Abstract
Genomic analyses have been applied extensively to analyze the process of transcription initiation in mammalian cells, but less to transcript 3′ end formation and transcription termination. We used a novel approach to prepare 3′ end fragments from polyadenylated RNA, and mapped the position of the poly(A) addition site using oligonucleotide arrays tiling 1% of the human genome. This approach revealed more 3′ ends than had been annotated. The distribution of these ends relative to RNA polymerase II (PolII) and di- and trimethylated lysine 4 and lysine 36 of histone H3 was compared. A substantial fraction of unannotated 3′ ends of RNA are intronic and antisense to the embedding gene. Poly(A) ends of annotated messages lie on average 2 kb upstream of the end of PolII binding (termination). Near the termination sites, and in some internal sites, unphosphorylated and C-terminal domain (CTD) serine 2 phosphorylated PolII (POLR2A) accumulate, suggesting pausing of the polymerase and perhaps dephosphorylation prior to release. Lysine 36 trimethylation occurs across transcribed genes, sometimes alternating with stretches of DNA in which lysine 36 dimethylation is more prominent. Lysine 36 methylation decreases at or near the site of polyadenylation, sometimes disappearing before disappearance of phosphorylated RNA PolII or release of PolII from DNA. Our results suggest that transcription termination loss of histone 3 lysine 36 methylation and later release of RNA polymerase. The latter is often associated with polymerase pausing. Overall, our study reveals extensive sites of poly(A) addition and provides insights into the events that occur during 3′ end formation.
Identification of the regions of the human genome that encode transcripts is essential for a complete functional understanding of the function of the genome. Studies over the last few years have found that many more regions are transcribed into RNA than can be accounted for by genes encoding known or predicted proteins (for reviews, see Rozowsky et al. 2006; Kapranov et al. 2007a), and noncoding RNAs that serve a number of functions have been identified (for reviews, see Mattick and Makunin 2006; Shamovsky and Nudler 2006; Carninci and Hayashizaki 2007; Kapranov et al. 2007b; Taft et al. 2007). Examples include the XIST RNA that is involved in X chromosome silencing, RNAs transcribed from portions of imprinted regions and functionally related to imprinting, precursors for small regulatory RNAs, RNA that can directly regulate transcription factors such as the steroid receptor, intergenic transcripts that appear to regulate the expression of adjacent coding genes such as the HOX genes, and cytoplasmic antisense RNAs from introns that may modulate the levels of expression of protein coding genes. However, the function of most noncoding RNAs is not known, and a substantial portion of these RNAs are intranuclear (Furuno et al. 2006; Gingeras 2007).
Our current understanding of the extent of transcriptionally active DNA has come primarily from massive application of established technology for cDNA and expressed sequence tag (EST) sequencing (Maeda et al. 2006) and more recently from newer technologies. These latter technologies include approaches for the display and sequence analysis of short sequences adjacent to sites of oligo(dT)-primed cDNA synthesis (Wei et al. 2004) and/or to cap sites at the 5′ end of mRNAs (Maruyama and Sugano 1994; Choi and Hagedorn 2003; Kodzius et al. 2006; Ng et al. 2006; Denoeud et al. 2007) as well as developments in the field of microarray analysis (Kapranov et al. 2002; Rinn et al. 2003; Bertone et al. 2004).
Studies employing genomic tiling arrays have been quite informative regarding the occurrence and distribution of transcriptionally active regions in large portions of the human genome. Early arrays consisted of PCR products derived from nonrepetitive portions of the genome. An early application of this approach was the study of the transcriptional activity of chromosome 22. This study showed the presence of substantial amounts of intergenic transcription as well as accumulation of transcripts from within introns, often in an antisense direction (Rinn et al. 2003). However, with advances in technology, the PCR product arrays have been replaced by microarrays containing very large numbers of oligonucleotides covering nonrepetitive regions of large portions of the genome such as entire chromosomes (Kapranov et al. 2002, 2005; Cheng et al. 2005) or the regions studied intensively by The ENCODE Project Consortium (2004). Whole-genome oligonucleotide tiling arrays have also been applied to transcript identification (Bertone et al. 2004; Cheng et al. 2005), and the advent of high-density oligonucleotide microarrays is expected to make the cost of whole-genome scanning generally affordable in the future.
One of the most extensively applied approaches for identifying the 3′ ends of transcripts involves generating short sequence tags from the ends of RNA by the addition of oligonucleotides that allow restriction site cleavage 21 bases from the 3′ end (Saha et al. 2002). This leads to short sequence tags that can be concatemerized and sequenced. Extensive sequencing is required in order to obtain enough tag sequences to identify and quantify less abundant RNA species, and the wide application of these approaches requires advances in economy and scale of sequencing that are only now becoming feasible. In addition, the short sequence tags may be challenging to align to unique regions of the genome, particularly if they are derived from repeat-containing regions, and they are rather short to be used for analysis with genome tiling microarrays.
The relationship between polyadenylation signals and transcription termination in higher cells is complex (for review, see Buratowski 2005). Studies of nascent transcripts in a few selected highly transcribed loci have shown that transcription may proceed well beyond the site of poly(A) addition, supporting a model in which elongated transcripts are cleaved at poly(A) addition sites (for review, see West et al. 2006) and the remaining 3′ ends of the transcripts degraded by mechanisms that are still under study. The presence of an accessible polyadenylation signal (AAUAAA in this case) was found to be necessary for efficient transcription termination (Nag et al. 2006) and transcription termination without precursor RNA cleavage has been observed in vivo in Drosophila (Osheim et al. 2002) as well as in vitro. However, our overall understanding of the relationship between poly(A) addition and transcription termination is limited, particularly for genes expressed at a low level.
Chromatin immunoprecipitation experiments can directly or indirectly indicate the presence of elongating RNA polymerase and potentially be used to map transcription units relative to the positions of poly(A) addition. POLR2A, the largest subunit of RNA polymerase II (PolII), contains a C-terminal domain consisting of many repeats (52 in humans) of the heptapeptide YSPTSPS that is variably phosphorylated (Buratowski 2003; Phatnani and Greenleaf 2006). PolII initially associates with the promoter in a form devoid of CTD phosphorylation. Early elongating forms of PolII are phosphorylated at serine 5 of the CTD principally by kinase activity of the general transcription factor TFIIH. The later stages of transcript elongation are associated with CTD serine 2 phosphorylation, mediated at least partly by CDK9. Therefore the presence of PolII phosphorylated on CTD serine 2 is a mark of progression of PolII, and presumably transcription, along the DNA template.
In addition to direct detection of PolII on DNA, specific histone modifications are associated with the presence of elongating PolII. Histone 3 lysine 4 is di- and trimethylated around the site of transcription initiation, mediated by the Set1 histone methylase (Barski et al. 2007; Heintzman et al. 2007). Subsequent phosphorylation of CTD on serine 2 recruits another histone methylase, KMT3 (also known as Set2) (Kizer et al. 2005; Morillon et al. 2005). KMT3 adds methyl groups to form trimethylated lysine 36 on histone 3. This mark has been suggested to be part of a process that ensures deacetylation of chromatin in the wake of PolII and thereby prevent internal reinitiation of transcription (for discussions, see Sims et al. 2004; Li et al. 2007).
In this study we have used a method for preparing collections of fragments from the 3′ ends of cDNA fragments that are of sufficient length for array hybridization. These fragments were analyzed with ENCODE genomic tiling arrays to map the sites of polyadenylation of stable RNAs in five cell types. We have compared these results with chromatin immunoprecipitation experiments using genomic tiling arrays and antibodies against various forms of the largest subunit of PolII and against di- and trimethylated lysines 4 and 36 of histone 3. The results confirm that a large fraction of poly(A) sites of mRNA do not correspond to the 3′ end of known mRNAs and do not represent sites at which PolII is removed from the DNA template. RNA polymerase CTD serine 5 phosphorylation is largely concentrated at promoter proximal regions that also show histone 3 lysine 4 di- and trimethylation. Serine 2 phosphorylation occurs across the body of most genes and initiates more or less concordantly with lysine 36 di- and trimethylation within genes. However, serine 2 phosphorylated and unphosphorylated RNA polymerases tend to accumulate at sites averaging about 2 kb beyond the site of transcript cleavage and polyadenylation and often well beyond the area of detectable histone 3 lysine 36 trimethylation. The results suggest a relatively complex mechanism of transcription termination that involves reduction of histone 3 lysine 36 trimethylation prior to RNA polymerase dephosphorylation and release from DNA.
Results
We previously established a procedure for the selective amplification of the 3′ ends of cDNAs and used it to study patterns of mRNA expression in human cells (Prashar and Weissman 1996; Subrahmanyam et al. 2001). In this procedure cDNA synthesis was primed with an oligo(dT) primer that had a PCR primer binding sequence attached to its 5′ end. The cDNA was then cut with a restriction enzyme and ligated to a Y-shaped adapter. Amplification was accomplished by PCR using one primer complementary to the oligo(dT) primer site and a second primer that had the same sequence as one arm of the Y-shaped adapter. In these studies, we sequenced over 1000 individual bands excised from gels and found that nearly all were derived from the 3′ ends of mRNA species. The only exceptions were those in which the oligo(dT) had primed from an internal adenine (A)-rich sequence in RNA.
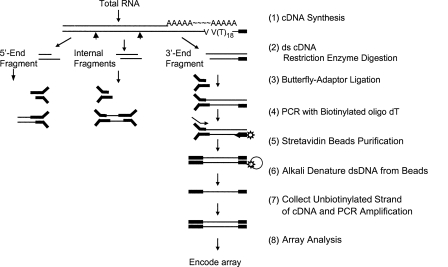
We have now modified this procedure to further improve its specificity by using a biotinylated primer to selectively capture the desired 3′ end fragments prior to further amplification and to prepare libraries of 3′ end fragments from polyadenylated RNA (Fig. 1). The resulting library of 3′ end fragments was used with genomic tiling arrays to map sites of poly(A) addition after normalization against total cDNA signals. The 3′ end cDNA fragments were prepared by cutting total cDNA with either Sau3AI or NlaIII restriction enzymes. An advantage of this approach is that the signal obtained from such fragments generally begins at a position corresponding to the restriction site and ends at the site of poly(A) addition, so that the orientation of the first-strand cDNA can often be deduced from the signal on genomic tiling arrays (Fig. 2).
Figure 1.
Schematic of the procedure used for preparation of 3′ end fragments from cDNA. The thin black bars indicate the positions of cDNA sequences, and the Y-shaped solid black bars show the butterfly adapters.
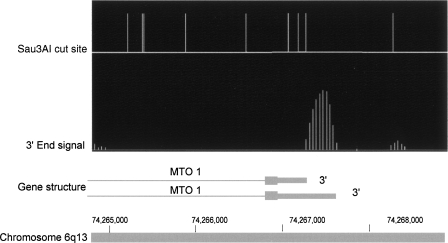
Figure 2.
