Abstract
While there are now extensive databases of human genomic sequences from both private and public efforts to catalog human nucleotide variation, there are very few large-scale surveys designed for the purpose of analyzing human population history. Demographic inference from patterns of SNP variation in current large public databases is complicated by ascertainment biases associated with SNP discovery and the ways that populations and regions of the genome are sampled. Here, we present results from a resequencing survey of 40 independent intergenic regions on the autosomes and X chromosome comprising ∼210 kb from each of 90 humans from six geographically diverse populations (i.e., a total of ∼18.9 Mb). Unlike other public DNA sequence databases, we include multiple indigenous populations that serve as important reservoirs of human genetic diversity, such as the San of Namibia, the Biaka of the Central African Republic, and Melanesians from Papua New Guinea. In fact, only 20% of the SNPs that we find are contained in the HapMap database. We identify several key differences in patterns of variability in our database compared with other large public databases, including higher levels of nucleotide diversity within populations, greater levels of differentiation between populations, and significant differences in the frequency spectrum. Because variants at loci included in this database are less likely to be subject to ascertainment biases or linked to sites under selection, these data will be more useful for accurately reconstructing past changes in size and structure of human populations.
With recent advances in molecular genotyping technology, we now have an unprecedented amount of information about human genetic diversity at the single nucleotide scale. The The International HapMap Consortium, for example, has genotyped four million single nucleotide polymorphisms (SNPs) in a panel of 270 individuals from four populations (The International HapMap Consortium 2005; Frazer et al. 2007). While large databases such as the HapMap are invaluable resources for genetic association studies, they have severe limitations for answering questions related to human demography (i.e., the history of changes in effective population size, population structure, and distribution). One limitation results from the fact that large-scale genotyping studies rely on SNPs that are typically discovered by full resequencing of individuals in small SNP discovery panels. Because the probability that a SNP is identified in the discovery panel is a function of its allele frequency, SNPs with large frequency differences between populations are often missing in the public databases when they are rare in the populations used for SNP discovery. This gives rise to a bias in aspects of the data that rely on the site frequency spectrum (e.g., nucleotide diversity, Tajima’s D, FST, and linkage disequilibrium) (Clark et al. 2005; Weir et al. 2005). Moreover, for SNPs that are discovered in one population and typed in a different one, the introduced ascertainment bias cannot be corrected accurately unless information on population history is available. This warrants caution when interpreting ascertained SNP data of this type for the purpose of demographic inference (Keinan et al. 2007; Jakobsson et al. 2008; Li et al. 2008).
Another complication arises from different strategies used to sample human populations. Some databases make use of admixed populations such as Europeans that were collected from the U.S. and who traced their ancestry to a variety of northern and western European countries and African Americans (Akey et al. 2004; Livingston et al. 2004; The ENCODE Project Consortium 2004; Crawford et al. 2005; Hinds et al. 2005; The International HapMap Consortium 2005). This is problematic for inferring demographic history as sampling strategies that pool individuals from many diverse populations and, to a lesser extent, that use admixed groups have been shown to confound signals of population structure and population growth (Ptak and Przeworski 2002). Data sets that contain only one African population (The International HapMap Consortium 2005) cannot be used to consider population dynamics within Africa.
Finally, many databases are enriched for genic regions (Akey et al. 2004; Livingston et al. 2004; The ENCODE Project Consortium 2004), which makes them less useful for analyses of demographic history because the results may be confounded by the effects of natural selection (Voight et al. 2005). We set out to develop a database with the intended purpose of inferring human demographic history by initiating a large-scale DNA sequencing survey of noncoding regions that lie far from genes in a collection of six populations from Africa, Europe, Asia, and Oceania. In particular, we focus on single-copy intergenic regions in areas of medium or high recombination at least 50 kb from the nearest gene (100 kb for the autosomes). By fully resequencing every locus in every sample, and by using well-defined population samples, we obtain less-biased estimates of the site-frequency spectrum. Our database is also unique in that it contains nuclear resequencing data from Melanesians and multiple African populations, including Central (Biaka) and Southern (San) Africans practicing a hunting–gathering lifestyle. By also including a European (French Basque), Asian (Han Chinese), and African (Senegalese Mandenka) food-producing population, we are able to compare levels of human genetic variation both within and between different continental groups and among groups practicing different subsistence strategies. Here, we present results from the first phase of our study, consisting of sequence data from 40 regions (20 autosomal and 20 X-linked) and compare levels and patterns of nucleotide variability among our population samples and with those gleaned from large public databases, such as the HapMap, ENCODE, and National Institute of Environmental Health Sciences projects (Livingston et al. 2004; The ENCODE Project Consortium 2004, 2007; The International HapMap Consortium 2005; Frazer et al. 2007).
Results
Nucleotide diversity and divergence
We resequenced a total of 112 kb from the autosomes and 98 kb from the X chromosome in 90 individuals. Initially, we identified a total of 1658 SNPs and 86 indel polymorphisms. During the course of our study, we discovered that some of the sampled individuals are closely related to each other (see Methods). To avoid any potential complication that this may cause, we consider only SNPs from unrelated individuals (see Supplementary material), yielding 1604 biallelic SNPs for further analysis (1094 on the autosomes and 512 on the X chromosome). Only 0.3% of polymorphic nucleotide positions contained missing data. We note that, because most of our samples come from males, there are fewer X chromosomes (∼86) than autosomes (∼158) in our sample.
