binding to PS as opposed to PA (see Discussion); therefore, this mutation was not studied further.
FIG. 3.
Characterization of DAG binding to the PKC Apl II-C1b mutated constructs. (A) Confocal fluorescence images of Sf9 cells expressing eGFP-PKC Apl II-C1b, eGFP-PKC Apl II-C1b-R273H, and eGFP-PKC Apl II-C1b-R256G/R257S at different points of the time-lapse experiment (Pre DOG being 0 s and Post DOG being 180 s). Cells were treated with DOG (2.5 or 5 μg/ml), which was added to the dish after 30 s of imaging. The white arrowheads point to unidentified internal membranous compartments in the cell. (B) Comparison of the translocation ratios of eGFP-PKC Apl II-C1b, eGFP-PKC Apl II-C1b-R273H, and eGFP-PKC Apl II-C1b-R256G/R257S at a DOG concentration of 2.5 μg/ml. The translocation ratio presented is an average of the translocation ratios at 120, 150, and 180 s, since translocation to DOG was maximal at these time points; n > 8 for each construct.
DiC8-PA synergizes with DOG for translocation.
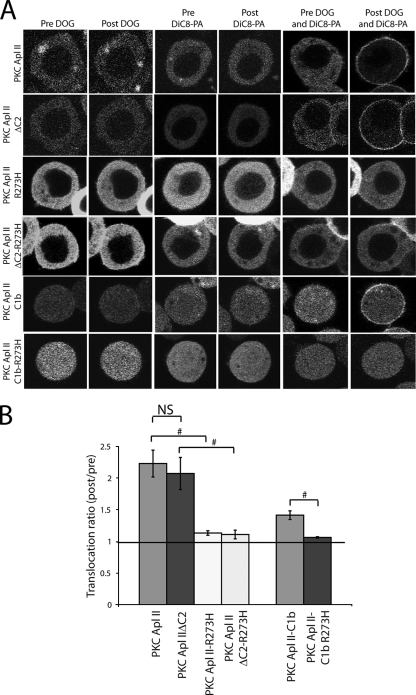
If PA interacts directly with the C1b domain next to the DAG/PE binding domain, it suggests that the two lipids act synergistically to translocate PKC Apl II. Indeed, when Sf9 cells were transfected with eGFP-PKC Apl II or eGFP-PKC Apl II ΔC2 and treated with a combination of small amounts of DOG and DiC8-PA that caused no translocation on their own, translocation was observed (Fig. 4A and B). Consistent with the synergism being due to the PA interaction site in C1b, DOG and DiC8-PA also could synergistically translocate the isolated eGFP-PKC Apl II-C1b domain (Fig. 4A). Similarly to the binding to PA alone, the R273H mutation abolished synergism in both the isolated C1b domain and the full-length enzymes (Fig. 4A and B). Furthermore, consistent with the specific PA-dependent translocation of eGFP-PKC Apl II, no synergism was observed for eGFP-PKC Apl I (data not shown).
FIG. 4.
DiC8-PA synergizes with DOG for the translocation of PKC Apl II and PKC Apl II ΔC2, and this synergism is a property of the C1b domain. (A) Confocal fluorescence images of Sf9 cells expressing eGFP- or mRFP-PKC Apl II, eGFP- or mRFP-PKC Apl II ΔC2, eGFP-PKC Apl II-R273H, eGFP-PKC Apl II ΔC2-R273H, the eGFP-PKC Apl II-C1b domain, and eGFP-PKC Apl II-C1b-R273H at different points of the time-lapse experiment (Pre DOG being 0 s and Post DOG being 180 s). Cells were treated with DOG alone, DiC8-PA alone, or a combination of DOG and DiC8-PA, which were added to the dish after 30 s of imaging. The DOG concentration used varied between constructs and corresponded to the highest concentration at which no translocation was observed. For PKC Apl II and PKC Apl II ΔC2, this concentration was 0.25 μg/ml. For PKC Apl II-R273H, PKC Apl II ΔC2-R273H, and PKC Apl II-C1b, the DOG concentration used was 0.03 μg/ml. For PKC Apl II-C1b-R273H, the concentration of DOG was 0.1 μg/ml. For all of the constructs shown, the concentration used for DiC8-PA was 5 μg/ml, which corresponded to the highest concentration at which no translocation was observed. (B) The translocation ratio (which corresponds to an average of the translocation ratios at 120, 150, and 180 s) of the constructs shown in panel A is presented in the presence of a combination of DiC8-PA and DOG at the concentrations cited above (#, P ≤ 0.005; two-tailed unpaired Student's t test). n > 7 for each observation. NS, not significant.
Interconversion between DiC8-PA and DOG was not important for synergism, since the presence of propranolol, an inhibitor of phosphatidate phosphohydrolase, or that of R59022, a DAG kinase inhibitor (12), in the media did not affect synergism (data not shown).
Both PI-PLC and PLD contribute to the translocation of PKC Apl II by 5HT in Aplysia sensory neurons.
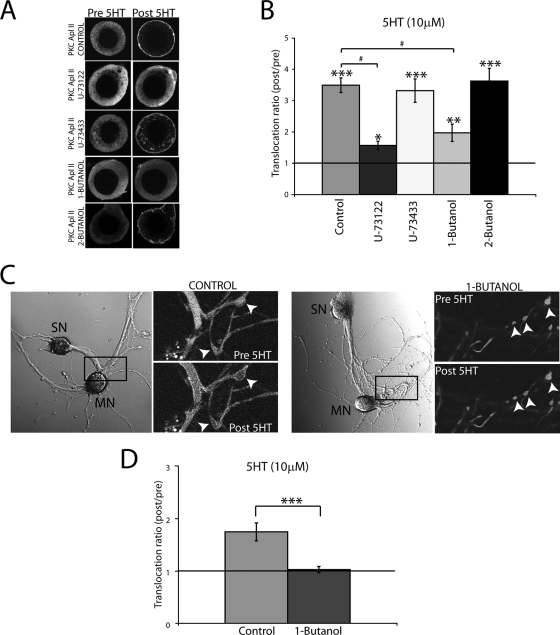
5HT application leads to the translocation of PKC Apl II but not PKC Apl I in isolated sensory neurons (53). To investigate the contribution of DAG and PA to the plasma membrane localization of PKC Apl II following the treatment of sensory neurons with 5HT, we used inhibitors of the signaling pathways involved in the generation of DAG and PA. Thus, we used U-73122, which is known to inhibit phosphatidylinositol-specific phospholipase C (PI-PLC) (4) and to impede DAG synthesis from phosphatidylinositol and 1-butanol, which can replace water in the reaction catalyzed by PLD and can inhibit the production of PA (9). Control experiments were performed with 2-butanol, which is an isomer of 1-butanol that does not have an effect on PLD (1), and with U-73433, which is an inactive analog of U-73122. It should be noted that the amount of translocation of PKC Apl II significantly varied with sensory neurons isolated from different batches of animals; thus, comparisons in the figures described below were made only between experiments performed on sensory cells isolated from the same batch of animals.