Example of poly(A) signal. Vertical lines in the top portion of the figure correspond to Sau3AI recognition sites in genomic DNA. Closely spaced vertical lines in the second row represent the normalized signals comparing 3′ end fragments to total cDNA. The lower horizontal line represents the 3′ end of the MTO1 gene. Thick bars are exons and thin lines are introns. The figure shows that the orientation of the transcript can be distinguished because the signal begins at a Sau3AI cutting site and extends part way toward the next cutting site. This shows that the major 3′ end of the MTO1 transcript in these HeLa cells is coincident with the shorter 3′ end in the literature, with a very weak second signal corresponding to the longer 3′ end. Note that computer smoothing of data causes some signal spread upstream of the cutting site.
To establish our procedures, we first prepared 3′ end libraries from HeLa cells using Sau3AI and analyzed these samples using Affymetrix U133 oligonucleotide chips that were designed for measuring mRNA levels. To compare the sensitivity of the 3′ end tiling chip display with that of commercial oligonucleotide chips designed for measuring cDNA levels, we prepared 3′ end libraries from HeLa cells and analyzed these on two types of arrays: (1) genomic tiling arrays covering the ENCODE regions of the genome and (2) Affymetrix U133 chips. The Affymetrix arrays revealed that 107 mRNAs were present in this region from HeLa cells. Of these, 96 had a definite poly(A) signal from the ENCODE array data, as observed manually on an IGB browser (http://www.affymetrix.com/support/developer/tools/download_igb.affx) display of Sau3AI-generated 3′ end fragments. An additional seven showed weak or diffuse signals, and only four gave no signals. Statistically this is somewhat more signals than would be expected for a random distribution of Sau3AI sites, since a fragment less than about 35 base pairs in length would not give a significant signal on the arrays.
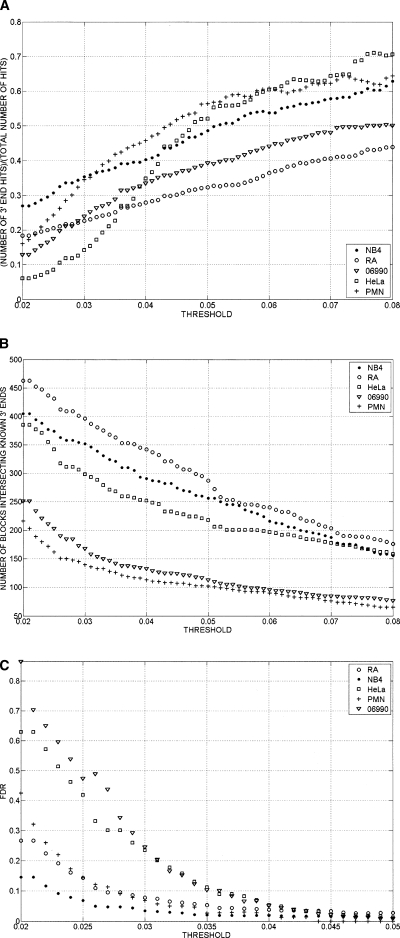
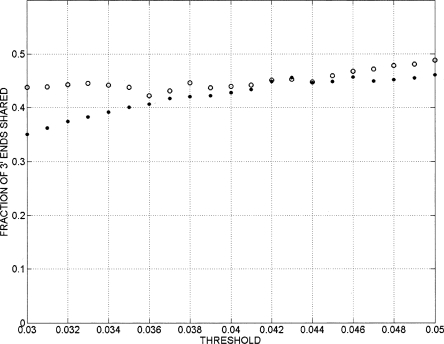
A second group of two samples was prepared in which the cDNA was cut with NlaIII rather than Sau3AI. Two of the genes that did not show a 3′ end RNA signal with Sau3AI cut fragments now showed a 3′ end. The absence of 3′ end signals for two mRNAs that were reported positive by the Affymetrix chips could stem from a variety of causes including limits of sensitivity, diffuse or far downstream 3′ ends, or false-positive calls by the Affymetrix chip. Conversely, of 23 genes represented on the U133 chip and reported as absent, 15 showed a poly(A) signal and two others had weak signals, suggesting that, overall, the display is more sensitive than the total cDNA analysis on 25-mer oligonucleotides. Of 42 genes not represented on the U133 chip, 30 showed a definite poly(A) tail and two showed a weak poly(A) signal. As the threshold for calling a positive signal was lowered, a greater fraction of the calls were from regions not adjacent to the 3′ ends of known genes (Fig. 3A). However, the number of signals associated with known genes also increased throughout the range as thresholds were lowered (Fig. 3B). More signals were observed associated with known genes than the number of such genes themselves, due to the use of multiple minor 3′ ends by many genes.
Figure 3.
(A) Ratio of 3′ end signals for annotated genes to total 3′ end signals as a function of the threshold value used to call 3′ ends. After quantile normalization, data from replicates were combined by averaging the signals of the replicates. Signal intensity yi = log(Ri) − log(Gi) was assigned to each probe, where log(Ri) and log(Gi) are Cy5 and Cy3 channels’ intensities after the aforementioned transformations were performed. Contiguous segments (bars) due to the signal coming from the enriched regions were obtained by joining probes with intensities yi above the threshold separated by less than a certain distance (max-gap of 114 bp). Only segments whose length was greater than a particular size (min-run of 114 bp) were selected. As the threshold for calling a positive signal was lowered, a greater fraction of the calls were from regions not adjacent to the 3′ ends of known genes. Results are shown for five cell types. GENCODE annotation was used for the analysis. (B) Total number of 3′ end signals associated with annotated genes as a function of threshold. Signals are considered as intersecting a known 3′ end if they lie within 2500 bases downstream from the known end. Signals were calculated as in A. The number of signals associated with known genes increased throughout the range as thresholds were lowered. Results are shown for five cell types. 06990 and PMN are end stage differentiated cells (a lymphoblastoid cell and a normal neutrophil); therefore, it is not surprising to see that a smaller number of 3′ ends are present in these cell lines. GENCODE annotation was used for the analysis. (C) False-discovery rate (FDR) as a function of the threshold used to call 3′ ends. Results are shown for five cell types. For each data set the genomic locations of the probes on the microarray were randomly shuffled. The max-gap and min-run procedures described in “Bioinformatic Analysis” section were applied to the randomized data. The FDR was computed as FDR(threshold) = N1(threshold)/N2(threshold), where N1 is the number of discovered blocks for the randomized data and N2 is the number of blocks for the nonrandomized data. FDR increases as the threshold for calling a positive signal decreases.
We performed similar analyses of the ENCODE region for triplicate RNA samples prepared from the NB4 promyelocytic cell line, from NB4 cells induced to neutrophil differentiation by treatment with retinoic acid, from normal human neutrophils, and from GM06990 lymphoblastoid cells, as well as a single analysis from K562 erythroleukemic cells. To deal with this large amount of data, we bioinformatically detected 3′ end fragments at a range of thresholds of sensitivity.
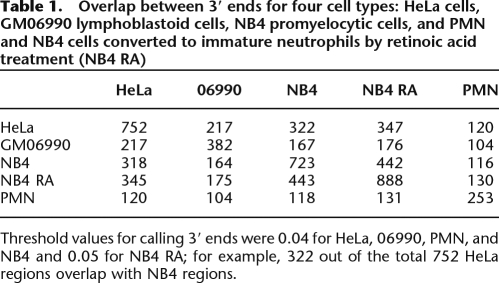
For each cell type, between 40% and 60% of the signals intersected regions showing a signal from the other cell types in this study when using a threshold of 0.04 for calling 3′ ends. This threshold was chosen because it gave a false-discovery rate of less than 5% for every cell type (Fig. 3C). Somewhat unexpectedly, the remaining 3′ ends often appeared to be cell type specific (Fig. 3B; Table 1). These did not reflect variability in the method as comparison of two randomly chosen HeLa experiments showed over 98% agreement in the location of 3′ ends detected with the same threshold. As shown in Figure 3B, the cell type specific 3′ end signals spanned the full range of intensities, with signals shared between cell types only modestly more abundant at high intensities than at low intensities, so that the cell type specificity was not due only to low-intensity signals. Furthermore, attempts to compare two cell types—using a different threshold for one cell than for the other—did not result in more than 50% overlap over the threshold range of 0.04–0.05 (Fig. 4).
Table 1.
Overlap between 3′ ends for four cell types: HeLa cells, GM06990 lymphoblastoid cells, NB4 promyelocytic cells, and PMN and NB4 cells converted to immature neutrophils by retinoic acid treatment (NB4 RA)
Threshold values for calling 3′ ends were 0.04 for HeLa, 06990, PMN, and NB4 and 0.05 for NB4 RA; for example, 322 out of the total 752 HeLa regions overlap with NB4 regions.
Figure 4.
Fraction of 3′ ends shared between HeLa and NB4 cells as a function of the threshold used for identifying 3′ end signals. Open circles represent the fraction of NB4 3′ end signals that are also present in HeLa, and filled circles represent the fraction of HeLa 3′ ends that are also present in NB4 cells.
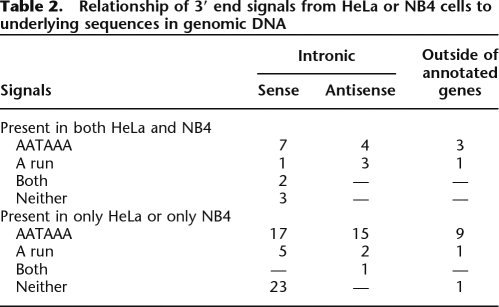
Approximately 100 of the HeLa or NB4 regions that gave 3′ end signals but were not within 2.5 kb downstream from the annotated 3′ ends of known or predicted genes were chosen randomly (Table 2). These signals were manually compared against the genomic sequence embedding them. As shown in Table 2, 58 of the signals were preceded by an AATAAA or ATTAAA polyadenylation sequence, and 12 signals were probably attributable to A (or T)-rich sequences in the genomic DNA. These sequences are presumably in transcripts extending through this region and represent either incompletely spliced mRNA, excised introns that had not been degraded, or longer transcripts passing through this region. In three cases, an A-rich sequence was preceded by a potential poly(A) addition signal lying about 20 bases upstream. This suggests that these sequences represent fragments of re-inserted cDNA copies of other genes. In some cases, there was no obvious reason for the signal in terms of poly(A) addition sites or runs. These could represent polyadenylation at atypical or noncanonical poly(A) addition signals (Venkataraman et al. 2005), regions spliced into distant fragments that were linked to poly(A), or transcripts terminating in repetitive regions not represented on the microarray.
Table 2.