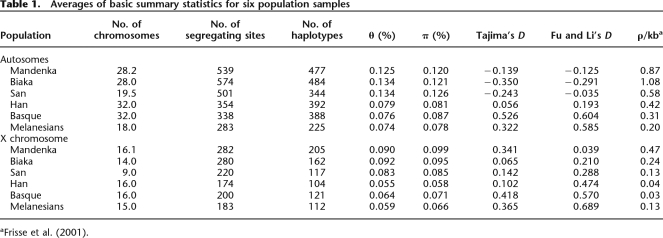
Table 1 provides some basic summaries of the data, including measures of nucleotide diversity (θ, π), the frequency spectrum of segregating mutations (Tajima’s D, Fu and Li’s D), and linkage disequilibrium (ρ). Autosomal nucleotide diversity levels (π), which average 0.116%, range from a high value of 0.191% at 18qMB73 to a low value of 0.044% at 5pMB4. Values for the X chromosome tend to be lower, with an average π of 0.100% (range: 0.047%–0.199%). Mean (±SEM) levels of human–chimpanzee sequence divergence are 1.35% ± 0.081% for the autosomes and 1.01% ± 0. 053% for the X chromosome. To test the null hypothesis that polymorphism and divergence fit the expectations of a standard neutral model allowing for interlocus variation in the underlying neutral mutation rate, we performed two 20-locus HKA tests: one for the autosomes and one for the X chromosome. Neither test rejected the null model (autosomal P = 0.755; X chromosomal P = 0.432) (Supplemental Tables 3 and 4).
Table 1.
Averages of basic summary statistics for six population samples
Most loci exhibit a pattern of reduced nucleotide diversity in non-Africans relative to Africans. For example, mean levels of autosomal diversity (as summarized by π) are 0.128% ± 0.011% for Africans compared with a mean value of 0.089% ± 0.008% for non-Africans. Similarly, mean levels of π for the X chromosome are 0.104% ± 0.008 for Africans and 0.076% ± 0.009% for non-Africans. This corresponds to a statistically significant reduction in nucleotide diversity of ∼30% for both the autosomes and X chromosome in our non-African sample (Mann–Whitney two-sample test, P = 0.012 and P = 0.014 for the autosomes and X chromosome, respectively). Separate HKA tests on African and non-African population groups also do not reject the null model for the autosomes (African P = 0.932; non-African P = 0.696) or X chromosome (African P = 0.586; non-African P = 0.635). Similarly, none of the HKA tests performed on the six individual populations rejects the null model (data not shown).
Nucleotide diversity and recombination rate
To explore the relationship between nucleotide diversity and recombination, we calculated recombination rates for a 1 Mb region encompassing each locus trio. For the 20 autosomal loci, the mean (±SEM) sex-averaged recombination rate is 2.18 ± 0.16 cM/Mb, with a range of 0.9–3.5 cM/Mb. When we plot diversity (as summarized by θ) versus recombination rates for each locus (Supplemental Fig. 2a), we find that diversity increases weakly, but significantly, with recombination rate (Spearman rank correlation, two-tailed t-test, R2 = 0.261, P = 0.021). This association could be caused by either positive or negative selection at linked sites (Maynard-Smith and Haigh 1974; Charlesworth et al. 1993), by variation in underlying mutation rate, or by some combination of these factors. A simple test of the hypothesis that variation in underlying mutation rate is responsible for the correlation between nucleotide diversity and recombination rate is to compare recombination rate with interspecific divergence (Hellmann et al. 2003). While we do not observe a significant positive correlation between recombination rate and human–chimpanzee divergence (DA) (Spearman rank correlation, two-tailed t-test, R2 = 0.109, P = 0.155), there is a positive trend (Supplemental Fig. 2b). Interestingly, θ/DA no longer increases with recombination rate (R2 = 0.001, P = 0.88) (Supplemental Fig. 2c). Thus, the weak association between nucleotide diversity and recombination rate is consistent with a neutral explanation (i.e., recombination-associated variation in mutation rates) (Hellmann et al. 2003). We do not see a positive correlation between recombination rate and X-linked diversity (R2 = 0.058, P = 0.322), which may be due, in part, to the smaller number of SNPs recovered from the X chromosome.
Frequency spectra
All 20 autosomal loci have a negative Tajima’s D (TD) value when considering the sample of pooled chromosomes from all populations (TD = −1.07 ± 0.09) (mean ± SEM). While the mean TD value for the X chromosome is also negative in the global sample (−0.09 ± 0.23), locus-specific values range from −1.50 to +2.00. When we examine TD values in individual population samples to control for the effects of fine-scale population structure (Ptak and Przeworski 2002), we find slightly negative mean autosomal values in the sub-Saharan African populations (−0.350, −0.243, and −0.139 for the Biaka, San, and Mandenka, respectively), and positive mean values in the non-African populations (0.056, 0.322, and 0.526 for the Han, Melanesians, and Basque, respectively). This is not true for the X chromosome data where all population samples have slightly positive TD values (i.e., ranging from 0.065 in the Biaka to 0.418 in the Basque). Of the 240 tests of individual TD values for the autosomes and X chromosome, we find ∼17% of the tests (16 autosomal and 24 X-linked) reject the standard neutral model at the 5% significance level (i.e., they have TD values that are too high or low based on 104 simulations of a neutral, panmictic Wright–Fisher population with recombination). Nine loci reject neutrality in more than a single population: two on the autosomes (7pMB8 and 10qMB128) and seven on the X chromosome (XpMB33, XpMB35, XpMB39, XqMB120, XqMB124, XqMB139, and XqMB143). Most outliers occur in non-African populations (32/40) and involve TD values that are significantly positive (25/32). This contrasts significantly (Fisher’s exact test, P = 0.04) with the African outliers where only 3/8 significant tests were due to positive TD values. We note, though, that these tests are not independent due to the shared ancestry of the sampled populations.