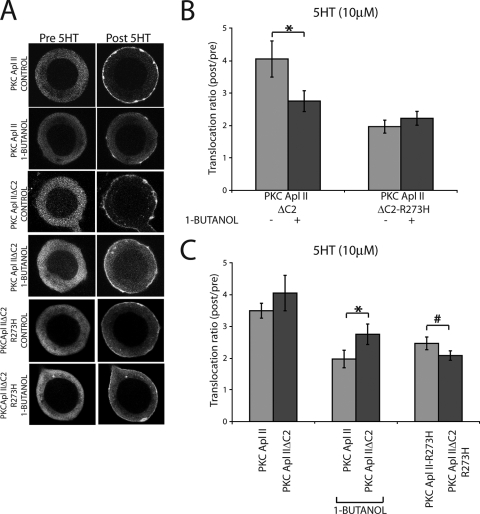
Aplysia sensory neurons were microinjected with plasmid DNA encoding eGFP-PKC Apl II, and the experiments were performed on the next day (53). In resting cells, eGFP-PKC Apl II was localized in the cytosol (Fig. 5A). Upon the treatment of the cells with 5HT, the fluorescence shifts from the cytosol to the membrane (Fig. 5A). This translocation reverts to control conditions when 5HT is removed (data not shown). When the neurons are treated for 1 min with the PI-PLC inhibitor U-73122 prior to 5HT treatment, translocation is significantly reduced (Fig. 5A and B). Similar results are obtained when the cells are treated for 10 min with the PLD inhibitor 1-butanol prior to 5HT treatment (Fig. 5A and B). However, U-73433 and 2-butanol did not inhibit translocation (Fig. 5A and B). These results suggest that both DAG generated through the PI-PLC pathway and PA generated through the PLD pathway are required for the translocation of PKC Apl II to the plasma membrane in response to 5HT in Aplysia sensory neurons.
FIG. 5.
Both PI-PLC and PLD contribute to the translocation of eGFP-PKC Apl II by 5HT in living sensory neurons. (A) Representative confocal fluorescence images of Aplysia sensory neurons expressing eGFP-PKC Apl II before (Pre 5HT) or 5 min following treatment with 5HT (Post 5HT) in the presence or absence of 10 μM U-73122 (PLC inhibitor), 10 μM U-73433 (inactive analog of U-73122), 1% 1-butanol (PLD alternative substrate), and 1% 2-butanol (inactive analog of 1-butanol). (B) The translocation ratio (Post 5HT/Pre 5HT) is shown for the different conditions cited in panel A. Error bars represent standard errors of the means; n ≥ 8 for each condition. One-way analysis of variance was used to compare the treatments. The translocation ratio of control cells was significantly higher than that of the U-73122-treated cells and the 1-butanol-treated cells (#, P < 0.001). Similarly, the translocation ratio of the U-73433-treated cells and that of the 2-butanol-treated cells also were significantly higher than that of the U-73122-treated cells and the 1-butanol-treated cells (P < 0.05). We also used one-tailed paired Student's t tests to determine if there was a difference between translocation after 5HT treatment compared to that before 5HT treatment for each condition (***, P ≤ 0.0001; **, P = 0.003; and *, P = 0.05; one-tailed paired Student's t test). (C) Translocation of eGFP-PKC Apl II in synaptic regions. The left panels show the bright-field images of cocultures of sensory neurons (SN) expressing eGFP-PKC Apl II and motor neurons (MN). The right panel shows a detail of confocal fluorescence images before and 1 min after the application of 5HT in the absence (control) or presence of 1-butanol (1%). (D) Summary plot showing the translocation ratio of eGFP-PKC Apl II at synaptic regions measured in the absence or presence of 1-butanol (1%) (***, P = 0.0005; two-tailed unpaired Student's t test). Error bars represent standard errors of the means; n = 15 varicosites from >3 cocultures for each condition.
We also could detect translocation in sensory neuron varicosities adjoining motor neuron processes in cocultures (Fig. 5C and D). These varicosities have been shown to have a high probability of being synapses (17). When the cocultures are treated with 1-butanol prior to 5HT treatment, translocation is inhibited (Fig. 5C and D), demonstrating that PA also is required for translocation at synapses.
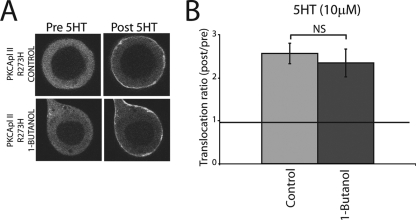
To confirm that 1-butanol was acting through the production of PA, we examined the translocation of eGFP-PKC Apl II-R273H in the absence or presence of 1-butanol. As shown in Fig. 6A and B, the translocation of this isoform was completely resistant to 1-butanol, consistently with 1-butanol acting through the inhibition of PA production. Thus, while PA is involved in the translocation of PKC Apl II, the mutant that does not interact with PA translocates in a PA-independent manner.
FIG. 6.
Blocking PA production has no effect on PKC Apl II-R273H translocation. (A) Confocal fluorescence images of the translocation of eGFP-PKC Apl II-R273H in the absence or presence of 1-butanol (1%). Pictures shown represent translocation after 5 min of treatment with 5HT. (B) Summary plot showing the translocation ratio of eGFP-PKC Apl II-R273H measured in the absence or presence of 1-butanol (1%). Error bars represent standard errors of the means; n ≥ 7 for each condition; NS, not significant.
To further investigate the translocation mechanism of the PKC Apl II-R273H mutant, below we examine the effects of this mutation on (i) the C2 domain-mediated inhibition of translocation and (ii) the translocation of the kinase to DAG.