Relationship of 3′ end signals from HeLa or NB4 cells to underlying sequences in genomic DNA
Overall, almost 60% of the cases examined were good candidates to represent the 3′ polyadenylated ends of unannotated RNAs. Twelve 3′ ends were in regions upstream and more than 2.5 kb downstream from any annotated gene. These might represent 3′ ends of entirely unannotated transcripts or very long 3′ extensions of known transcripts (Moucadel et al. 2007). Of the 3′ ends within introns that could be evaluated, 26 of the poly(A) signals that could be oriented arose from transcripts in the same orientation as the embedding gene while 20 were in the antisense orientation. This compares well with an earlier estimate that 10 of 25 intronic transcripts detected from a PCR fragment arrays representing chromosome 22 were in the antisense orientation with respect to the embedding gene (Rinn et al. 2003).
As a further check of the method, we selected 19 of the 3′ end HeLa signals that did not lie within 2.5 kb of any 3′ end annotated in RefSeq. For each such signal, we designed a pair of PCR primers with one upstream and the other downstream from the estimated position of the poly(A) site and performed 3′ RACE on HeLa cell cDNA. In 18 of the 19 cases, a band was seen with one or the other of these primers that was substantially clearer and more prominent than any band seen with the other primer of the pair. These bands were sequenced and in 17 cases gave satisfactory reads. Of these, 16 bands showed a poly(A) signal (either AAUAAA or, in one case, ATTAAA) in the DNA ∼20 bases upstream of the poly(A). In the remaining case, there was a poly(A) tract encoded in the genomic DNA, and oligo(dT) priming presumably occurred from a transcript embedding this genomic sequence. These results indicated that a somewhat higher percentage of 3′ end signals were associated with poly(A) signal hexanucleotides in the DNA than might have been anticipated from the manual estimates.
Methylation of histone H3 at lysine 36 is mediated by the enzyme KMT3/NSD1. This is a histone methylase that directly binds to the PolII serine 2 phosphorylated C-terminal domain of the largest subunit of PolII (Li et al. 2007). This has been suggested to be part of a mechanism that recruits the histone deacetylase RPD3 to remove histone acetylation behind the progressing RNA polymerase and therefore prevents random transcription reinitiations within the body of the gene (Cuthbert et al. 2004; Carrozza et al. 2005; Joshi and Struhl 2005; Keogh et al. 2005). To the extent that this mechanism accounts for histone 3 lysine 36 methylation, the methylation pattern might be expected to reflect the presence of elongating RNA polymerase.
In a few cases of actively transcribed mammalian genes, it has been experimentally confirmed that transcription extends well downstream from the polyadenylation site. To study the relationship between polyadenylation sites, chromatin modifications, and termination of transcription on a global basis, we performed ChIP–chip experiments across the ENCODE regions with material from HeLa, NB4, and K562 cells. These studies were performed using various antibodies directed against specific histone modifications or different forms of PolII, as described in the Methods section.
Strong signals of lysine 36 methylation were detected across many actively transcribed genes, beginning downstream from the transcription initiation site and extending generally up to or beyond the polyadenylation site. Reduction in the signal often occurred near the site of polyadenylation. In a number of cases, it was not possible to determine where the signal ended for the reasons mentioned above. In the remaining genes, the signal for histone 3 lysine 36 trimethylation commonly declined and then ended at distances, ranging from −2.5 to +6 (average +1.8) kb downstream from the poly(A) addition site. The decline in signal beyond the poly(A) site was not always continuous and often appeared to occur in several steps.
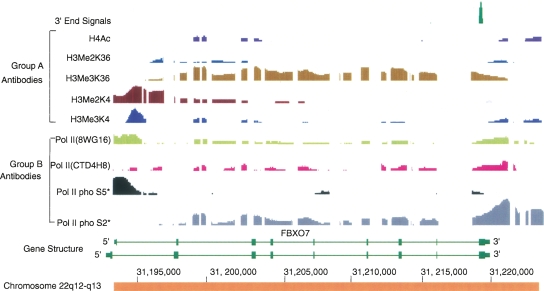
Figure 5 presents a fairly typical pattern for an actively transcribed gene. Lysine 4 di- and trimethylation signals were strong around sites of initiation and often biphasic with a gap at the actual site of initiation (Barski et al. 2007). This gap is presumably due to a combination of binding by PolII and other factors, resulting in nucleosome displacement. Dimethylation of histone 3 lysine 4 extended somewhat further downstream than trimethylation in many cases (Fig. 5).
Figure 5.
Example of the most common pattern for histone and polymerase modifications at the 3′ ends of expressed genes. Chromatin IP results from the chromosome 22q12-q13 region are displayed with the Integrated Genome Browser (IGB) on Human Assembly 7. FBXO7 gene is annotated at this locus. Two antibody groups were used: Group A, human histone modifications; Group B, various phosphorylation states of human PolII. All the chromatin immunoprecipitates were prepared from HeLa cells unless starred to indicate the results were from K562 cells. The information about antibodies used for the data in Figs. 5, 6, 7, 8 is listed in Table 3.The first row shows the RNA polyadenylation signal, which was detected by the 3′ end enrichment method. This 3′ end signal corresponds exactly to the reported 3′ end of the mRNA (RefSeq). Dimethylation of histone 3 lysine 4 extends over a broader region than trimethylation of lysine 4. PolII serine 5 phosphorylation always shows a peak at the 5′ end of the gene. As expected, serine 2 phosphorylation extends through the body of the gene and beyond the poly(A) site. The signal increases just before disappearance of the polymerase from DNA, indicating RNA polymerase pausing occurs before release. Histone 3 K36 trimethylation tracks with PolII serine 2 through the body of the gene; however, just beyond the poly(A) site, trimethylation begins to decrease and apparently becomes uncoupled from serine 2 phosphorylation.
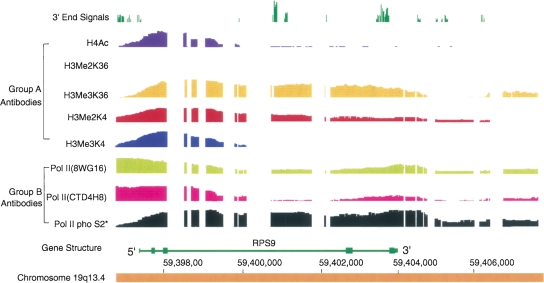
There were a number of genes that showed substantial variation from the common pattern of lysine 36 methylation. A few genes or genetic areas showed very low levels of lysine 36 methylation but did show histone 3 lysine 4 dimethylation (Figs. 6 7) spread beyond the promoter into or across the body of a gene. Some, often short genes or genes transcribed at relatively low levels, had minimal or no detectable lysine 36 trimethylation (Fig. 7).
Figure 6.
Chromatin IP results for a region containing the gene RPS9, an example of another pattern of histone and PolII modification. The analyses and data are as described in the legend for Figure 5. RPS9 is a highly transcribed gene. The results show that almost all the antigens are detectable throughout the entire region except for the absence of histone 3 dimethyl lysine 36.
Figure 7.
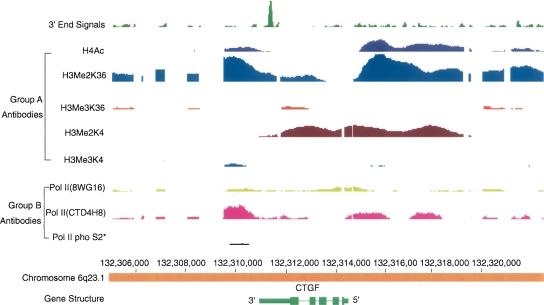
Example of another type of deviation from the standard pattern. In this figure, the dimethylation of both histone 3 lysine 36 (K36) and lysine 4 (K4) extends across the entire gene. Interestingly, histone dimethyl lysine 36 is also enriched for some distance upstream of the 5′ end of the CTGF gene and then decreased to lower levels over most of the body of the gene. Histone dimethyl lysine 4 shows almost the inverse pattern of dimethyl lysine 36, increasing later than lysine 36, mainly retained over the body of the gene, and dropping just before the 3′ end signal appears. It seems that PolII (4H8) and acetylated histone 4 were similarly distributed to histone dimethyl lysine 36 throughout this whole region. Unlike typical genes, trimethylations at lysine 4 and lysine 36 sites were essentially absent. Note that transcription of this gene proceeds right to left.
To test whether the reduction of trimethylated lysine 36 beyond the polyadenylation site was due to partial removal of methyl groups, we also performed chromatin immunoprecipitation with an antibody directed to dimethyl lysine 36 of histone 3. We observed a signal of dimethyl lysine 36 at the position where the trimethylation waned. Unexpectedly, there was often a strong signal of dimethylation of lysine 36 upstream and around the initiation site for transcription (Figs. 7, 8). Some genes, especially large ones, showed broad stretches of dimethylation of lysine 36, extending tens of kilobases, preceding or following another broad zone in which trimethylation of lysine 36 was more predominant (see Fig. 8); also, there were blocks of dimethylated lysine 36 that did not overlap with known transcripts.
Figure 8.
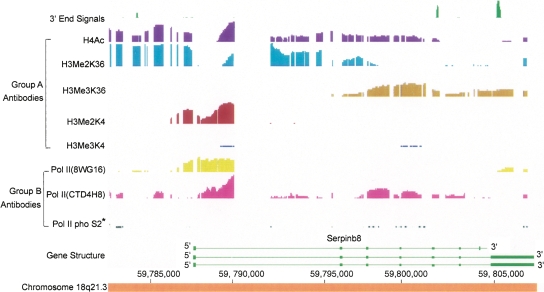
An example of another type of deviation from the standard pattern. In several large genes, such as SERPINB8, there are stretches of histone lysine 36 (K36) dimethylation extending over tens of kilobases and alternating with stretches of trimethylation. In some other genes, the pattern is revered with the 5′ end portion showing trimethylation. The functional significance of these patterns remains to be determined.
Measurement of DNA-associated RNA polymerase would, in principle, provide a direct assessment of the relationship between sites of poly(A) addition and sites of transcription termination or at least the release of DNA from the RNA polymerase. However, PolII undergoes a series of phosphorylations during mRNA transcription. The enzyme initially binds to promoters in an unphosphorylated state, then becomes phosphorylated first on serine 5 of the YSPTSPS repeat motif, then subsequently on serine 2. To more directly study the relationship between the ends of PolII binding regions and mRNA polyadenylation sites, we employed antibodies reported to interact selectively with unphosphorylated CTD of the large subunit, with CTD phosphorylated on serine 2, CTD phosphorylated on serine 5 or total RNA polymerase regardless of its phosphorylation state (Table 3).
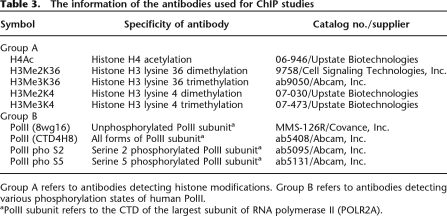
Table 3.
The information of the antibodies used for ChIP studies
Group A refers to antibodies detecting histone modifications. Group B refers to antibodies detecting various phosphorylation states of human PolII.
aPolII subunit refers to the CTD of the largest subunit of RNA polymerase II (POLR2A).