Within- and between-group differences
Wright’s FST values for our global sample are 0.158 for the autosomes and 0.257 for the X chromosome. These values are higher than for other large published databases, which range from 0.098 to 0.130 for the autosomes (Akey et al. 2002; Clark et al. 2005; The International HapMap Consortium 2005; Weir et al. 2005) and from 0.195 to 0.210 for the X chromosome (Akey et al. 2002; The International HapMap Consortium 2005). We also calculated FST between all pairs of populations (Supplemental Table 5) and used these values to construct population trees showing the degree of genetic similarity between populations (Fig. 1). The greatest FST values are between sub-Saharan African and non-African groups. Equivalently, the longest internal branch in Figure 1 separates sub-Saharan African populations from non-African populations. For these between-group comparisons, pairwise FST values range from 0.154 to 0.283 for the autosomes and from 0.167 to 0.403 for the X chromosome. However, there is also a substantial amount of differentiation within sub-Saharan African and non-African groups, with within-group FST values as high as 0.119 for the autosomes (between Melanesians and Han) and 0.242 for the X (between Melanesians and the French Basque).
Figure 1.
Neighbor-joining trees based on FST distances between populations for autosomal and X chromosome data.
Haplotype diversity
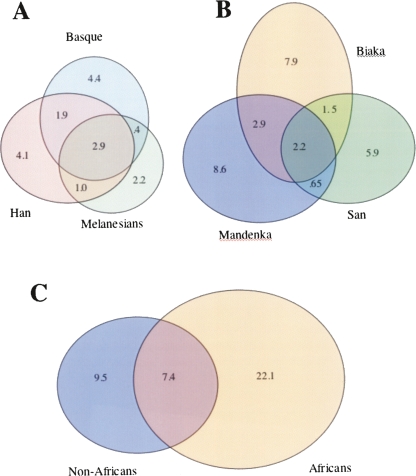
For the autosomal data, we estimated phase and categorized each haplotype as one that was shared among multiple populations or unique to a single population (see Supplemental material). The results of averaging the number of haplotypes in different categories across the 20 regions are shown in Figure 2. Each oval represents a single population (Fig. 2A,B) or group of populations (Fig. 2C). The area of the oval is proportional to the average number of haplotypes found within the population/group, and the area of the intersection of two or more ovals is proportional to the average number of haplotypes that are shared between each of the underlying groups. Figure 2 shows that sub-Saharan African populations tend to have substantially more haplotypes than do non-African populations and that haplotype sharing across populations is more common in non-African populations than in sub-Saharan African populations. Also, it appears that non-African haplotype diversity is not a simple subset of sub-Saharan African haplotype diversity, as had been claimed (Tishkoff et al. 1996). Similarly, we find that non-African SNPs are not a simple subset of African SNP diversity. For example, of those SNPs that are exclusive to one continental group (i.e., not shared between Africans and non-Africans), about 25% are unique to non-Africans. This result does not change when we consider only those SNPs with a MAF of ≥5%.
Figure 2.
Haplotype Venn diagrams based on phased autosomal sequence data. Numbers refer to the average number of distinct haplotypes for each of the 20 regions. See Methods for details.
Comparison with HapMap SNPs
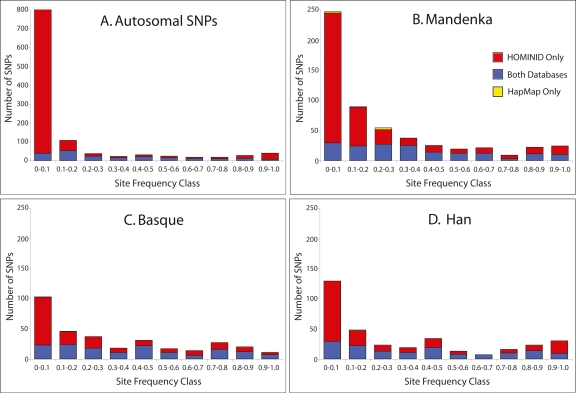
To examine the extent to which HapMap contained SNPs identified in our sequencing survey, we downloaded HapMap Phase II (August 22, 2006) SNP data from the http://www.hapmap.org and mapped the exact locations of these SNPs relative to our own data using build hg18 of the human genome. We then examined the proportion of our SNPs that are found in the HapMap (1) for the X chromosome and the autosomes separately, (2) for all SNPs, (3) for SNPs with MAF ≥ 0.1, and (4) for each of our six study populations (see Supplemental Table 6). The proportion of our SNPs contained in the HapMap database (“coverage”) varied from a low of 2% (i.e., 1 out of 57 SNPs; region 16pMB17) to a high of 64% (i.e., nine out of 14 SNPs; region XqMB139). Figure 3 shows coverage for all autosomal SNPs as well as for our three population samples that are comparable to those in the HapMap database (e.g., our Mandenka sample corresponds to HapMap Yorubans, our Basque sample to HapMap CEU, and our Han sample to HapMap Han). Overall, only 18.2% of our autosomal SNPs are also found in HapMap, with most of our low-frequency SNPs missing from HapMap (Fig. 3). This pattern holds for individual populations, including the Han (where only 41.8% of the SNPs identified here are contained in HapMap). This is striking given that the two Han samples presumably represent the same population.
Figure 3.
Site-frequency spectra of polymorphisms identified at 20 autosomal regions sequenced herein. Current data set is compared with HapMap SNPs (A) globally and for similar populations: (B) Mandenka (Yorubans), (C) French Basque (CEU), and (D) Han Chinese (Han Chinese). SNPs are classed into those found both in the current and HapMap databases (blue), in the current database only (red), or in the HapMap database only (yellow). The number of SNPs identified per population is scaled proportionally; note that Africans contain more SNPs than non-Africans.