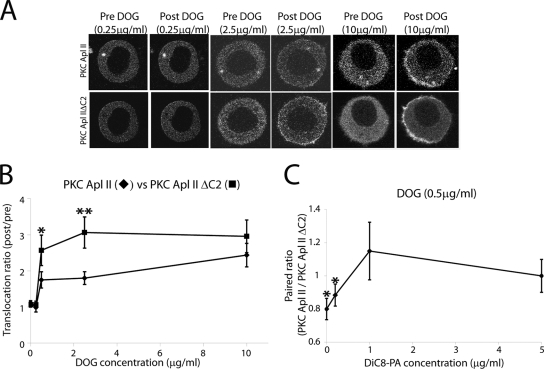
C2 domain-mediated inhibition is removed in the R273H mutant, and the binding affinity to DAG is increased.
Earlier we had reported that the C2 domain of PKC Apl II lowers the affinity of phorbol esters to bind to the C1 domains (37). A comparison of the translocation of PKC Apl II and PKC Apl II ΔC2 in Sf9 cells revealed that at low concentrations of DOG (0.5 and 2.5 μg/ml), PKC Apl II ΔC2 translocated more efficiently than PKC Apl II, demonstrating that the negative effect of the C2 domain also could be observed in translocation experiments (Fig. 7A and B). At higher concentrations of DOG (10 μg/ml), the difference between the kinases was not significant (Fig. 7A and B). In these experiments, we compared the translocation of different tagged forms (e.g., eGFP-PKC Apl II and mRFP-PKC Apl II ΔC2) in the same cell. Differences between the two kinases were not due to the type of fluorescent tag, since similar results were obtained with mRFP-PKC Apl II and eGFP-PKC Apl II ΔC2 (data not shown). The results also are not due to competition between the two kinases, since similar results are seen when the kinases are translocated individually (data not shown). Furthermore, translocation to DOG did not require DOG to be converted to PA, because the addition of DOG in the presence of the DAG kinase inhibitor R59022 did not affect translocation (data not shown).
FIG. 7.
PA removes C2 domain-mediated inhibition. (A) Confocal fluorescence images of Sf9 cells coexpressing mRFP-PKC Apl II and eGFP-PKC Apl II ΔC2 at different points of the time-lapse experiment (Pre DOG being 0 s and Post DOG being 60 s). DOG (0.25, 2.5, and 10 μg/ml) was added to the dish after 30 s of recording. (B) Dose-response of PKC Apl II (diamonds) translocation and PKC Apl II ΔC2 (squares) translocation at different concentrations of DOG. The translocation ratio of PKC Apl II ΔC2 is significantly higher than that of PKC Apl II at DOG concentrations of 0.5 and 2.5 μg/ml, respectively (*, P ≤ 0.04; **, P = 0.005; one-tailed paired Student's t test; n > 8 for each concentration of DOG). (C) The paired ratio (at 60 s), which represents the translocation ratio for PKC Apl II divided by the translocation ratio for PKC Apl II ΔC2, is shown at different concentrations of DiC8-PA in the presence of DOG (0.5 μg/ml). The translocation ratio of PKC Apl II ΔC2 was significantly higher than that of PKC Apl II when DOG (0.5 μg/ml) was combined with DiC8-PA (0.2 μg/ml). *, P ≤ 0.05 by one-tailed paired Student's t test. This difference was no longer significant when DOG (0.5 μg/ml) was combined with either DiC8-PA (1 μg/ml) or DiC8-PA (5 μg/ml); n > 8 for each concentration of DiC8-PA.
We next examined the effect of different concentrations of DiC8-PA on translocation using low concentrations of DOG (0.5 μg/ml) in which the difference between the translocation of PKC Apl II and that of PKC Apl II ΔC2 was maximal (Fig. 7B). We measured the paired ratio, defined as the Im/Ic ratio for PKC Apl II (normalized to time zero) divided by the Im/Ic ratio for PKC Apl II ΔC2 (normalized to time zero), in the same cell. DiC8-PA concentrations of 1 μg/ml were sufficient to remove any difference between the translocation of PKC Apl II and PKC Apl II ΔC2, bringing the paired ratio close to 1 (Fig. 7C). Thus, in the presence of DiC8-PA, C2 domain-mediated inhibition was not observed.
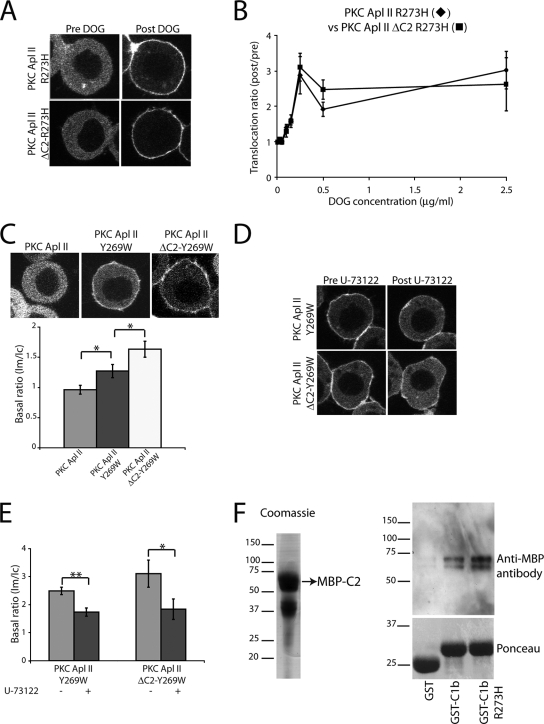
Since mutating R273H removed the effects of exogenous DiC8-PA (Fig. 1), we initially expected to see a similar or even larger difference in comparisons of eGFP-PKC Apl II-R273H to eGFP-PKC Apl II ΔC2-R273H because of C2 domain-mediated inhibition. Figure 8A shows a representative example of the translocation of eGFP-PKC Apl II-R273H and eGFP-PKC Apl II ΔC2-R273H in response to DOG at 0.25 μg/ml. At this concentration of DOG and all of the other concentrations tested, these proteins translocated similarly (Fig. 8B). Thus, this mutation has the same effect as exogenous PA, removing the effect of C2 domain-mediated inhibition.
FIG. 8.