We explored the use of four different antibodies reported to detect different phosphorylation states of PolII (Table 3). Monoclonal antibody 8wg16 is directed against the unphosphorylated form of PolII CTD. Antibody PolII pho S5 reacts with CTD phosphorylated at serine 5. An antibody to PolII phos2 that has been used in earlier studies has been reported to react with serine 2–phosphorylated CTD, but more recent studies have shown that it can also react with serine 5–phosphorylated CTD and especially with the doubly-phosphorylated CTD (Phatnani and Greenleaf 2006). We used a newer antibody (reported to be specific for serine 2 phosphorylation). Antibody 4H8 is stated to react with all forms of CTD, but the chromatin immunoprecipitation studies reported here suggest that it may inefficiently recognize serine 2–phosphorylated polymerase during transcript elongation.
Unphosphorylated PolII typically showed a peak of DNA binding that overlaps the site of transcription initiation found at the center of the prominent biphasic peaks of histone 3 lysine 4 di- and trimethylation that occur flanking the transcription initiation site. In addition, in a few actively transcribed genes unphosphorylated PolII signals occurred throughout the gene body, and in many cases there was a peak of signal coinciding with the 3′ termination site of RNA polymerase binding, as estimated by comparison of patterns with each of the antibodies. Furthermore, there was a curious unevenness or lumpiness of signals across many genes that did not necessarily correlate closely with the positions of known exons. Similar signals were seen in each of three biological replicate samples and in analyses performed on different cell types and by either of two investigators in our group (J.L. and S.H.) using either the same antibody or a second antibody directed against unphosphorylated RNA polymerase CTD. The signals for serine 5 phosphorylated PolII were usually confined to a region of 1–2 kb overlapping and extending slightly downward from the transcription initiation site. Of note, in most genes there was no significant signal from this antibody across the bulk of the transcribed region. The signals for serine 2 RNA polymerase were distributed in a complementary fashion, being weak or absent at the initiation site and stronger within a couple of kilobases downstream from the initiation site. When present, significant signals extended (e.g., see Fig. 5) for an average distance of about 2 kb downstream from the poly(A) addition site but with considerable variability between genes. Conversely, there were several genes in which the histone modification could be detected in the absence of detectable signals for phosphorylated RNA polymerase. As with the signals for unphosphorylated RNA polymerase, the intensity of the signals for serine 2-phosphorylated RNA polymerase was irregular across the transcribed regions of genes, the pattern of intensity did not always match that of histone 3 lysine 36 methylation, and there was frequently a prominently increased signal that began well after the poly(A) addition site and immediately preceded the region where all PolII signals disappeared. The latter is strongly suggestive of RNA polymerase pausing prior to release (Birse et al. 1997). The alternative possibility that this signal represents a second initiation site for PolII is unlikely because there was no mark of initiation of transcription such as trimethyl or dimethyl histone 3 lysine 4, serine 5 phosphorylated PolII, or excess of unphosphorylated PolII.
Except at the site of transcription initiation, there was commonly a correlation between signals for serine 2 phosphorylated and unphosphorylated RNA polymerase. One caveat in interpreting the data is that, because the CTD contains 52 copies of the heptapeptide motif in which phosphorylation occurs, we cannot distinguish between single molecules of RNA polymerase large subunit with partially phosphorylated CTDs and the presence of two populations of molecules, one completely phosphorylated and the other unphosphorylated. This makes it particularly difficult to judge whether or to what extent dephosphorylation necessarily occurs prior to RNA polymerase release.
To study the relationship between histone 3 lysine 36 methylation and the presence of PolII at the 3′ end of transcription units, we first had to exclude genes where: (1) there was no signal for PolII and/or for lysine 36 methylation; (2) there were large blocks of repetitive sequences downstream from the poly(A) addition site; (3) there was another gene whose 3′ end was within 4 kb or whose 5′ end was within 3 kb of the gene of interest; and (4) the polyadenylation site was within 2 kb of the end of the ENCODE region.
These criteria were used to eliminate genes that could not be evaluated. Forty-five genes were not excluded, and these were manually inspected. For 35 of these genes, it was possible to compare the furthest extent of lysine 36 methylation to the presence of PolII. For 23 of these genes, the polymerase clearly extended beyond (3′ to) the region marked by lysine 36 trimethylation. In 10 of the remaining genes the result was ambiguous, and in 12 genes lysine 36 trimethylation and PolII binding disappeared at the same position. No case showed lysine 36 methylation extending downstream beyond regions associated with PolII.
Discussion
Characterization of the structure and function of RNA transcripts in mammalian cells has become a large and active area of research. Approaches that have been used include hybridization of RNA to genomic tiling arrays and extensive sequencing of ESTs or of short 3′ end tags. The extent of the transcriptionally active genome seems to regularly expand the more sensitive the detection method or the more exhaustive the analysis.
The approach described herein has considerable sensitivity, as it detects evidence for more transcripts from known genes than are reported as present by the relatively sparse 35-mer oligonucleotide arrays used commonly for cDNA detection and quantitation. With a fairly stringent threshold we detected a large number of transcripts with 3′ ends in introns or extragenic regions. Most of these had hallmarks of sites of initiation of reverse transcription by oligo(dT), including appropriately located upstream poly(A) addition signals and/or A-rich tracks in DNA at one edge of the positive signal. The putative ends lacking these signatures could have derived from cross-hybridization, fragments linked to poly(A) fragments through splicing, poly(A) sites located within repetitive sequences not represented on the arrays, polyadenylation at variant or alternative poly(A) signals (Beaudoing et al. 2000; Venkataraman et al. 2005; Cheng et al. 2006), or mispriming during PCR amplification. Polyadenylation has also been suggested to occur on RNA fragments derived from transcription extending beyond the polyadenylation site on known genes, but most of these would fall within the 2.5-kb region downstream from known polyadenylation sites that were scored as being associated with known genes in our analyses. It is curious that a substantial fraction of the 3′ ends that did not match known genes were apparently cell type specific.
Poly(A) sites corresponding to the 3′ ends of transcripts of known genes are generally presumed to correspond to one or more splice variants of the coding regions of the gene (Le Texier et al. 2006). However, recent evidence shows that a much more complex situation may exist. Hybridization of 5′ RACE products to genomic tiling arrays have indicated the occurrence of many transcripts whose 5′ end lies far upstream of known promoters for a gene (Kapranov et al. 2005). In one extensively studied and informative situation, spliced transcripts of the beta globin locus have been identified that originate in sequences far upstream of the coding portions of the gene (Xiang et al. 2006). Analysis of some of these transcripts has shown that they can arise from a repetitive element lying more than 200 kb upstream of the embryonic epsilon globin gene and be spliced into multiple forms. These apparently noncoding RNAs are spliced into the second exon of the globin gene and thereby give rise to transcripts that will have identical 3′ end sequences to those of the globin coding mRNA.
Polyadenylated noncoding RNA transcripts have become increasingly interesting as their prevalence has become more appreciated (Goodrich and Kugel 2006; Mattick and Makunin 2006; Pang et al. 2006; Kapranov et al. 2007a, b; Pauler et al. 2007; Prasanth and Spector 2007; Roma et al. 2007). These transcripts include precursors of small regulatory RNAs but also are related to a number of other processes. Some may act in cis to regulate X chromosome silencing or autosomal imprinting, whereas others act in trans, either as activators or inhibitors of certain transcription factors. Transcription extending from upstream and across known genes may inhibit expression of the gene without regard to the structure of the noncoding RNA (De Gobbi et al. 2006).
Antisense transcripts arising within known genes could represent intronic genes, but antisense intronic transcription is a more general phenomenon (De Gobbi et al. 2006; Hayashizaki and Carninci 2006; Kapranov et al. 2007a). For example, Rinn et al. (2003) used synthetic oligonucleotide probes directed against previously undescribed intronic transcripts that had been detected with a PCR fragment array tiling chromosome 22; 10 of the 25 oligonucleotide pairs detected antisense intronic transcription. In the present experiments, of those intronic transcripts whose termini lay downstream from a polyadenylation signal, about 40% were oriented in an antisense direction compared to the embedding gene. These antisense RNAs may be important in gene regulation, such as in the parental origin-specific silencing of imprinted genes (Pauler et al. 2007). In another particularly informative case, Hoogenkamp and colleagues found two antisense transcripts originating in introns of the gene for the transcription factor PU.1, a transcription factor important for determining lineage differentiation in hematopoietic development. These transcripts were found in the cytoplasm, and down-regulation of the transcripts with siRNA led to up-regulation of PU.1 mRNA (Ebraliddze and Tenen 2006). Studies such as these suggest that antisense intronic transcripts may represent a rather common mechanism for regulating the level of expression of specific genes.
Intronic transcripts with 3′ sequences in the sense orientation with respect to known genes are of uncertain significance. They could hypothetically arise from incompletely spliced mRNA or retained introns that include A-rich sequences. Alternatively, these transcripts could represent novel RNAs that extend across all or portions of known genes. Transcription from upstream sites has been observed to either be required for (Ho et al. 2006; Zhao et al. 2006) or to inhibit transcription of nearby genes (Kelley and Kuroda 2000; Yang and Kuroda 2007) so that such RNAs might also have modulatory roles in the control of gene expression.
The current model for changes in PolII CTD phosphorylation during transcriptions is that PolII with an unphosphorylated CTD is bound to the initiation complex. The RNA polymerase then is phosphorylated on CTD serine 5 by kinases associated with the general transcription factor TFIIH. As RNA polymerase traverses the gene, it becomes phosphorylated at serine 2 by CDK9 or related kinases. This phosphorylated RNA polymerase then binds KMT3, an enzyme that methylates lysine 36 of histone 3. RNA polymerase continues transcription for some distance beyond the poly(A) addition signal before it is released from the DNA. The present data provide more specificity to several aspects of this model as well as show phenomenological heterogeneity between genes.
The pattern of histone 3 lysine 36 trimethylation in the majority of genes is relatively simple. This modification begins some distance downstream from the transcription initiation site and continues across the gene template. The modification decreases in what is sometimes more of a stepwise than a continuous fashion near or beyond the end of the gene, often near the site of cleavage and polyadenylation, and some distance upstream of the furthest regions where RNA polymerase serine 2 phosphorylation can be detected. This is consistent with models for multiple transcription termination signals in the DNA that are not closely linked to the site of polyadenylation (West et al. 2006), in addition to effects of the polyadenylation signals on RNA polymerase pausing or termination. They also indicate that either demethylation becomes more active or KMT3 becomes less active prior to removal of the RNA polymerase from the DNA template.
In some genes, principally some of the less actively transcribed genes, histone 3 lysine 36 methylation is seen without RNA polymerase serine 2 phosphorylation. This presumably represents the longer persistence of histone methylation in comparison to the dwell time of phosphorylated RNA polymerase. On the other hand, there were very few examples where substantial serine 2 phosphorylation of RNA polymerase was detected in the absence of histone 3 lysine 36 methylation in the body of the gene, consistent with the direct relationship between the phosphorylation and the association of KMT3 with DNA. In one class of exceptional genes, histone 3 lysine 4 dimethylation was found across much or all of the body of the gene. This is reminiscent of studies in yeast in which inactivation of a kinase thought to be responsible for serine 2 phosphorylation of PolII resulted in spreading of histone 3 lysine 4 di-/trimethylation into the body of genes.