Discussion
In total, we sequenced 112 kb from the autosomes and 98 kb from the X chromosome in each of 90 individuals, a substantially larger data set than reported in previous population-based intergenic resequencing studies (Frisse et al. 2001; Yu et al. 2002; Voight et al. 2005). Over half of the SNPs that we found are private to a single population sample, and of these, just 3% (21 out of 825) are included in the HapMap project. Overall, while 98% of the HapMap SNPs are found in our resequencing study (320 out of 327), only 20% of our SNPs (320 out of 1604) are contained in the HapMap database. When we confine our analysis to SNPs with minor allele frequency >10%, we find 56% of our SNPs in the HapMap database (Supplemental Table 6). This suggests that a substantial proportion of human nucleotide variation is not represented in current public databases and may be localized to regional populations (Fig. 3) (see below for additional discussion).
While our estimates of autosomal nucleotide diversity (Table 1) are similar to estimates from other studies of intergenic regions (Frisse et al. 2001; Yu et al. 2002; Voight et al. 2005), they are higher than those from large public databases that include genic regions. For example, we find that π = 0.120% for our sample of Mandenka, a population that practices a farming lifestyle similar to Yorubans. Estimates of nucleotide diversity in Yorubans (π = 0.076%) and African Americans (π = 0.092%) are substantially lower for 135 environmental response genes (Livingston et al. 2004; Plagnol and Wall 2006) and for 300 inflammatory response genes in the Seattle SNPs project (Akey et al. 2004; Crawford et al. 2005), respectively (Supplemental Fig. 3). Similarly, our estimate of π (0.087%) in the French Basque is higher than for comparable estimates of π in the CEPH Utah sample in the Environmental Genome Project (0.059%) and in the Seattle SNPs project (π = 0.071%) (Supplemental Fig. 3). Our estimates of nucleotide diversity in Table 1 are statistically significantly higher than estimates from comparable samples in the NIEHS database for Asia (Asian π = 0.055% ± 0.003%; Mann–Whitney two-sample test, P < 0.0001), for Europe (CEPH π = 0.060% ± 0.003%; P < 0.0001), and for Africa (Yoruban π = 0.078% ± 0.003%; P < 0.0001). Less than 10% of the bases sequenced by the Seattle SNP and Environmental Genome Project studies are at nonsynonymous sites, so direct selection against deleterious alleles is not a sufficient explanation for the observed differences in nucleotide diversity between our study and genic resequencing studies. It is likely that the well-known effects of variation-reducing selection (Hill and Robertson 1966) are more relevant in genic regions than in intergenic regions.
Our sampling of multiple sub-Saharan African and non-African populations also provides us with a more comprehensive view of genetic differences between populations. The FST values reported here for the autosomes (0.16) and X chromosome (0.26) are slightly higher than published values for genotyped SNPs (0.12 and 0.21 for autosomes and the X chromosome, respectively) (The International HapMap Consortium 2005) (Supplemental Table 7). Another notable observation is the greater levels of population differentiation on the X chromosome compared with the autosomes. Our findings of higher within-group diversity and FST values are likely attributable to two main causes: our sampling of a more diverse collection of human populations and our resequencing design, which allow us to find more of the rare and private SNPs that are missing from the HapMap. In Figure 1, the longest external branches lead to the San and the Melanesians. The additional human diversity that these populations represent was unavailable to previous studies with more restricted study populations. Moreover, because our study focuses on intergenic regions, which are less likely to be affected by natural selection, we expect that our higher estimates of nucleotide diversity more accurately reflect the underlying mutation rate and population size. This is supported by multilocus HKA tests, which show that levels of polymorphism within humans are correlated to human–chimpanzee divergence, as expected under a neutral model of molecular evolution.
We find that a small number of loci in our database reject neutrality tests based on the frequency spectrum. Interestingly, most of these outliers occur in non-African populations and involve positive TD values on the X chromosome. This provides a different picture of variability than previously observed in a study of 15 X-linked introns where outliers also tended to occur in non-African populations, but they exhibited an excess of rare polymorphisms (Hammer et al. 2004). These authors also found a statistically significant positive correlation between Fu and Li’s D values and recombination rate in their non-African (but not African) sample. Such a relationship is not expected under a neutral, equilibrium model (Przeworski et al. 2001) and was explained by the effects of diversity-reducing selection, leading to an excess of singletons at loci in regions of lower recombination on the X chromosomes of non-Africans (Hammer et al. 2004). Interestingly, their non-African sample also rejected the null model in a multilocus HKA test. Here, we see no relationship between FLD values and recombination rates for the entire data set of autosomes or X chromosomes for either Africans or non-African populations (data not shown). While there are a number of sampling factors that differ between this study and that of Hammer et al. (2004), the absence of such a relationship for X-linked loci in our non-African samples (Supplemental Fig. 4) is consistent with the hypothesis that these loci are unlinked to sites under recent diversity-reducing selection. We suggest that the observed pattern of outliers on the X chromosome with respect to TD more likely reflects a history of recent population bottlenecks in non-African groups (Voight et al. 2005; Garrigan and Hammer 2006).
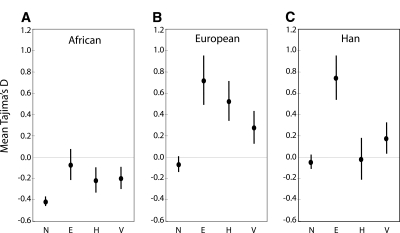
To see what effect study design might have on the frequency spectrum of segregating mutations, we compared the distribution of TD values from our study with the comparable distributions from the NIEHS (Livingston et al. 2004), ENCODE (The International HapMap Consortium 2005), and Voight et al. (2005) studies. We first subsampled the data to make the size of each locus comparable across studies. We then tabulated the mean TD and the standard error of the mean for each study. Figure 4 shows the results for West African, European, and Asian samples. There are significant differences in mean TD across studies (Fig. 4A, F = 3.12, P = 0.027; Fig. 4B, F = 5.80, P < 0.001; Fig. 4C, F = 4.60, P = 0.004). A cursory examination of Figure 4 suggests that the NIEHS data set is an outlier to the other three studies, with substantially lower TD values in both the African and European populations. One possible explanation is that the NIEHS data set, which is primarily from genic regions, is subject to more purifying selection than regions from the other studies, leading to a skew toward rare variants. This hypothesis is consistent with the observation that the NIEHS sequence data have significantly less variation than do our study and the Voight et al. (2005) study, both of which examined noncoding regions. We should note, though, that differences in laboratory protocols and sequencing error rates across studies may also explain our observed pattern. In addition, it is not clear whether the publicly available databases are sufficient for reconstructing the full ENCODE resequencing data, so it is unknown whether the ENCODE TD values are directly comparable to the TD values from other studies. Additional analyses of the frequency spectrum across studies can be found in the Supplemental material.