Characterization of DAG binding to the R273H and Y269W mutants. (A) Confocal fluorescence images of Sf9 cells expressing eGFP-PKC Apl II-R273H and eGFP-PKC Apl II ΔC2-R273H, respectively, at different points of the time-lapse experiment (Pre DOG being 0 s and Post DOG being 180 s). Cells were treated with DOG (0.25 μg/ml), which was added to the dish after 30 s of imaging. (B) Comparison of the dose-response of eGFP-PKC Apl II-R273H (diamonds) to that of eGFP-PKC Apl II ΔC2-R273H (squares) at different concentrations of DOG. The translocation ratio shown is an average of the translocation ratios at 120, 150, and 180 s. The translocation ratios of eGFP-PKC Apl II-R273H and eGFP-PKC Apl II ΔC2-R273H are not significantly different at any time point examined; n > 8 for each concentration of DOG. (C) Top, confocal fluorescence images of Sf9 cells expressing eGFP-PKC Apl II, eGFP-PKC Apl II-Y269W, and eGFP-PKC Apl II ΔC2-Y269W, respectively. Bottom, summary plot showing the basal translocation ratio (Im/Ic) of eGFP-PKC Apl II, eGFP-PKC Apl II-Y269W, and eGFP-PKC Apl II ΔC2-Y269W (*, P ≤ 0.04 by two-tailed unpaired Student's t test). Error bars represent standard errors of the means; n ≥ 15 for each condition. (D) Confocal fluorescence images of Sf9 cells expressing eGFP-PKC Apl II-Y269W and eGFP-PKC Apl II ΔC2-Y269W, respectively, at different points of the time-lapse experiment (Pre U-73122 being 0 min and Post U-73122 being 5 min). Only cells with a strong basal translocation for eGFP-PKC Apl II-Y269W and eGFP-PKC Apl II ΔC2-Y269W were used for this experiment; thus, the initial translocation is larger than that seen in panel C. (E) Summary plot showing the basal translocation ratio (Im/Ic) of eGFP-PKC Apl II-Y269W and eGFP-PKC Apl II ΔC2-Y269W in the absence or presence of U-73122 (**, P = 0.001; *, P = 0.01; two-tailed paired Student's t test). Error bars represent standard errors of the means; n = 8 for each condition. (F) Left, purified MBP-C2 protein (∼69 kDa) was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the gel was stained with Coomassie. The band shown between the 37- and 50-kDa molecular mass standards is the ovalbumin that was used to avoid nonspecific binding to the beads. Right, MBP-C2 was incubated with GST alone, GST-C1b, and GST-C1b-R273H bound to glutathione-Sepharose beads as described in Materials and Methods. The presence of the GST proteins was confirmed by staining the membrane with Ponceau red (bottom). The presence of the MBP-C2 domain was revealed using an antibody against MBP (clone MBP-17).
During the course of these experiments, we also noted that eGFP-PKC Apl II-R273H required less DOG for translocation than did eGFP-PKC Apl II. The minimal concentration required for translocation was lowered from 0.5 to 0.10 μg/ml (Fig. 8B). This also was true for eGFP-PKC Apl II ΔC2-R273H (Fig. 8B) and, thus, cannot be explained by removing C2 domain-mediated inhibition. To determine if the increased affinity for DAG could explain the lack of C2 domain-mediated inhibition, we tested another mutant of PKC Apl II Y269W. This residue is found in chordate nPKCs (Fig. 2A) and has been reported to increase affinity for DAG (13). Indeed, we found that this mutation in the context of PKC Apl II was sufficient to increase association with membranes even in the absence of added DOG or PA (Fig. 8C). However, even with the higher affinity for membranes, there still was significantly more membrane association with eGFP-PKC Apl II ΔC2-Y269W than with eGFP-PKC Apl II-Y269W (Fig. 8C). The higher membrane affinity of the Y269W kinase was at least partly due to higher affinity for DAG, since U-73122, an inhibitor of PI-PLC that impedes DAG production in cells (4), decreased the basal translocation of both eGFP-PKC Apl II-Y269W and eGFP-PKC Apl II ΔC2-Y269W (Fig. 8D and E). Thus, increasing affinity for DAG is not sufficient to remove C2 domain-mediated inhibition, and the loss of C2 domain-mediated inhibition in the R273H PKC cannot be attributed to its higher affinity for DOG.
C2-mediated inhibition has been shown to be due to direct intermolecular interactions between the C2 domain and the C1 domain (40). To verify this in PKC Apl II, we constructed MBP-C2, GST-C1b, and GST-C1b-R273H fusion proteins. The proteins were expressed in bacteria and purified as described in Materials and Methods. There was significantly more binding of the MBP-C2 domain to the GST-C1b domain compared to that of GST alone (Fig. 8F). However, the introduction of the R273H mutation into this fusion protein did not reduce this binding (Fig. 8F). Thus, the removal of C2 domain-mediated inhibition by this mutation is not due to a decreased interaction between the C1b and the C2 domains.
PA both removes C2 domain-mediated inhibition and synergizes with DAG to assist in the translocation of PKC Apl II in sensory neurons.
We have shown that PA can activate PKC Apl II by two mechanisms: (i) the removal of C2 domain-mediated inhibition and (ii) the synergistic activation with DAG. To determine the contribution of these two mechanisms in sensory neurons, we first examined the 5HT-mediated translocation of eGFP-PKC Apl II and eGFP-PKC Apl II ΔC2 in the absence and presence of 1-butanol. Since PKC Apl II ΔC2 is not affected by C2 domain-mediated inhibition, any effect of 1-butanol must be due to a synergistic role of PA. 1-Butanol inhibited the translocation of eGFP-PKC Apl II ΔC2, which is consistent with an important role for the synergism of DAG and PA in the activation of PKC Apl II and independent of an effect of the C2 domain (Fig. 9A and B). 1-Butanol had no effect on the translocation of eGFP-PKC Apl II ΔC2-R273H, which is consistent with 1-butanol acting through reducing levels of PA (Fig. 9A and B). There also was a role for PA in removing C2 domain-mediated inhibition in sensory cells. There was no significant difference between the translocation of eGFP-PKC Apl II and eGFP-PKC Apl II ΔC2 by 5HT in sensory cells (Fig. 9C), when PA presumably was present. However, in the presence of 1-butanol, eGFP-PKC Apl II ΔC2 translocated better than PKC Apl II (Fig. 9C), consistent with an inhibitory contribution of the C2 domain that is normally removed by PA. Thus, our results suggest that PA acts in sensory neurons both to remove C2 domain-mediated inhibition and to synergize with DAG to translocate PKC Apl II to the membrane.
FIG. 9.