There is a certain analogy between the distribution of di- versus trimethylation of histone 3 lysine 4 and lysine 36, although they are largely nonoverlapping in their location. In both cases, the trimethylated form of the modified histone is more narrowly distributed. In particular, histone 3 trimethylated lysine 4 is generally limited to the region proximate to the transcription initiation site methylase(s) (Santos-Rosa et al. 2002; Schneider et al. 2004; Bernstein et al. 2005; Barski et al. 2007; Koch et al. 2007). Of possible relevance, specific demethylases for histone 3 trimethylated lysine 4 have been identified recently (Liang et al. 2007; Secombe et al. 2007). Lysine 36 trimethylation is generally limited to the body of the gene, presumably reflecting recruitment of KMT3 by CTD serine 2 phosphorylated RNA polymerase. On the other hand, both dimethyl lysine 4 and dimethyl lysine 36 also occur in large blocks of chromatin not necessarily connected to sites where trimethylation occurs, and the functional significance of these blocks is currently under investigation.
Interpretations of the presence and phosphorylation state of RNA polymerase are somewhat complicated by potential limitations in the specificity of the antibodies. Nevertheless, there is a clear distinction in patterns detected with the antibodies directed against serine 2, serine 5, and unphosphorylated RNA polymerase, with most of the regions detected by the serine 2 antibody not showing any reaction with serine 5. The serine 5 antibody generally showed a clear peak at or near the transcription start site where reactivity with serine 2 antibody was low or absent. Serine 5 signals were commonly absent over most of the body of the gene. When present, they were often weak and did not correlate with the signals from serine 2 phosphorylated RNA polymerase or unphosphorylated RNA polymerase. This could occur because of dephosphorylation of serine 5 in the body of the gene, because of presence of serine 5 phosphorylation at only a limited subset of potential sites, or because of blocking of the doubly phosphorylated sites on the enzyme by tight association with other proteins, such as KMT3. While the first is the simplest explanation for the complete lack of reactivity of progressing RNA polymerase to serine 5 antibody, other possibilities cannot be excluded.
The antibody directed against dephosphorylated RNA polymerase is at least partially specific as demonstrated by the lack of correlation with serine 2 detection at the origin and incomplete correlation with serine 5 detection in the body of the gene. The pattern of intensities of dephosphorylated or total RNA polymerase detection across genes sometimes showed regions of strong signal sometimes alternating with regions of extremely weak signal. Similar patterns may be seen with the serine 2 directed antibody so that the pattern does not merely reflect a change in the ratio of phosphorylated to dephosphorylated enzyme. Presumably these observations reflect differences in the rate of progression of RNA polymerase through the gene. While regions of RNA polymerase density sometimes correlate with the presence of exons (Brodsky et al. 2005), this is often not the case in the present studies. RNA polymerase slowing or pausing has been implicated both in the generation of alternate spliced forms of mRNA and in transcription termination. The accumulation of RNA polymerase at some internal regions of genes is prominent, and the basis and significance of the differences in progression rate of the RNA polymerase remains a subject for further investigation.
Another recurrent feature of the distribution of different phosphorylation states of RNA polymerase is the accumulation of dephosphorylated RNA polymerase at the 3′ end of the transcription unit, often concurrent with the accumulation of serine 2 RNA polymerase. This suggests both a pile-up of RNA polymerase molecules preceding the point of release and also the possibility that RNA polymerase may become dephosphorylated prior to or concurrent with release from the DNA. A third observation with respect to dephosphorylated RNA polymerase is that, in some very active genes, strong signals for this form of the enzyme are detected across the entire body of the gene, relative to the signals for serine 2 phosphorylated enzyme. Perhaps when the gene is densely populated with RNA polymerases, the unphosphorylated enzyme can progress through the body without modification, or dephosphorylation occurs secondary to crowding of RNA polymerase molecules on DNA. Finally, the description of CTD as either phosphorylated at serine 2, serine 5, or both serines 2 and 5 is a major simplification of the potential for alternative sites or levels of phosphorylation and ignores potential conformational changes in the CTD (Phatnani and Greenleaf 2006).
In summary, sensitive detection of polyadenylation sites in cellular RNA has confirmed the existence of a number of antisense and intergenic transcripts as well as more transcripts from known genes than are detectable by some of the standard approaches. Combining this data with ChIP–chip data has provided a more detailed picture of the relationship between RNA polymerase occupation of DNA and phosphorylation in relation to the 3′ ends of mature transcripts as well as revealing gene specific variations in the general pattern. Termination of transcription is a complex process involving several nonsynchronous steps in addition to recognition of transcribed polyadenylation signals in RNA (Nag et al. 2006). Histone 3 lysine36 trimethylation did not always parallel the levels of serine 2 phosphorylated PolII and often diminished near the site of polyadenylation and disappeared before serine 2-phosphorylated PolII CTD disappeared.
Near the 3′ end of transcription units there frequently was an increased concentration of both serine 2 phosphorylated and unphosphorylated RNA polymerase, suggesting that polymerase progression slowed or paused prior to release from the DNA and that dephosphorylation of the RNA polymerase CTD may precede polymerase release from the DNA.
Methods
Sample preparation
Four cell lines as well as normal human neutrophils were used in this study. In general, three biological replicates were used for the analyses. Cervical adenocarcinoma (HeLa-S3) total RNA was ordered from Ambion. Acute promyelocytic leukemia (APL) cell line NB4 (Lanotte et al. 1991) was used directly or cultured with all-trans-retinoic acid (RA) for 48 h to differentiate cells to neutrophils (Khanna-Gupta et al. 1994). Human B-lymphocyte cell line GM06990 was obtained from Coriell Cell Repositories. Human erythroblast cell line K562 (Migliaccio et al. 2002) was obtained from American Type Culture Collection (ATCC). The cell lines were cultured by their respective standard protocols. Human primary neutrophil cells (Tsukahara et al. 2003) were prepared from healthy donors as previously described (Subrahmanyam et al. 2001). The total RNA was extracted and purified with TRIzol reagents (Invitrogen Corp.) and QIA Quick PCR Purification Kit (QIAGEN). An aliquot of the RNA obtained from each cell preparation was analyzed by electrophoresis on a formaldehyde agarose gel, to ensure sample quality and integrity based on the relative intensity of the 28S and 18S ribosomal RNA bands (requiring a ratio of 1.7:1).
cDNA synthesis
To 10 μg of total RNA in 10 μL of water in a 0.5-mL “no-stick” microcentrifuge tube (USA Scientific) we added 1 μL (200 ng) of two-base anchored oligo(dT) primer with a heel: 5′-TAGAAGCCGAGACGTCGGTCG-T(18)NN-3′, N = A, C, G, T. The contents were mixed on ice, heated to 65°C for 5 min, and chilled on ice for 5 min. This denaturation and annealing step was repeated, and the tubes were held on ice. The first-strand cDNA synthesis reaction was set up as follows: 4 μL of 5× first-strand buffer, 1 μL of 0.1 M DTT, and 1 μL of RNase inhibitor (40 U/μL) were mixed and warmed to 45°C. cDNA synthesis was initiated by adding 1 μL (200 U) of SuperScript II, RNase H-reverse transcriptase (Invitrogen), and incubation continued (at this stage the final reaction volume was 20 μL) at 45°C for 1 h. This step was performed in a humidified incubator instead of in a water bath to avoid evaporation.
After first strand synthesis, the tube was chilled on ice and centrifuged briefly to collect all the contents, and second-strand synthesis reaction was set up on ice in the same tube as follows: 20 μL of first strand reaction, 91 μL of water, 30 μL of 5× second strand buffer, 3 μL of 10 mM dNTPs, 4 μL of Escherichia coli DNA polymerase I (10 U/μL), 1 μL of E. coli DNA ligase (10 U/μL), and 1 μL of RNase H (3 U/μL). The total volume of the reaction was 150 μL. The tubes were incubated at 16°C for 2 h.
The reaction was stopped by adding 10 μL of 0.5 M EDTA (pH 8.0). The mixture was extracted once with phenol/chloroform (1:1 v/v) and once with chloroform (presaturated with nuclease-free water). cDNA was precipitated by adding 0.5 volume of 7.5 M ammonium acetate and 2.5 volumes of ethanol (−20°C). At this stage, the sample could be left overnight at −20°C. Prior to precipitation of the cDNA, 1 μL (20 μg) of glycogen was added as a carrier.
The cDNA was precipitated by centrifugation in an Eppendorf centrifuge at top speed for 15 min. Without disturbing the pellet, the ethanol was carefully removed. The pellet was washed with 70% ethanol and centrifuged again for 15 min. The supernatant was removed, and the pellet was dried at room temperature. The cDNA was dissolved in 20 μL of water or TE buffer.
cDNA restriction enzyme cutting
To the 20 μL of cDNA prepared described above, 3 μL of Sau3AI buffer (New England Biolabs), 0.3 μL of 10× BSA, 3.7 μL of water, and 3 μL of Sau3AI (10 U/μL) were added, and the sample was gently mixed and incubated for 2 h at 37°C. The enzyme was heat-inactivated for 20 min at 65°C and the tubes were kept on ice.
Butterfly adaptor ligation
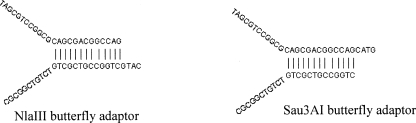
The enzyme-cut fragments were ligated to synthetic Y-shaped adaptors (butterfly adapters) with a complementary overhang as described: We added 4 μL of the butterfly adaptor with an overhang complementary to the ends of the restriction enzyme-digested sample (30 μL), then added 4 μL of 10× T4 DNA ligase buffer and 2 μL of T4 DNA ligase, mixed well, and incubated the sample at 16°C overnight. The enzyme was inactivated at 65°C for 15 min. Below are the sequences and structures of the butterfly adapters for NlaIII and Sau3AI fragments (Scheme 1).
Scheme 1.
PCR amplification of 3′ end fragments (Fig. 1)
The ligated cDNA was used as the template for the 3′ end fragment enrichment at this stage. The PCR mixture (50 μL) consisted of 2 μL of the template, 5 μL of 10× PCR buffer (100 mM Tris HCl at pH 8.3, 500 mM KCl), 2 μL of 15 mM MgCl2, 200 μM dNTPs, 500 nM each of the 5′ and 3′ PCR primers, and 1 U of Platinum Taq DNA Polymerase High-Fidelity system (Invitrogen) and was heated for 2 min to 94°C in the first PCR cycle. PCR consisted of 28–30 cycles of 30 sec at 94°C, 2 min at 56°C , and 30 sec at 72°C.