Figure 4.
Mean and standard error of Tajima’s D for African, European, and Han samples across different studies; (N) NIEHS; (E) ENCODE; (H) data presented herein; (V) Voight et al. (2005).
Conclusion
As a consequence of intensive studies of human diversity over the past two decades, the broad brush strokes of human demographic history are now apparent (Garrigan and Hammer 2006). Still there are many unanswered questions concerning past changes in population size and structure. For example, how many and how severe were bottlenecks associated with human migrations out of Africa; when did human populations begin to grow dramatically; and to what extent, if any, did anatomically modern humans interbreed with archaic forms? These and other questions relating to human demographic history can only be answered by collecting large amounts of data from many unlinked regions of the genome in a diverse array of human populations. While large databases such as the HapMap are invaluable resources for genetic association studies, the present database was specifically designed for the purpose of reconstructing human demographic history. Toward this end we endeavored to construct a database that (1) focuses entirely on noncoding regions that are at least 50 kb away from the nearest gene/functional unit, (2) fully resequences every locus in every sample, (3) surveys a wide range of human populations, including indigenous populations unrelated to populations sampled in previous large-scale surveys, and (4) systematically examines variation on both the X chromosome and the autosomes.
Many of the population-level patterns of variation observed here have been noted before, such as greater diversity, more rare variants and lower levels of linkage disequilibrium in sub-Saharan African populations (Frisse et al. 2001; Garrigan and Hammer 2006). We have broadened these observations by surveying variation in a wider range of African and non-African populations. In this regard, it is interesting to note that the San and the Biaka, food-gathering populations that were marginalized after the spread of agriculture in Africa (Excoffier and Schneider 1999), harbor as much (or slightly more) genetic variation as do the Mandenka, a large food-producing West African population. This observation was not obvious from an analysis of pre-ascertained SNP variation in the same populations (Conrad et al. 2006), thus highlighting the advantages of collecting resequencing data. Indeed, our study suggests that there may be many higher frequency (MAF ≥ 0.05) variants segregating in non-HapMap populations. Future association studies (especially those involving individuals with a substantial amount of sub-Saharan African ancestry) might be improved by widespread screening for SNPs involving large, multi-ethnic ascertainment panels, similar to the resequencing effort described here. Almost all of the DNA samples that we used came from the CEPH Human Genome Diversity Panel (Cann et al. 2002). This was a deliberate decision to ensure that other researchers could use the same samples for complementary studies (e.g., of natural selection in genic regions). We hope that our data will serve as the core of a new database of human sequence variation for answering the many open evolutionary and historical questions about our species. All of our data and analyses are publicly available at http://hammerlab.biosci.arizona.edu.
Methods
Samples
The initial 90 DNA samples used in this study come from publicly available cell lines administered by the CEPH Human Genome Diversity Panel (Cann et al. 2002). Individual identifiers for each of these samples are given in Supplemental Table 1. The seven San individuals in the CEPH-HGDP are listed along with three additional San samples that are part of the YCC collection (The Y Chromosome Consortium 2002). Subsequently, it was discovered that the CEPH-HGDP contains several pairs of close relatives (Rosenberg 2006). We restricted our analyses to panels of unrelated individuals by disregarding the following samples: no. 451 (Biaka), no. 919 (Mandenka), no. 3043 (San), and nos. 490, 658, 664, 789, 823, 824, and 825 (Melanesian). For loci 31–40, we replaced the samples from Bouganville (for whom there were only nine unrelated individuals) with 15 unrelated Papua New Guineans (Stoneking et al. 1990). We could detect no population structure between the two groups (Results not shown), and all subsequent analyses present the combined data as from a “Melanesian” population.
Regions sequenced
The regions used for sequencing were chosen to minimize any potential confounding effects of natural selection. Specifically, we identified 40 different ∼20-kb regions (20 on the X chromosome and 20 on the autosomes) of primarily single-copy noncoding (i.e., putatively nonfunctional) DNA in regions of medium or high recombination (r ≥ 0.9 cM/Mb) (Kong et al. 2002). Ten of the autosomal regions were chosen to encompass the locus pairs sequenced by Frisse et al. (2001) (see below). The remaining regions were chosen in the following manner: Each region was at least 50 kb (100 kb for the autosomes) away from the nearest gene, with “gene” defined as the union of both stringent (“Known Genes”) and broad (“Gene Bounds”) gene-prediction definitions (Burge and Karlin 1997, 1998; Hsu et al. 2006). Within each region, we gathered ∼4–6 kb of sequence data from three or four discrete subsections that spanned most of the distance of each region (locus trio). Our resequencing scheme is similar to those of Frisse et al. (2001) and Voight et al. (2005) who developed the locus pair design, which surveys pairs of tightly, but not completely, linked segments. By omitting intervening segments, this design provides a cost-effective strategy to survey many independent loci. Our locus trio design expands upon the locus pair approach by gathering three times as much sequence data per locus and by allowing for joint estimates of levels of polymorphism, allele frequency spectra, and linkage disequilibrium over ∼15–20 kb at each locus (see Garrigan et al. 2005 for a figure illustrating the locus trio design). Subsections were chosen to minimize the number of repetitive elements after alignment with orthologous sequences in the chimp genome. To avoid noncoding functional DNA, we rejected candidate regions that had human–chimp divergence less than the average human–chimp exonic divergence rate (∼0.76%; exons defined by the CCDS track on UCSC, which restricts its genes to those that are highly characterized protein-encoding genes). We used this as a benchmark to exclude regions that were putatively under strong constraint. The chosen locus trios are also far from the nearest ultraconserved element (Bejerano et al. 2004) (i.e., ∼6.4 Mb). After scanning the whole genome for regions that met the above search criteria, we ranked these regions based on additional criteria, including (1) the quality and quantity of nearby ESTs, (2) the distance to the nearest gene beyond the minimum requirements, (3) the number of base pairs of non-repeat masked sequence included in the locus trio, and (4) the number of homo-/hetero-polymers included in the final target region. In addition to the 10 autosomal regions chosen by Frisse et al. (2001), we selected the top 10 autosomal and 20 X-linked regions to sequence, making sure that no two regions were within 1 Mb of each other. Detailed resequencing methods are presented in Supplemental materials and summarized in Supplemental Figure 1. Information about each region is provided in Supplemental Table 2; exact locations and primer sequences are available from the authors upon request.