PA removes C2 domain-mediated inhibition and synergizes with DAG to assist the translocation of PKC Apl II in sensory neurons. (A) Confocal fluorescence images of the translocation of eGFP-PKC Apl II, eGFP-PKC Apl II ΔC2, and eGFP-PKC Apl II ΔC2-R273H in the absence or presence of 1-butanol (1%). Pictures shown represent translocation after 5 min of treatment with 5HT. (B) Summary plot showing the translocation ratios for control cells expressing eGFP-PKC Apl II ΔC2 and eGFP-PKC Apl II ΔC2-R273H. The ratios were measured in the absence or presence of the PLD inhibitor 1-butanol (1%). Error bars represent standard errors of the means; n ≥ 7 for each condition. eGFP-PKC Apl II translocation ratios used in this plot are the same as those used for Fig. 5. As mentioned above, the translocation of eGFP-PKC Apl II was significantly inhibited in the presence of 1-butanol (***, P = 0.0001 by two-tailed unpaired Student's t test). There was also an inhibition of eGFP-PKC Apl II ΔC2 translocation in the presence of 1-butanol (*, P = 0.05 by two-tailed unpaired Student's t test). There was no effect of 1-butanol on the translocation of eGFP-PKC Apl II ΔC2-R273H. (C) Summary plot showing the translocation ratios of cells expressing eGFP-PKC Apl II ΔC2 compared to those of cells expressing eGFP-PKC Apl II in the absence or presence of 1-butanol (1%). The translocation ratios of cells expressing eGFP-PKC Apl II-R273H and eGFP-PKC Apl II ΔC2-R273H in the absence or presence of 1-butanol were pooled together, since there was no effect of 1-butanol on the mutants. Error bars represent standard errors of the means; n ≥ 7 for each condition. Translocation ratios shown in this plot are the same as those shown in Fig. 5. There is a significant difference between the translocation ratios of eGFP-PKC Apl II and those of eGFP-PKC Apl II ΔC2 in the presence of 1-butanol (*, P = 0.03 by one-tailed unpaired Student's t test). There is also a small difference between the translocation ratios of eGFP-PKC Apl II-R273H and those of eGFP-PKC Apl II ΔC2-R273H, but this is of borderline significance (#, P = 0.11 by two-tailed unpaired Student's t test).
The C2 domain has been postulated to increase the translocation of the PKC Apl II orthologue, PKCɛ, through binding to lipids or through binding to receptors for activated C kinase. The R273H mutation allows us to determine whether there is any positive role for the C2 domain, since this mutation removes C2 domain-mediated inhibition but the C2 domain is still present. A comparison of eGFP-PKC Apl II-R273H to eGFP-PKC Apl II ΔC2-R273H suggests a small positive effect of the C2 domain, since there is a slightly better translocation of eGFP-PKC Apl II-R273H than eGFP-PKC Apl II ΔC2-R273H (Fig. 9C), but this is of borderline significance (P = 0.11 by a two-tailed unpaired Student's t test).
DISCUSSION
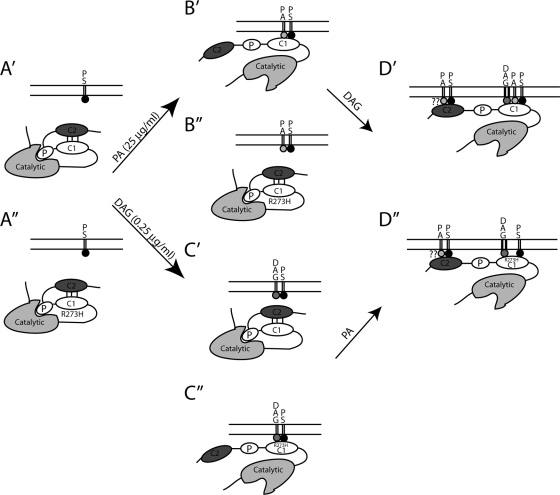
The present results lead us to propose a model in which PA binding to the C1b domain is important for two purposes: (i) to remove C2 domain-mediated inhibition and (ii) to facilitate the binding of DAG to the C1 domain, allowing for the synergistic translocation of the protein (Fig. 10A′, B′, and D′). The R273H mutation both removes C2 domain-mediated inhibition and blocks the ability of the kinase to interact with PA in the plasma membrane (Fig. 10A", C", and D"). This is consistent with PA and the C2 domain normally competing for binding to the region around R273 and explains how PA can remove C2 domain-mediated inhibition. However, since the GST-C1b-R273H protein still binds to the C2 domain, it suggests that binding can be dissociated from inhibition (Fig. 10B"). One possibility is that the binding of the C1b domain to one region of the C2 domain is important for allowing the inhibitory interaction through a distinct domain. The R273H mutation does not block the binding, but the inhibitory interaction is no longer present. The R273H mutation also strengthens the interaction of the C1 domain with DAG in the plasma membrane (Fig. 10D"), either by altering the DAG binding site or perhaps by allowing stronger interactions with other lipids, such as PS. It is possible that once binding to the C1 domain is removed, the C2 domain also contributes to membrane binding (Fig. 10D′ and D").
FIG. 10.
Model for nPKC membrane translocation and activation. Our present results lead us to propose a model in which the C2 domain of PKC Apl II binds to the C1b domain in the wild type (A′) and the R273H mutant (A") but only inhibits DAG binding to the C1b domain in the wild type. P, pseudosubstrate. (B′) PA binding to the C1 domain would remove the C1-C2 domain interactions in the wild-type protein and allow translocation in conjunction with PS at high concentrations. (B") PA does not interact with the R273H mutant. On the other hand, low concentrations of DAG would be sufficient to translocate the R273H mutant (C") in the absence of PA but not the wild-type protein (C′) due to the higher affinity of the R273H kinase for DAG and the loss of C2 domain-mediated inhibition. For the wild-type protein, PA would remove C2 domain inhibition and synergize with DAG to translocate the enzyme (D′). The R273H mutant does not bind to PA but has a higher binding affinity to DAG (D"). It is not clear from the present study whether the C2 domain contributes to membrane binding at this step (??).
PA binding to the C1 domain and not to the C2 domain.
Our results suggest that PA does not act by binding to the C2 domain directly but by binding to the C1b domain. This is consistent with in vitro data with isolated C1 and C2 domains of PKC Apl II, in which PA binds to the isolated C1 domains better than to the C2 domain (36, 37). The vertebrate homologue of PKC Apl II, PKCɛ, also has been shown to be synergistically activated by DAG and PA, but in this case it was suggested that synergism was due to PA binding to the C2 domain (18). The main justification for this model was the in vitro binding of the C2 domain of PKCɛ to PA and the lack of translocation when residues in the C2 domain responsible for PA binding were mutated. While these mutations decreased the affinity of the C2 domain for PA, they also could have strengthened C2-C1 domain interactions, and this could be the reason for their effect on translocation. It is striking that in these two very different systems, PKCɛ and PKC Apl II both demonstrated a requirement for the combination of DAG and PA for physiological activation, suggesting a highly conserved role for PA in the activation of this class of PKC isoform. Moreover, R273 is conserved in PKCɛ (Fig. 2A). Nevertheless, it is still possible that the enzymes are activated differently; indeed, there are significant differences between the translocation of PKC Apl II and that of PKCɛ (see below).