The following sets of primers were used for PCR amplification of the adapter ligated 3′-end cDNAs: Biotin-TAGAAGCCGAGACGTCGGTCG was used as 3′ primer; while TAGCGTCCGG CGCAGCGAC served as the 5′ primer.
Purification of PCR products
3′ end PCR-amplified products prepared with a biotinylated primer were captured on magnetic porous glass (MPG) particles coated with streptavidin (CPG). A total of 0.1 mg of beads (in 100 μL) were used per reaction. Beads coated with streptavidin (CPG) were blocked by adding 1/10 vol of 40 mg/mL DNA-free tRNA and incubating on ice for 1 h with occasional gentle vortexing. Just before nucleic acid capture, the beads were separated using a magnetic stand, and the supernatant was removed by pipetting. All subsequent capture, washing, and release procedures were performed with the help of a magnetic stand. After the blocking step, the binding reaction was performed by adding the biotinylated PCR products (50 μL) and 8 μL of 3 M NaCl to the sample followed by addition of the beads. The reaction was carried out at room temperature for 30 min with occasional gentle mixing to avoid bead sedimentation. After removal of unbound cDNA, the beads were washed three times with washing buffer (20% glycerol, 10 mM NaCl, 0.2 mM EDTA, 10 mM Tris-HCl at pH 7.5), three times with nuclease-free water containing 50 mg/mL tRNA, and three times with nuclease-free water.
To remove the single-stranded cDNA, alkaline hydrolysis was performed in the presence of 50 μL of Tris-formate buffer (pH 9.0) (obtained by combining 100 mM Tris base with 16.6 mM formic acid and 0.016 mM EDTA, final concentrations) at 65°C for 10 min. After the incubation, 100 μL of 0.5 M Tris-EDTA (pH 7.4) were added before the separation of the liquid phase (containing cDNA) from the beads. The cycle of alkaline elution was repeated three times. The alkaline-treated fractions were removed and pooled together, and single-stranded cDNA was subsequently precipitated with ethanol under standard conditions.
PCR amplification of 3′ end fragments
The 3′ end enrichment fragments released from streptavidin beads were PCR amplified to obtain sufficient material for array analysis, by the PCR protocol described in step 5.
ENCODE arrays
PCR products from each sample were hybridized to NimbleGen’s ENCODE microarray. This array is available via the NimbleGen standard service model to life science researchers. The single array contains more than 384,000 unique 50-mer probes selected from 30 megabases of human sequence data specified by the ENCODE Project Consortium (2007). These probes are spaced apart every 38 bases on the average, thus creating a 12-base overlap between probes. No probes were included for interspersed repetitive DNA, thus there are inevitable gaps in genome tiling paths on the array. Data is presented in comparison to human genome build HG17.
Antibodies, nuclear extracts, and immunoprecipitations
The antibodies used in the present study are listed in Table 3. For each ChIP-chip assay, 1 × 108 cells were cross-linked with formaldehyde at a final concentration of 1% for 10 min followed by addition of glycine in PBS at a final concentration of 125 mM. Cells were collected by centrifugation and washed twice in cold 1× PBS, and nuclear-enriched extracts were prepared as described elsewhere (Bernstein et al. 2005). The lysate was sonicated with a Branson 250 Sonifier to shear the chromatin (output 20%, 100% duty cycle, five 30-sec pulses), and the samples were clarified by centrifugation. Factor–DNA complexes were immunoprecipitated with their unique antibodies overnight at 4°C. Each immunoprecipitation sample was incubated with protein A–agarose (Upstate Biotechnology) for 1 h at 4°C followed by three washes with RIPA buffer and one wash with 1× PBS. The antibody–DNA complexes were eluted from the beads by addition of 1% SDS, 1× TE (10 mM Tris-HCl at pH 7.6, 1 mM EDTA at pH 8), incubation for 10 min at 65°C, addition of 0.67% SDS in 1× TE, incubation for another 10 min at 65°C, and finally gentle vortexing at room temperature for 10 min. The beads were removed by centrifugation, and the supernatants were incubated overnight at 65°C to reverse the cross-linking. To purify the DNA, proteinase K solution (400 μg of proteinase K/mL, 1× TE) was added, and the samples were incubated for 2 h at 45°C, followed by a phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation to recover the DNA. Immunoprecipitated DNA was analyzed by PCR for the presence of enriched factor binding at target sequences. Reactions used 2× Taq Mastermix (QIAGEN) under the following reaction conditions: 5 min at 94°C, 30 cycles of 30 sec at 94°C, 30 sec at 53°C, 30 sec at 72°C, and 10 min at 72°C. PCR products were analyzed by gel electrophoresis.
DNA samples to be hybridized to microarrays were labeled by random priming with nonamer oligonucleotides attached to Cy3 or Cy5 dyes. Control samples for 3′ end cDNA were prepared by labeling total cDNA with Cy3 and controls for the chromatin immunoprecipitation experiments were total genomic DNA prepared from chromatin cross-inked and precipitated by the same procedure as this test sample but with non-specific IgG rather than factor specific antibodies. These controls were also labeled with Cy3. Test samples were labeled with Cy5 and applied to the same chip as the Cy3 labeled control sample.
Bioinformatic analysis
NimbleGen ENCODE tiling arrays with probe length equal to 50 bp and probe separation of 38 bp were used in the current study for analysis of 3′ ends as well as for chromatin immunoprecipitation experiments. Signals from Cy5 and Cy3 channels of the microarray scans were averaged within the moving window of width of 114 bp containing three probes.
Locally weighted linear regression (LOESS) was applied to log-transformed data after averaging of the signals. Cy5 and Cy3 dyes perform differently at different average signal intensities, but LOESS regression compensates for these intensity-dependent effects. Normalization between replicates was performed using quantile normalization. The signal intensities in the Cy5 and Cy3 channels of the replicates were quantile-normalized against Cy5 and Cy3 channels of an arbitrarily chosen array. This procedure forced the signals in each channel to have identical distributions.
After quantile normalization, data from replicates were combined by averaging the signals of the replicates. Signal intensity, yi = log(Ri) − log(Gi), was assigned to each probe, where log(Ri) and log(Gi) are Cy5 and Cy3 channels’ intensities after performing the aforementioned transformations. Contiguous segments (bars) due to the signal coming from the enriched regions were obtained by joining probes with intensities yi above the threshold separated by less than a certain distance (maximum gap = 114 bp, “max-gap”; Kampa et al. 2004). Only segments whose length was greater than a particular size (minimum run = 114 bp; “min-run”) were selected. Analysis of chromatin immunoprecipitation experiments utilized similar procedures with quantile normalization and a window of 1000 base pairs (Zhang et al. 2007). The false-discovery rate (FDR) in our experiments was estimated in the following way. For each data set, the genomic locations of the probes on the microarray were randomly shuffled. The max-gap and min-run procedures described above were applied to the randomized data. The FDR was computed as FDR(threshold) = N1(threshold)/N2(threshold), where N1 is the number of discovered blocks for the randomized data and N2 is the number of blocks for the nonrandomized data. FDR as a function of threshold for the different cell lines data sets is shown in Fig. 3C. GENCODE annotation was used for the all the analysis in this study, and all of our manual analyses are based on the RefSeq gene set.
Acknowledgments
This work was supported by CEGS NIH grant 1P50HG02357-01 and UMASS NIH grant R01 DK54369. We thank Janet Hernandez, who did great editorial work for this manuscript.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.075804.107.
References
- Ahn S.H., Kim M., Buratowski S., Kim M., Buratowski S., Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K., Wang Z., Wei G., Chepelev I., Zhao K., Wei G., Chepelev I., Zhao K., Chepelev I., Zhao K., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Beaudoing E., Freier S., Wyatt J.R., Claverie J.M., Gautheret D., Freier S., Wyatt J.R., Claverie J.M., Gautheret D., Wyatt J.R., Claverie J.M., Gautheret D., Claverie J.M., Gautheret D., Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., Karlsson E.K., Kulbokas E.J., Gingeras T.R., Kulbokas E.J., Gingeras T.R., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Tongprasit W., Samanta M., Weissman S., Samanta M., Weissman S., Weissman S., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Birse C.E., Lee B.A., Hansen K., Proudfoot N.J., Lee B.A., Hansen K., Proudfoot N.J., Hansen K., Proudfoot N.J., Proudfoot N.J. Transcriptional termination signals for RNA polymerase II in fission yeast. EMBO J. 1997;16:3633–3643. doi: 10.1093/emboj/16.12.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky A.S., Meyer C.A., Swinburne I.A., Hall G., Keenan B.J., Liu X.S., Fox E.A., Silver P.A., Meyer C.A., Swinburne I.A., Hall G., Keenan B.J., Liu X.S., Fox E.A., Silver P.A., Swinburne I.A., Hall G., Keenan B.J., Liu X.S., Fox E.A., Silver P.A., Hall G., Keenan B.J., Liu X.S., Fox E.A., Silver P.A., Keenan B.J., Liu X.S., Fox E.A., Silver P.A., Liu X.S., Fox E.A., Silver P.A., Fox E.A., Silver P.A., Silver P.A. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol. 2005;6:R64. doi: 10.1186/gb-2005-6-8-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. The CTD code. Nat. Struct. Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Carninci P., Hayashizaki Y., Hayashizaki Y. Curr. Opin. Genet. Dev. Vol. 17. 2007. Noncoding RNA transcription beyond annotated genes; pp. 139–144. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., Shia W.J., Anderson S., Yates J., Washburn M.P., Anderson S., Yates J., Washburn M.P., Yates J., Washburn M.P., Washburn M.P., et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Patel S., Long J., Stern D., Tammana H., Helt G., Long J., Stern D., Tammana H., Helt G., Stern D., Tammana H., Helt G., Tammana H., Helt G., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Miura R.M., Tian B., Miura R.M., Tian B., Tian B. Prediction of mRNA polyadenylation sites by support vector machine. Bioinformatics. 2006;22:2320–2325. doi: 10.1093/bioinformatics/btl394. [DOI] [PubMed] [Google Scholar]