Haplotype estimation
Diploid genotype data were computationally phased using Phase 2.1 (Stephens and Donnelly 2003). To examine the distribution of haplotypes within and among populations, we constructed haplotypes excluding sites with overall minor allele frequency (MAF) of less than 0.05 and counted the number of distinct haplotypes for each of the 20 autosomal regions. We tabulated which populations contained each haplotype and constructed Venn diagrams. We did not perform a similar analysis with the X chromosome data because of the smaller sample size in most of the populations.
Statistical analyses
The genetic data were summarized with a battery of summary statistics: θ, π, Tajima’s D (TD), Fu and Li’s D (FLD), and ρ (Watterson 1975; Tajima 1983, 1989; Fu and Li 1993; Frisse et al. 2001). Under neutral equilibrium conditions both π and θ estimate the neutral parameter 3Neμ for X-linked loci and 4Neμ for autosomal loci, where Ne is the effective population size and μ is the neutral mutation rate. We calculated FST with sample size corrections following the method of Weir (1996). To test for deviations from a neutral equilibrium frequency distribution, Tajima’s D (Tajima 1989) and Fu and Li’s D (FLD) with an outgroup (Fu and Li 1993) were also calculated using the population genetics library Libsequence (K. Thornton; http://molpopgen.org/software/libsequence.html). P values were determined by 104 Monte Carlo replicates of the coalescent process under a neutral panmictic model with recombination and Ne = 104. Ratios of polymorphism to divergence were compared with expectations under a neutral, equilibrium model using a multilocus HKA test (Hudson et al. 1987) with the software HKA (J. Hey; http://lifesci.rutgers.edu/∼heylab/). Divergence data were derived for each of these loci by estimating the net divergence (DA) (Nei 1987) between homologous sequences from a common chimpanzee sequence and all human sequences. Estimated sex-averaged recombination rates were taken from the University of California, Santa Cruz genome browser (UCSC; http://www.genome.ucsc.edu) (Kent et al. 2002) using the March 2006 freeze of the Human Genome Project Working draft (hg18). Recombination rates represent averages for a window of 1 Mb encompassing each locus and were estimated from the deCODE Genetics map (Kong et al. 2002), which is based on 5136 microsatellite markers in 146 families, representing a total of 1257 meioses.
Acknowledgments
We thank Ryan Sprissler and Laurel Johnstone of the Genomic Analysis and Technology Core at the University of Arizona for aid in development of the DNA sequencing pipeline, and John D. Morelli, Brittany Tamarkin, Kimiko Della Croce, and Samina Makda for their dedicated computational and laboratory assistance. We also thank Howard Cann at the Foundation Jean Dausset (CEPH) in Paris for providing DNA samples. This research was funded by a National Science Foundation HOMINID grant (BCS-0423670) to M.F.H. and J.D.W.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.075630.107.