Comparison between PKCɛ and PKCθ.
Giorgione and coworkers reported that the deletion of the C2 domain did not affect the ability of PKCɛ to bind to PS/DAG membranes (16). However, in this study, saturating amounts of DAG were used, and this might have compensated for the lack of the C2 domain. Another study by Stahelin and coworkers reported that the deletion of the C2 domain of PKCɛ induced a faster membrane translocation in HEK293 cells (45). Furthermore, the C2-deleted construct had a higher affinity (∼60%) for PS/DAG membranes and a higher level of activity (<50%) than the PKCɛ wild type. These results suggest that C2-mediated inhibition is conserved during evolution.
C2 domain-mediated inhibition is a general feature of nPKCs. One recent study by Melowic and colleagues (27) reported that the deletion of the C2 domain of PKCθ greatly enhanced its affinity to PS/DAG-containing membranes. Furthermore, the authors proposed that the C2 domain of PKCθ is involved in keeping the enzyme in an inactive conformation, presumably by interacting with the C1a and C1b domains (27). Interestingly, PKCθ also has an arginine in position 273 (Fig. 2A).
PS specificity may be encoded in the C1b domain of PKC Apl II.
PKCɛ does not specifically translocate to the plasma membrane and is reported not to show specificity for PS (11, 25, 45). Indeed, specificity for PS in both cPKCs and nPKCs has been attributed to C1-C2 domain interactions (3, 8, 27, 45-47). PKC Apl II shows specificity for PS in in vitro kinase assays (36) and specifically translocates to the plasma membrane after 5HT or DOG addition. In contrast to other studies that indicate that specificity for PS is conferred by the C1-C2 domain interactions (3, 8, 27, 45-47), the C1b domain of PKC Apl II appears to be sufficient to confer PS specificity, at least as far as specific translocation to the plasma membrane is an assay for specificity for PS. Interestingly, mutating Arg256/257 to the residues present in PKC Apl I Gly/Ser allowed DOG-dependent translocation to internal membranes, suggesting that these residues were involved in the specificity for PS. Interestingly, these residues are not conserved in PKCɛ, in which the arginines are replaced by lysine and valine (Fig. 2).
PKC Apl II translocates better than PKC Apl I to DOG alone (53). In mammalian PKCs, the higher affinity of nPKCs for DAG has been attributed to a tryptophan residue in their C1b domains as opposed to a tyrosine residue in cPKCs (13). However, PKC Apl I and PKC Apl II both contain a tyrosine residue in this position (Fig. 2A), suggesting that other differences in the C1b or C1a domain also mediate the different responses to DAG. Interestingly, the tryptophan residue is present in all chordate nPKCs of both the delta and the epsilon family but not in most nonchordate nPKCs (Fig. 2A), including prechordate deuterostomes such as Ciona intestinalis and sea urchins (data not shown). The presence of this change in both the delta and epsilon family must be due to convergent evolution in chordates, since these two kinases initially diverged before the bilaterian ancestor (42). Introducing this mutant into PKC Apl II greatly increased its basal translocation, and this was due to increased affinity for DAG.
Source of PA in Aplysia neurons.
We show that both DAG produced by PI-PLC and PA produced by PLD are required for the translocation of PKC Apl II to the plasma membrane in sensory neurons. The production of DAG probably is induced by acting through Gq protein-coupled 5HT receptors, leading to the activation of PLC and the production of DAG from phosphatidylinositol. However, the signaling pathway leading to the production of PA is less clear. 5HT might be activating members of the Rho family GTPases, leading to the activation of PLD (7, 34). Indeed, Aplysia Rho GTPases (ApRho, ApRac, and ApCdc42) were shown to be highly conserved and expressed in sensory neurons (49). It also was shown that 5HT can activate ApCdc42 in Aplysia sensory neurons (49). It also is possible that PLD is constitutively active in Aplysia neurons and that 1-butanol inhibits this constitutive activity. The cloning of Aplysia PLDs and determining their mechanism of activation will be required to answer this question.
Summary.
In summary, we propose a model in which PA is required in addition to DAG for the translocation of PKC Apl II. Moreover, we show that PA acts through the C1b domain. These results suggest that the regulation of PLD is important for the physiological activation of PKC Apl II and possibly other nPKCs. Moreover, while in sensory neurons 5HT is sufficient to activate PKC Apl II, our results suggest that in some cases the requirement for both DAG and PA allows these kinases to act as coincidence detectors between stimuli that activate PLC and those that activate PLD, similarly to cPKCs, such as PKC Apl I, that act as coincidence detectors for stimuli that activate PLC and those that cause calcium entry.
Acknowledgments
This work was supported by Canadian Institutes of Health Research grant MOP 12046 (W.S.S.). C.A.F. is the recipient of a postdoctoral fellowship from the Fonds de la Recherche en Santé du Québec (FRSQ), and W.S.S. is a William Dawson Scholar and an FRSQ Chercheur National. D.W. is supported by a fellowship from the FRSQ.
We thank Peter McPherson for helpful comments.
Footnotes
Published ahead of print on 27 May 2008.