- Choi Y.H., Hagedorn C.H., Hagedorn C.H. Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3′ poly(A) end phenotype. Proc. Natl. Acad. Sci. 2003;100:7033–7038. doi: 10.1073/pnas.1232347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbert G.L., Daujat S., Snowden A.W., Erdjument-Bromage H., Hagiwara T., Yamada N., Schneider R., Gregory P.D., Tempst P., Bannister A.J., Daujat S., Snowden A.W., Erdjument-Bromage H., Hagiwara T., Yamada N., Schneider R., Gregory P.D., Tempst P., Bannister A.J., Snowden A.W., Erdjument-Bromage H., Hagiwara T., Yamada N., Schneider R., Gregory P.D., Tempst P., Bannister A.J., Erdjument-Bromage H., Hagiwara T., Yamada N., Schneider R., Gregory P.D., Tempst P., Bannister A.J., Hagiwara T., Yamada N., Schneider R., Gregory P.D., Tempst P., Bannister A.J., Yamada N., Schneider R., Gregory P.D., Tempst P., Bannister A.J., Schneider R., Gregory P.D., Tempst P., Bannister A.J., Gregory P.D., Tempst P., Bannister A.J., Tempst P., Bannister A.J., Bannister A.J., et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- De Gobbi M., Viprakasit V., Hughes J.R., Fisher C., Buckle V.J., Ayyub H., Gibbons R.J., Vernimmen D., Yoshinaga Y., de Jong P., Viprakasit V., Hughes J.R., Fisher C., Buckle V.J., Ayyub H., Gibbons R.J., Vernimmen D., Yoshinaga Y., de Jong P., Hughes J.R., Fisher C., Buckle V.J., Ayyub H., Gibbons R.J., Vernimmen D., Yoshinaga Y., de Jong P., Fisher C., Buckle V.J., Ayyub H., Gibbons R.J., Vernimmen D., Yoshinaga Y., de Jong P., Buckle V.J., Ayyub H., Gibbons R.J., Vernimmen D., Yoshinaga Y., de Jong P., Ayyub H., Gibbons R.J., Vernimmen D., Yoshinaga Y., de Jong P., Gibbons R.J., Vernimmen D., Yoshinaga Y., de Jong P., Vernimmen D., Yoshinaga Y., de Jong P., Yoshinaga Y., de Jong P., de Jong P., et al. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312:1215–1217. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]
- Denoeud F., Kapranov P., Ucla C., Frankish A., Castelo R., Drenkow J., Lagarde J., Alioto T., Manzano C., Chrast J., Kapranov P., Ucla C., Frankish A., Castelo R., Drenkow J., Lagarde J., Alioto T., Manzano C., Chrast J., Ucla C., Frankish A., Castelo R., Drenkow J., Lagarde J., Alioto T., Manzano C., Chrast J., Frankish A., Castelo R., Drenkow J., Lagarde J., Alioto T., Manzano C., Chrast J., Castelo R., Drenkow J., Lagarde J., Alioto T., Manzano C., Chrast J., Drenkow J., Lagarde J., Alioto T., Manzano C., Chrast J., Lagarde J., Alioto T., Manzano C., Chrast J., Alioto T., Manzano C., Chrast J., Manzano C., Chrast J., Chrast J., et al. Prominent use of distal 5′ transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res. 2007;17:746–759. doi: 10.1101/gr.5660607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebraliddze A., Tenen D.G., Tenen D.G. Regulation of the PU.1 gene by sense and functional antisense RNAs generated through the same chromatin architecture. Blood. 2006;108 (Suppl.):234a. [Google Scholar]
- The ENCODE Project Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuno M., Pang K.C., Ninomiya N., Fukuda S., Frith M.C., Bult C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Pang K.C., Ninomiya N., Fukuda S., Frith M.C., Bult C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Ninomiya N., Fukuda S., Frith M.C., Bult C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Fukuda S., Frith M.C., Bult C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Frith M.C., Bult C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Bult C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Kai C., Kawai J., Carninci P., Hayashizaki Y., Kawai J., Carninci P., Hayashizaki Y., Carninci P., Hayashizaki Y., Hayashizaki Y., et al. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T.R. Origin of phenotypes: Genes and transcripts. Genome Res. 2007;17:682–690. doi: 10.1101/gr.6525007. [DOI] [PubMed] [Google Scholar]
- Goodrich J.A., Kugel J.F., Kugel J.F. Non-coding-RNA regulators of RNA polymerase II transcription. Nat. Rev. Mol. Cell Biol. 2006;7:612–616. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- Hayashizaki Y., Carninci P., Carninci P. Genome network and FANTOM3: Assessing the complexity of the transcriptome. PLoS Genet. 2006;2:e63. doi: 10.1371/journal.pgen.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., Barrera L.O., Van Calcar S., Qu C., Ching K.A., Van Calcar S., Qu C., Ching K.A., Qu C., Ching K.A., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Ho Y., Elefant F., Liebhaber S.A., Cooke N.E., Elefant F., Liebhaber S.A., Cooke N.E., Liebhaber S.A., Cooke N.E., Cooke N.E. Locus control region transcription plays an active role in long-range gene activation. Mol. Cell. 2006;23:365–375. doi: 10.1016/j.molcel.2006.05.041. [DOI] [PubMed] [Google Scholar]
- Hoogenkamp M., Krysinska H., Ingram R., Huang G., Barlow R., Clarke D., Ebralidze A., Zhang P., Tagoh H., Cockerill P.N., Krysinska H., Ingram R., Huang G., Barlow R., Clarke D., Ebralidze A., Zhang P., Tagoh H., Cockerill P.N., Ingram R., Huang G., Barlow R., Clarke D., Ebralidze A., Zhang P., Tagoh H., Cockerill P.N., Huang G., Barlow R., Clarke D., Ebralidze A., Zhang P., Tagoh H., Cockerill P.N., Barlow R., Clarke D., Ebralidze A., Zhang P., Tagoh H., Cockerill P.N., Clarke D., Ebralidze A., Zhang P., Tagoh H., Cockerill P.N., Ebralidze A., Zhang P., Tagoh H., Cockerill P.N., Zhang P., Tagoh H., Cockerill P.N., Tagoh H., Cockerill P.N., Cockerill P.N., et al. The Pu.1 locus is differentially regulated at the level of chromatin structure and noncoding transcription by alternate mechanisms at distinct developmental stages of hematopoiesis. Mol. Cell. Biol. 2007;27::7425–7438. doi: 10.1128/MCB.00905-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A.A., Struhl K., Struhl K. Eaf3 chromodomain interaction with methylated H3–K36 links histone deacetylation to Pol II elongation. Mol. Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Kampa D., Cheng J., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Cheng J., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Piccolboni A., Bekiranov S., Helt G., Bekiranov S., Helt G., Helt G., et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Strausberg R.L., Fodor S.P., Gingeras T.R., Fodor S.P., Gingeras T.R., Gingeras T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Drenkow J., Cheng J., Long J., Helt G., Dike S., Gingeras T.R., Drenkow J., Cheng J., Long J., Helt G., Dike S., Gingeras T.R., Cheng J., Long J., Helt G., Dike S., Gingeras T.R., Long J., Helt G., Dike S., Gingeras T.R., Helt G., Dike S., Gingeras T.R., Dike S., Gingeras T.R., Gingeras T.R. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Willingham A.T., Gingeras T.R., Willingham A.T., Gingeras T.R., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007a;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermüller J., Hofacker I.L., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermüller J., Hofacker I.L., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermüller J., Hofacker I.L., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermüller J., Hofacker I.L., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermüller J., Hofacker I.L., Willingham A.T., Stadler P.F., Hertel J., Hackermüller J., Hofacker I.L., Stadler P.F., Hertel J., Hackermüller J., Hofacker I.L., Hertel J., Hackermüller J., Hofacker I.L., Hackermüller J., Hofacker I.L., Hofacker I.L., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007b;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kelley R.L., Kuroda M.I., Kuroda M.I. Noncoding RNA genes in dosage compensation and imprinting. Cell. 2000;103:9–12. doi: 10.1016/s0092-8674(00)00099-4. [DOI] [PubMed] [Google Scholar]
- Keogh M.C., Kurdistani S.K., Morris S.A., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J., Kurdistani S.K., Morris S.A., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J., Morris S.A., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J., Schuldiner M., Chin K., Punna T., Thompson N.J., Chin K., Punna T., Thompson N.J., Punna T., Thompson N.J., Thompson N.J., et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Khanna-Gupta A., Kolibaba K., Zibello T.A., Berliner N., Kolibaba K., Zibello T.A., Berliner N., Zibello T.A., Berliner N., Berliner N. NB4 cells show bilineage potential and an aberrant pattern of neutrophil secondary granule protein gene expression. Blood. 1994;84:294–302. [PubMed] [Google Scholar]
- Kizer K.O., Phatnani H.P., Shibata Y., Hall H., Greenleaf A.L., Strahl B.D., Phatnani H.P., Shibata Y., Hall H., Greenleaf A.L., Strahl B.D., Shibata Y., Hall H., Greenleaf A.L., Strahl B.D., Hall H., Greenleaf A.L., Strahl B.D., Greenleaf A.L., Strahl B.D., Strahl B.D. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C.M., Andrews R.M., Flicek P., Dillon S.C., Karaöz U., Clelland G.K., Wilcox S., Beare D.M., Fowler J.C., Couttet P., Andrews R.M., Flicek P., Dillon S.C., Karaöz U., Clelland G.K., Wilcox S., Beare D.M., Fowler J.C., Couttet P., Flicek P., Dillon S.C., Karaöz U., Clelland G.K., Wilcox S., Beare D.M., Fowler J.C., Couttet P., Dillon S.C., Karaöz U., Clelland G.K., Wilcox S., Beare D.M., Fowler J.C., Couttet P., Karaöz U., Clelland G.K., Wilcox S., Beare D.M., Fowler J.C., Couttet P., Clelland G.K., Wilcox S., Beare D.M., Fowler J.C., Couttet P., Wilcox S., Beare D.M., Fowler J.C., Couttet P., Beare D.M., Fowler J.C., Couttet P., Fowler J.C., Couttet P., Couttet P., et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodzius R., Kojima M., Nishiyori H., Nakamura M., Fukuda S., Tagami M., Sasaki D., Imamura K., Kai C., Harbers M., Kojima M., Nishiyori H., Nakamura M., Fukuda S., Tagami M., Sasaki D., Imamura K., Kai C., Harbers M., Nishiyori H., Nakamura M., Fukuda S., Tagami M., Sasaki D., Imamura K., Kai C., Harbers M., Nakamura M., Fukuda S., Tagami M., Sasaki D., Imamura K., Kai C., Harbers M., Fukuda S., Tagami M., Sasaki D., Imamura K., Kai C., Harbers M., Tagami M., Sasaki D., Imamura K., Kai C., Harbers M., Sasaki D., Imamura K., Kai C., Harbers M., Imamura K., Kai C., Harbers M., Kai C., Harbers M., Harbers M., et al. CAGE: Cap Analysis of Gene Expression. Nat. Methods. 2006;3:211–222. doi: 10.1038/nmeth0306-211. [DOI] [PubMed] [Google Scholar]