Reference
- Akey J.M., Zhang G., Zhang K., Jin L., Shriver M.D., Zhang G., Zhang K., Jin L., Shriver M.D., Zhang K., Jin L., Shriver M.D., Jin L., Shriver M.D., Shriver M.D. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey J.M., Eberle M.A., Rieder M.J., Carlson C.S., Shriver M.D., Nickerson D.A., Kruglyak L., Eberle M.A., Rieder M.J., Carlson C.S., Shriver M.D., Nickerson D.A., Kruglyak L., Rieder M.J., Carlson C.S., Shriver M.D., Nickerson D.A., Kruglyak L., Carlson C.S., Shriver M.D., Nickerson D.A., Kruglyak L., Shriver M.D., Nickerson D.A., Kruglyak L., Nickerson D.A., Kruglyak L., Kruglyak L. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2004;2:e286. doi: 10.1371/journal.pbio.0020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Stephen S., Kent W.J., Mattick J.S., Haussler D., Kent W.J., Mattick J.S., Haussler D., Mattick J.S., Haussler D., Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Burge C., Karlin S., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Karlin S., Karlin S. Finding the genes in genomic DNA. Curr. Opin. Struct. Biol. 1998;8:346–354. doi: 10.1016/s0959-440x(98)80069-9. [DOI] [PubMed] [Google Scholar]
- Cann H.M., de Toma C., Cazes L., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., de Toma C., Cazes L., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., Cazes L., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., Legrand M.F., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., Morel V., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., Piouffre L., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., Bodmer J., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., Bodmer W.F., Bonne-Tamir B., Cambon-Thomsen A., Bonne-Tamir B., Cambon-Thomsen A., Cambon-Thomsen A., et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Morgan M.T., Charlesworth D., Morgan M.T., Charlesworth D., Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A.G., Hubisz M.J., Bustamante C.D., Williamson S.H., Nielsen R., Hubisz M.J., Bustamante C.D., Williamson S.H., Nielsen R., Bustamante C.D., Williamson S.H., Nielsen R., Williamson S.H., Nielsen R., Nielsen R. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 2005;15:1496–1502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad D.F., Jakobsson M., Coop G., Wen X., Wall J.D., Rosenberg N.A., Pritchard J.K., Jakobsson M., Coop G., Wen X., Wall J.D., Rosenberg N.A., Pritchard J.K., Coop G., Wen X., Wall J.D., Rosenberg N.A., Pritchard J.K., Wen X., Wall J.D., Rosenberg N.A., Pritchard J.K., Wall J.D., Rosenberg N.A., Pritchard J.K., Rosenberg N.A., Pritchard J.K., Pritchard J.K. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat. Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- Crawford D.C., Akey D.T., Nickerson D.A., Akey D.T., Nickerson D.A., Nickerson D.A. The patterns of natural variation in human genes. Annu. Rev. Genomics Hum. Genet. 2005;6:287–312. doi: 10.1146/annurev.genom.6.080604.162309. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Schneider S., Schneider S. Why hunter–gatherer populations do not show signs of pleistocene demographic expansions. Proc. Natl. Acad. Sci. 1999;96:10597–10602. doi: 10.1073/pnas.96.19.10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., Boudreau A., Hardenbol P., Leal S.M., Hardenbol P., Leal S.M., Leal S.M., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisse L., Hudson R.R., Bartoszewicz A., Wall J.D., Donfack J., Di Rienzo A., Hudson R.R., Bartoszewicz A., Wall J.D., Donfack J., Di Rienzo A., Bartoszewicz A., Wall J.D., Donfack J., Di Rienzo A., Wall J.D., Donfack J., Di Rienzo A., Donfack J., Di Rienzo A., Di Rienzo A. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am. J. Hum. Genet. 2001;69:831–843. doi: 10.1086/323612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.X., Li W.H., Li W.H. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D., Hammer M.F., Hammer M.F. Reconstructing human origins in the genomic era. Nat. Rev. Genet. 2006;7:669–680. doi: 10.1038/nrg1941. [DOI] [PubMed] [Google Scholar]
- Garrigan D., Mobasher Z., Kingan S.B., Wilder J.A., Hammer M.F., Mobasher Z., Kingan S.B., Wilder J.A., Hammer M.F., Kingan S.B., Wilder J.A., Hammer M.F., Wilder J.A., Hammer M.F., Hammer M.F. Deep haplotype divergence and long-range linkage disequilibrium at xp21.1 provide evidence that humans descend from a structured ancestral population. Genetics. 2005;170:1849–1856. doi: 10.1534/genetics.105.041095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M.F., Garrigan D., Wood E., Wilder J.A., Mobasher Z., Bigham A., Krenz J.G., Nachman M.W., Garrigan D., Wood E., Wilder J.A., Mobasher Z., Bigham A., Krenz J.G., Nachman M.W., Wood E., Wilder J.A., Mobasher Z., Bigham A., Krenz J.G., Nachman M.W., Wilder J.A., Mobasher Z., Bigham A., Krenz J.G., Nachman M.W., Mobasher Z., Bigham A., Krenz J.G., Nachman M.W., Bigham A., Krenz J.G., Nachman M.W., Krenz J.G., Nachman M.W., Nachman M.W. Heterogeneous patterns of variation among multiple human x-linked Loci: The possible role of diversity-reducing selection in non-Africans. Genetics. 2004;167:1841–1853. doi: 10.1534/genetics.103.025361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I., Ebersberger I., Ptak S.E., Paabo S., Przeworski M., Ebersberger I., Ptak S.E., Paabo S., Przeworski M., Ptak S.E., Paabo S., Przeworski M., Paabo S., Przeworski M., Przeworski M. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 2003;72:1527–1535. doi: 10.1086/375657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W.G., Robertson A., Robertson A. The effect of linkage on limits to artificial selection. Genet. Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Hinds D.A., Stuve L.L., Nilsen G.B., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R., Stuve L.L., Nilsen G.B., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R., Nilsen G.B., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R., Ballinger D.G., Frazer K.A., Cox D.R., Frazer K.A., Cox D.R., Cox D.R. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- Hsu F., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., Haussler D., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., Haussler D., Clawson H., Kuhn R.M., Diekhans M., Haussler D., Kuhn R.M., Diekhans M., Haussler D., Diekhans M., Haussler D., Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- Hudson R.R., Kreitman M., Aguade M., Kreitman M., Aguade M., Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M., Scholz S.W., Scheet P., Gibbs J.R., VanLiere J.M., Fung H.C., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R., Scholz S.W., Scheet P., Gibbs J.R., VanLiere J.M., Fung H.C., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R., Scheet P., Gibbs J.R., VanLiere J.M., Fung H.C., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R., Gibbs J.R., VanLiere J.M., Fung H.C., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R., VanLiere J.M., Fung H.C., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R., Fung H.C., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R., Degnan J.H., Wang K., Guerreiro R., Wang K., Guerreiro R., Guerreiro R., et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Keinan A., Mullikin J.C., Patterson N., Reich D., Mullikin J.C., Patterson N., Reich D., Patterson N., Reich D., Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat. Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Gudbjartsson D.F., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Gudbjartsson D.F., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., Barnard J., Hallbeck B., Masson G., Hallbeck B., Masson G., Masson G., et al. A high-resolution recombination map of the human genome. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Feldman M., Cavalli-Sforza L.L., Cavalli-Sforza L.L., et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Livingston R.J., von Niederhausern A., Jegga A.G., Crawford D.C., Carlson C.S., Rieder M.J., Gowrisankar S., Aronow B.J., Weiss R.B., Nickerson D.A., von Niederhausern A., Jegga A.G., Crawford D.C., Carlson C.S., Rieder M.J., Gowrisankar S., Aronow B.J., Weiss R.B., Nickerson D.A., Jegga A.G., Crawford D.C., Carlson C.S., Rieder M.J., Gowrisankar S., Aronow B.J., Weiss R.B., Nickerson D.A., Crawford D.C., Carlson C.S., Rieder M.J., Gowrisankar S., Aronow B.J., Weiss R.B., Nickerson D.A., Carlson C.S., Rieder M.J., Gowrisankar S., Aronow B.J., Weiss R.B., Nickerson D.A., Rieder M.J., Gowrisankar S., Aronow B.J., Weiss R.B., Nickerson D.A., Gowrisankar S., Aronow B.J., Weiss R.B., Nickerson D.A., Aronow B.J., Weiss R.B., Nickerson D.A., Weiss R.B., Nickerson D.A., Nickerson D.A. Pattern of sequence variation across 213 environmental response genes. Genome Res. 2004;14:1821–1831. doi: 10.1101/gr.2730004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard-Smith J., Haigh J., Haigh J. The hitch-hiking effect of a favourable gene. Genet. Res. 1974;23:23–35. [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. Columbia University Press; New York: 1987. [Google Scholar]
- Plagnol V., Wall J.D., Wall J.D. Possible ancestral structure in human populations. PLoS Genet. 2006;2:e105. doi: 10.1371/journal.pgen.0020105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M., Wall J.D., Andolfatto P., Wall J.D., Andolfatto P., Andolfatto P. Recombination and the frequency spectrum in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 2001;18:291–298. doi: 10.1093/oxfordjournals.molbev.a003805. [DOI] [PubMed] [Google Scholar]
- Ptak S.E., Przeworski M., Przeworski M. Evidence for population growth in humans is confounded by fine-scale population structure. Trends Genet. 2002;18:559–563. doi: 10.1016/s0168-9525(02)02781-6. [DOI] [PubMed] [Google Scholar]
- Rosenberg N.A. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann. Hum. Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Stephens M., Donnelly P., Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneking M., Jorde L.B., Bhatia K., Wilson A.C., Jorde L.B., Bhatia K., Wilson A.C., Bhatia K., Wilson A.C., Wilson A.C. Geographic variation in human mitochondrial DNA from Papua New Guinea. Genetics. 1990;124:717–733. doi: 10.1093/genetics/124.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff S.A., Dietzsch E., Speed W., Pakstis A.J., Kidd J.R., Cheung K., Bonne-Tamir B., Santachiara-Benerecetti A.S., Moral P., Krings M., Dietzsch E., Speed W., Pakstis A.J., Kidd J.R., Cheung K., Bonne-Tamir B., Santachiara-Benerecetti A.S., Moral P., Krings M., Speed W., Pakstis A.J., Kidd J.R., Cheung K., Bonne-Tamir B., Santachiara-Benerecetti A.S., Moral P., Krings M., Pakstis A.J., Kidd J.R., Cheung K., Bonne-Tamir B., Santachiara-Benerecetti A.S., Moral P., Krings M., Kidd J.R., Cheung K., Bonne-Tamir B., Santachiara-Benerecetti A.S., Moral P., Krings M., Cheung K., Bonne-Tamir B., Santachiara-Benerecetti A.S., Moral P., Krings M., Bonne-Tamir B., Santachiara-Benerecetti A.S., Moral P., Krings M., Santachiara-Benerecetti A.S., Moral P., Krings M., Moral P., Krings M., Krings M. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996;271:1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- Voight B.F., Adams A.M., Frisse L.A., Qian Y., Hudson R.R., Di Rienzo A., Adams A.M., Frisse L.A., Qian Y., Hudson R.R., Di Rienzo A., Frisse L.A., Qian Y., Hudson R.R., Di Rienzo A., Qian Y., Hudson R.R., Di Rienzo A., Hudson R.R., Di Rienzo A., Di Rienzo A. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc. Natl. Acad. Sci. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G.A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Weir B.S. Genetic data analysis II: Methods for discrete population genetic data. Sinauer Associates; Sunderland, MA: 1996. [Google Scholar]
- Weir B.S., Cardon L.R., Anderson A.D., Nielsen D.M., Hill W.G., Cardon L.R., Anderson A.D., Nielsen D.M., Hill W.G., Anderson A.D., Nielsen D.M., Hill W.G., Nielsen D.M., Hill W.G., Hill W.G. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 2005;15:1468–1476. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N., Chen F.C., Ota S., Jorde L.B., Pamilo P., Patthy L., Ramsay M., Jenkins T., Shyue S.K., Li W.H., Chen F.C., Ota S., Jorde L.B., Pamilo P., Patthy L., Ramsay M., Jenkins T., Shyue S.K., Li W.H., Ota S., Jorde L.B., Pamilo P., Patthy L., Ramsay M., Jenkins T., Shyue S.K., Li W.H., Jorde L.B., Pamilo P., Patthy L., Ramsay M., Jenkins T., Shyue S.K., Li W.H., Pamilo P., Patthy L., Ramsay M., Jenkins T., Shyue S.K., Li W.H., Patthy L., Ramsay M., Jenkins T., Shyue S.K., Li W.H., Ramsay M., Jenkins T., Shyue S.K., Li W.H., Jenkins T., Shyue S.K., Li W.H., Shyue S.K., Li W.H., Li W.H. Larger genetic differences within Africans than between Africans and Eurasians. Genetics. 2002;161:269–274. doi: 10.1093/genetics/161.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]