REFERENCES
- 1.Banno, Y., Y. Takuwa, Y. Akao, H. Okamoto, Y. Osawa, T. Naganawa, S. Nakashima, P. G. Suh, and Y. Nozawa. 2001. Involvement of phospholipase D in sphingosine 1-phosphate-induced activation of phosphatidylinositol 3-kinase and Akt in Chinese hamster ovary cells overexpressing EDG3. J. Biol. Chem. 27635622-35628. [DOI] [PubMed] [Google Scholar]
- 2.Benes, C. H., N. Wu, A. E. Elia, T. Dharia, L. C. Cantley, and S. P. Soltoff. 2005. The C2 domain of PKCdelta is a phosphotyrosine binding domain. Cell 121271-280. [DOI] [PubMed] [Google Scholar]
- 3.Bittova, L., R. V. Stahelin, and W. Cho. 2001. Roles of ionic residues of the C1 domain in protein kinase C-alpha activation and the origin of phosphatidylserine specificity. J. Biol. Chem. 2764218-4226. [DOI] [PubMed] [Google Scholar]
- 4.Bleasdale, J. E., N. R. Thakur, R. S. Gremban, G. L. Bundy, F. A. Fitzpatrick, R. J. Smith, and S. Bunting. 1990. Selective inhibition of receptor-coupled phospholipase C-dependent processes in human platelets and polymorphonuclear neutrophils. J. Pharmacol. Exp. Ther. 255756-768. [PubMed] [Google Scholar]
- 5.Byrne, J. H., and E. R. Kandel. 1996. Presynaptic facilitation revisited: state and time dependence. J. Neurosci. 16425-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrne, J. H., R. Zwartjes, R. Homayouni, S. D. Critz, and A. Eskin. 1993. Roles of second messenger pathways in neuronal plasticity and in learning and memory. Insights gained from Aplysia. Adv. Second Messenger Phosphoprotein Res. 2747-108. [PubMed] [Google Scholar]
- 7.Cazzolli, R., A. N. Shemon, M. Q. Fang, and W. E. Hughes. 2006. Phospholipid signalling through phospholipase D and phosphatidic acid. IUBMB Life 58457-461. [DOI] [PubMed] [Google Scholar]
- 8.Cho, W. 2001. Membrane targeting by C1 and C2 domains. J. Biol. Chem. 27632407-32410. [DOI] [PubMed] [Google Scholar]
- 9.Cissel, D. S., P. F. Fraundorfer, and M. A. Beaven. 1998. Thapsigargin-induced secretion is dependent on activation of a cholera toxin-sensitive and phosphatidylinositol-3-kinase-regulated phospholipase D in a mast cell line. J. Pharmacol. Exp. Ther. 285110-118. [PubMed] [Google Scholar]
- 10.Corbalán-García, S., and J. C. Gomez-Fernandez. 2006. Protein kinase C regulatory domains: the art of decoding many different signals in membranes. Biochim. Biophys. Acta 1761633-654. [DOI] [PubMed] [Google Scholar]
- 11.Corbalán-Garcia, S., S. Sanchez-Carrillo, J. Garcia-Garcia, and J. C. Gomez-Fernandez. 2003. Characterization of the membrane binding mode of the C2 domain of PKC epsilon. Biochemistry 4211661-11668. [DOI] [PubMed] [Google Scholar]
- 12.de Chaffoy de Courcelles, D. C., P. Roevens, and H. Van Belle. 1985. R 59 022, a diacylglycerol kinase inhibitor. Its effect on diacylglycerol and thrombin-induced C kinase activation in the intact platelet. J. Biol. Chem. 26015762-15770. [PubMed] [Google Scholar]
- 13.Dries, D. R., L. L. Gallegos, and A. C. Newton. 2007. A single residue in the C1 domain sensitizes novel protein kinase C isoforms to cellular diacylglycerol production. J. Biol. Chem. 282826-830. [DOI] [PubMed] [Google Scholar]
- 14.Fang, Y., M. Vilella-Bach, R. Bachmann, A. Flanigan, and J. Chen. 2001. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 2941942-1945. [DOI] [PubMed] [Google Scholar]
- 15.García-García, J., J. C. Gomez-Fernandez, and S. Corbalan-Garcia. 2001. Structural characterization of the C2 domain of novel protein kinase Cɛ. Eur. J. Biochem. 2681107-1117. [DOI] [PubMed] [Google Scholar]
- 16.Giorgione, J. R., J. H. Lin, J. A. McCammon, and A. C. Newton. 2006. Increased membrane affinity of the C1 domain of protein kinase Cdelta compensates for the lack of involvement of its C2 domain in membrane recruitment. J. Biol. Chem. 2811660-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glanzman, D. L., E. R. Kandel, and S. Schacher. 1989. Identified target motor neuron regulates neurite outgrowth and synapse formation of aplysia sensory neurons in vitro. Neuron 3441-450. [DOI] [PubMed] [Google Scholar]
- 18.Jose Lopez-Andreo, M., J. C. Gomez-Fernandez, and S. Corbalan-Garcia. 2003. The simultaneous production of phosphatidic acid and diacylglycerol is essential for the translocation of protein kinase Cɛ to the plasma membrane in RBL-2H3 cells. Mol. Biol. Cell 144885-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kandel, E. R. 2001. The molecular biology of memory storage: a dialogue between genes and synapses. Science 2941030-1038. [DOI] [PubMed] [Google Scholar]
- 20.Koul, O., and G. Hauser. 1987. Modulation of rat brain cytosolic phosphatidate phosphohydrolase: effect of cationic amphiphilic drugs and divalent cations. Arch. Biochem. Biophys. 253453-461. [DOI] [PubMed] [Google Scholar]
- 21.Kruger, K. E., W. S. Sossin, T. C. Sacktor, P. J. Bergold, S. Beushausen, and J. H. Schwartz. 1991. Cloning and characterization of Ca2+-dependent and Ca2+-independent PKCs expressed in Aplysia sensory cells. J. Neurosci. 112303-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, P., W. J. Fung, and A. M. Gilfillan. 1992. Phosphatidylcholine-specific phospholipase D-derived 1,2-diacylglycerol does not initiate protein kinase C activation in the RBL 2H3 mast-cell line. Biochem. J. 287325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majewski, H., and L. Iannazzo. 1998. Protein kinase C: a physiological mediator of enhanced transmitter output. Progr. Neurobiol. 55463-475. [DOI] [PubMed] [Google Scholar]
- 24.Manseau, F., X. Fan, W. S. Sossin, and V. F. Castellucci. 2001. Ca2+-independent PKC Apl II mediates the serotonin induced facilitation at depressed synapses in Aplysia. J. Neurosci. 211247-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medkova, M., and W. Cho. 1998. Differential membrane-binding and activation mechanisms of protein kinase C-alpha and -epsilon. Biochemistry 374892-4900. [DOI] [PubMed] [Google Scholar]
- 26.Medkova, M., and W. Cho. 1999. Interplay of C1 and C2 domains of protein kinase C-alpha in its membrane binding and activation. J. Biol. Chem. 27419852-19861. [DOI] [PubMed] [Google Scholar]
- 27.Melowic, H. R., R. V. Stahelin, N. R. Blatner, W. Tian, K. Hayashi, A. Altman, and W. Cho. 2007. Mechanism of diacylglycerol-induced membrane targeting and activation of protein kinase Cθ. J. Biol. Chem. 28221467-21476. [DOI] [PubMed] [Google Scholar]
- 28.Meshulam, T., M. M. Billah, S. Eckel, K. K. Griendling, and R. D. Diamond. 1995. Relationship of phospholipase C- and phospholipase D-mediated phospholipid remodeling pathways to respiratory burst activation in human neutrophils stimulated by Candida albicans hyphae. J. Leukoc. Biol. 57842-850. [DOI] [PubMed] [Google Scholar]
- 29.Nalefski, E. A., and J. J. Falke. 1996. The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci. 52375-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newton, A. C. 1995. Protein kinase C. Seeing two domains. Curr. Biol. 5973-976. [DOI] [PubMed] [Google Scholar]
- 31.Newton, A. C. 1995. Protein kinase C: structure, function, and regulation. J. Biol. Chem. 27028495-28498. [DOI] [PubMed] [Google Scholar]
- 32.Oancea, E., and T. Meyer. 1998. Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell 95307-318. [DOI] [PubMed] [Google Scholar]
- 33.Ochoa, W. F., J. Garcia-Garcia, I. Fita, S. Corbalan-Garcia, N. Verdaguer, and J. C. Gomez-Fernandez. 2001. Structure of the C2 domain from novel protein kinase Cɛ. A membrane binding model for Ca2+-independent C2 domains. J. Mol. Biol. 311837-849. [DOI] [PubMed] [Google Scholar]
- 34.Oude Weernink, P. A., L. Han, K. H. Jakobs, and M. Schmidt. 2007. Dynamic phospholipid signaling by G protein-coupled receptors. Biochim. Biophys. Acta 1768888-900. [DOI] [PubMed] [Google Scholar]
- 35.Pappa, H., J. Murray-Rust, L. V. Dekker, P. J. Parker, and N. Q. McDonald. 1998. Crystal structure of the C2 domain from protein kinase C-delta. Structure 6885-894. [DOI] [PubMed] [Google Scholar]
- 36.Pepio, A. M., X. Fan, and W. S. Sossin. 1998. The role of C2 domains in Ca2+-activated and Ca2+-independent protein kinase Cs in Aplysia. J. Biol. Chem. 27319040-19048. (Erratum, 273:22856.) [DOI] [PubMed] [Google Scholar]
- 37.Pepio, A. M., and W. S. Sossin. 1998. The C2 domain of the Ca2+-independent protein kinase C Apl II inhibits phorbol ester binding to the C1 domain in a phosphatidic acid-sensitive manner. Biochemistry 371256-1263. [DOI] [PubMed] [Google Scholar]
- 38.Pepio, A. M., and W. S. Sossin. 2001. Membrane translocation of novel protein kinase Cs is regulated by phosphorylation of the C2 domain. J. Biol. Chem. 2763846-3855. [DOI] [PubMed] [Google Scholar]
- 39.Sacktor, T. C., and J. H. Schwartz. 1990. Sensitizing stimuli cause translocation of protein kinase C in Aplysia sensory neurons. Proc. Natl. Acad. Sci. USA 872036-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slater, S. J., J. L. Seiz, A. C. Cook, C. J. Buzas, S. A. Malinowski, J. L. Kershner, B. A. Stagliano, and C. D. Stubbs. 2002. Regulation of PKC alpha activity by C1-C2 domain interactions. J. Biol. Chem. 27715277-15285. [DOI] [PubMed] [Google Scholar]
- 41.Songyang, Z., and L. C. Cantley. 1995. Recognition and specificity in protein tyrosine kinase-mediated signalling. Trends Biochem. Sci. 20470-475. [DOI] [PubMed] [Google Scholar]
- 42.Sossin, W. S. 2007. Isoform specificity of protein kinase Cs in synaptic plasticity. Learn Mem. 14236-246. [DOI] [PubMed] [Google Scholar]
- 43.Sossin, W. S., A. R. Diaz, and J. H. Schwartz. 1993. Characterization of two isoforms of protein kinase C in the nervous system of Aplysia californica. J. Biol. Chem. 2685763-5768. [PubMed] [Google Scholar]
- 44.Sossin, W. S., X. T. Fan, and F. Saberi. 1996. Expression and characterization of aplysia protein kinase C—a negative regulatory role for the E region. J. Neurosci. 1610-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stahelin, R. V., M. A. Digman, M. Medkova, B. Ananthanarayanan, H. R. Melowic, J. D. Rafter, and W. Cho. 2005. Diacylglycerol-induced membrane targeting and activation of protein kinase Cɛ: mechanistic differences between protein kinases Cδ and Cɛ. J. Biol. Chem. 28019784-19793. [DOI] [PubMed] [Google Scholar]
- 46.Stahelin, R. V., M. A. Digman, M. Medkova, B. Ananthanarayanan, J. D. Rafter, H. R. Melowic, and W. Cho. 2004. Mechanism of diacylglycerol-induced membrane targeting and activation of protein kinase Cδ. J. Biol. Chem. 27929501-29512. [DOI] [PubMed] [Google Scholar]
- 47.Stahelin, R. V., J. Wang, N. R. Blatner, J. D. Rafter, D. Murray, and W. Cho. 2005. The origin of C1A-C2 interdomain interactions in protein kinase Cα. J. Biol. Chem. 28036452-36463. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka, C., and Y. Nishizuka. 1994. The protein kinase C family for neuronal signaling. Annu. Rev. Neurosci. 17551-567. [DOI] [PubMed] [Google Scholar]
- 49.Udo, H., I. Jin, J. H. Kim, H. L. Li, T. Youn, R. D. Hawkins, E. R. Kandel, and C. H. Bailey. 2005. Serotonin-induced regulation of the actin network for learning-related synaptic growth requires Cdc42, N-WASP, and PAK in Aplysia sensory neurons. Neuron 45887-901. [DOI] [PubMed] [Google Scholar]
- 50.Xu, R. X., T. Pawelczyk, T. H. Xia, and S. C. Brown. 1997. NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions. Biochemistry 3610709-10717. [DOI] [PubMed] [Google Scholar]
- 51.Yaffe, M. B. 2002. Phosphotyrosine-binding domains in signal transduction. Nat. Rev. Mol. Cell Biol. 3177-186. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, G., M. G. Kazanietz, P. M. Blumberg, and J. H. Hurley. 1995. Crystal structure of the cys2 activator-binding domain of protein kinase C delta in complex with phorbol ester. Cell 81917-924. [DOI] [PubMed] [Google Scholar]
- 53.Zhao, Y., K. Leal, C. Abi-Farah, K. C. Martin, W. S. Sossin, and M. Klein. 2006. Isoform specificity of PKC translocation in living Aplysia sensory neurons and a role for Ca2+-dependent PKC APL I in the induction of intermediate-term facilitation. J. Neurosci. 268847-8856. [DOI] [PMC free article] [PubMed] [Google Scholar]