- Lanotte M., Martin-Thouvenin V., Najman S., Balerini P., Valensi F., Martin-Thouvenin V., Najman S., Balerini P., Valensi F., Najman S., Balerini P., Valensi F., Balerini P., Valensi F., Valensi F., et al. NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3) Blood. 1991;77:1080–1086. [PubMed] [Google Scholar]
- Le Texier V., Riethoven J.J., Kumanduri V., Gopalakrishnan C., Lopez F., Thanaraj T.A., Riethoven J.J., Kumanduri V., Gopalakrishnan C., Lopez F., Thanaraj T.A., Kumanduri V., Gopalakrishnan C., Lopez F., Thanaraj T.A., Gopalakrishnan C., Lopez F., Thanaraj T.A., Lopez F., Thanaraj T.A., Thanaraj T.A. AltTrans: Transcript pattern variants annotated for both alternative splicing and alternative polyadenylation. BMC Bioinformatics. 2006;7:169. doi: 10.1186/1471-2105-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Gogol M., Carey M., Lee D., Seidel C., Workman J.L., Gogol M., Carey M., Lee D., Seidel C., Workman J.L., Carey M., Lee D., Seidel C., Workman J.L., Lee D., Seidel C., Workman J.L., Seidel C., Workman J.L., Workman J.L. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- Liang G., Klose R.J., Gardner K.E., Zhang Y., Klose R.J., Gardner K.E., Zhang Y., Gardner K.E., Zhang Y., Zhang Y. Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nat. Struct. Mol. Biol. 2007;14:243–245. doi: 10.1038/nsmb1204. [DOI] [PubMed] [Google Scholar]
- Maeda N., Kasukawa T., Oyama R., Gough J., Frith M., Engström P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Kasukawa T., Oyama R., Gough J., Frith M., Engström P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Oyama R., Gough J., Frith M., Engström P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Gough J., Frith M., Engström P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Frith M., Engström P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Engström P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Aturaliya R.N., Batalov S., Beisel K.W., Batalov S., Beisel K.W., Beisel K.W., et al. Transcript annotation in FANTOM3: Mouse gene catalog based on physical cDNAs. PLoS Genet. 2006;2:e62. doi: 10.1371/journal.pgen.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K., Sugano S., Sugano S. Oligo-capping: A simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene. 1994;138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- Mattick J.S., Makunin I.V., Makunin I.V. Hum. Mol. Genet. Vol. 15. 2006. Non-coding RNA; pp. R17–R29. [DOI] [PubMed] [Google Scholar]
- Migliaccio G., Di Pietro R., di Giacomo V., Di Baldassarre A., Migliaccio A.R., Maccioni L., Galanello R., Papayannopoulou T., Di Pietro R., di Giacomo V., Di Baldassarre A., Migliaccio A.R., Maccioni L., Galanello R., Papayannopoulou T., di Giacomo V., Di Baldassarre A., Migliaccio A.R., Maccioni L., Galanello R., Papayannopoulou T., Di Baldassarre A., Migliaccio A.R., Maccioni L., Galanello R., Papayannopoulou T., Migliaccio A.R., Maccioni L., Galanello R., Papayannopoulou T., Maccioni L., Galanello R., Papayannopoulou T., Galanello R., Papayannopoulou T., Papayannopoulou T. In vitro mass production of human erythroid cells from the blood of normal donors and of thalassemic patients. Blood Cells Mol. Dis. 2002;28:169–180. doi: 10.1006/bcmd.2002.0502. [DOI] [PubMed] [Google Scholar]
- Morillon A., Karabetsou N., Nair A., Mellor J., Karabetsou N., Nair A., Mellor J., Nair A., Mellor J., Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Moucadel V., Lopez F., Ara T., Benech P., Gautheret D., Lopez F., Ara T., Benech P., Gautheret D., Ara T., Benech P., Gautheret D., Benech P., Gautheret D., Gautheret D. Beyond the 3′ end: Experimental validation of extended transcript isoforms. Nucleic Acids Res. 2007;35:1947–1957. doi: 10.1093/nar/gkm062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., Narsinh K., Kazerouninia A., Martinson H.G., Narsinh K., Kazerouninia A., Martinson H.G., Kazerouninia A., Martinson H.G., Martinson H.G. The conserved AAUAAA hexamer of the poly(A) signal can act alone to trigger a stable decrease in RNA polymerase II transcription velocity. RNA. 2006;12:1534–1544. doi: 10.1261/rna.103206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P., Tan J.J., Ooi H.S., Lee Y.L., Chiu K.P., Fullwood M.J., Srinivasan K.G., Perbost C., Du L., Sung W.K., Tan J.J., Ooi H.S., Lee Y.L., Chiu K.P., Fullwood M.J., Srinivasan K.G., Perbost C., Du L., Sung W.K., Ooi H.S., Lee Y.L., Chiu K.P., Fullwood M.J., Srinivasan K.G., Perbost C., Du L., Sung W.K., Lee Y.L., Chiu K.P., Fullwood M.J., Srinivasan K.G., Perbost C., Du L., Sung W.K., Chiu K.P., Fullwood M.J., Srinivasan K.G., Perbost C., Du L., Sung W.K., Fullwood M.J., Srinivasan K.G., Perbost C., Du L., Sung W.K., Srinivasan K.G., Perbost C., Du L., Sung W.K., Perbost C., Du L., Sung W.K., Du L., Sung W.K., Sung W.K., et al. Multiplex sequencing of paired-end ditags (MS-PET): A strategy for the ultra-high-throughput analysis of transcriptomes and genomes. Nucleic Acids Res. 2006;34:e84. doi: 10.1093/nar/gkl444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim Y.N., Sikes M.L., Beyer A.L., Sikes M.L., Beyer A.L., Beyer A.L. EM visualization of Pol II genes in Drosophila: Most genes terminate without prior 3′ end cleavage of nascent transcripts. Chromosoma. 2002;111:1–12. doi: 10.1007/s00412-002-0183-7. [DOI] [PubMed] [Google Scholar]
- Pang K.C., Frith M.C., Mattick J.S., Frith M.C., Mattick J.S., Mattick J.S. Rapid evolution of noncoding RNAs: Lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Pauler F.M., Koerner M.V., Barlow D.P., Koerner M.V., Barlow D.P., Barlow D.P. Silencing by imprinted noncoding RNAs: Is transcription the answer? Trends Genet. 2007;23:284–292. doi: 10.1016/j.tig.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani H.P., Greenleaf A.L., Greenleaf A.L. Phosphorylation and functions of the RNA polymerase II CTD. Genes & Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Prasanth K.V., Spector D.L., Spector D.L. Eukaryotic regulatory RNAs: An answer to the “genome complexity” conundrum. Genes & Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Prashar Y., Weissman S.M., Weissman S.M. Analysis of differential gene expression by display of 3′ end restriction fragments of cDNAs. Proc. Natl. Acad. Sci. 1996;93:659–663. doi: 10.1073/pnas.93.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J.L., Euskirchen G., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Euskirchen G., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Nelson F.K., Miller P., Gerstein M., Miller P., Gerstein M., Gerstein M. The transcriptional activity of human chromosome 22. Genes & Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roma G., Cobellis G., Claudiani P., Maione F., Cruz P., Tripoli G., Sardiello M., Peluso I., Stupka E., Cobellis G., Claudiani P., Maione F., Cruz P., Tripoli G., Sardiello M., Peluso I., Stupka E., Claudiani P., Maione F., Cruz P., Tripoli G., Sardiello M., Peluso I., Stupka E., Maione F., Cruz P., Tripoli G., Sardiello M., Peluso I., Stupka E., Cruz P., Tripoli G., Sardiello M., Peluso I., Stupka E., Tripoli G., Sardiello M., Peluso I., Stupka E., Sardiello M., Peluso I., Stupka E., Peluso I., Stupka E., Stupka E. Genome Res. Vol. 17. 2007. A novel view of the transcriptome revealed from gene trapping in mouse embryonic stem cells; pp. 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J., Wu J., Lian Z., Nagalakshmi U., Korbel J.O., Kapranov P., Zheng D., Dyke S., Newburger P., Miller P., Wu J., Lian Z., Nagalakshmi U., Korbel J.O., Kapranov P., Zheng D., Dyke S., Newburger P., Miller P., Lian Z., Nagalakshmi U., Korbel J.O., Kapranov P., Zheng D., Dyke S., Newburger P., Miller P., Nagalakshmi U., Korbel J.O., Kapranov P., Zheng D., Dyke S., Newburger P., Miller P., Korbel J.O., Kapranov P., Zheng D., Dyke S., Newburger P., Miller P., Kapranov P., Zheng D., Dyke S., Newburger P., Miller P., Zheng D., Dyke S., Newburger P., Miller P., Dyke S., Newburger P., Miller P., Newburger P., Miller P., Miller P., et al. Novel transcribed regions in the human genome. Cold Spring Harb. Symp. Quant. Biol. 2006;71:111–116. doi: 10.1101/sqb.2006.71.054. [DOI] [PubMed] [Google Scholar]
- Saha S., Sparks A.B., Rago C., Akmaev V., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E., Sparks A.B., Rago C., Akmaev V., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E., Rago C., Akmaev V., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E., Akmaev V., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E., Vogelstein B., Kinzler K.W., Velculescu V.E., Kinzler K.W., Velculescu V.E., Velculescu V.E. Using the transcriptome to annotate the genome. Nat. Biotechnol. 2002;20:508–512. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Schreiber S.L., Mellor J., Kouzarides T., Mellor J., Kouzarides T., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schneider R., Bannister A.J., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T., Bannister A.J., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T., Thorne A.W., Crane-Robinson C., Kouzarides T., Crane-Robinson C., Kouzarides T., Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Secombe J., Li L., Carlos L., Eisenman R.N., Li L., Carlos L., Eisenman R.N., Carlos L., Eisenman R.N., Eisenman R.N. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes & Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamovsky I., Nudler E., Nudler E. Gene control by large noncoding RNAs. Sci. STKE. 2006;2006:pe40. doi: 10.1126/stke.3552006pe40. [DOI] [PubMed] [Google Scholar]
- Sims R.J., Belotserkovskaya R., Reinberg D., Belotserkovskaya R., Reinberg D., Reinberg D. Elongation by RNA polymerase II: The short and long of it. Genes & Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Subrahmanyam Y.V., Yamaga S., Prashar Y., Lee H.H., Hoe N.P., Kluger Y., Gerstein M., Goguen J.D., Newburger P.E., Weissman S.M., Yamaga S., Prashar Y., Lee H.H., Hoe N.P., Kluger Y., Gerstein M., Goguen J.D., Newburger P.E., Weissman S.M., Prashar Y., Lee H.H., Hoe N.P., Kluger Y., Gerstein M., Goguen J.D., Newburger P.E., Weissman S.M., Lee H.H., Hoe N.P., Kluger Y., Gerstein M., Goguen J.D., Newburger P.E., Weissman S.M., Hoe N.P., Kluger Y., Gerstein M., Goguen J.D., Newburger P.E., Weissman S.M., Kluger Y., Gerstein M., Goguen J.D., Newburger P.E., Weissman S.M., Gerstein M., Goguen J.D., Newburger P.E., Weissman S.M., Goguen J.D., Newburger P.E., Weissman S.M., Newburger P.E., Weissman S.M., Weissman S.M. RNA expression patterns change dramatically in human neutrophils exposed to bacteria. Blood. 2001;97:2457–2468. doi: 10.1182/blood.v97.8.2457. [DOI] [PubMed] [Google Scholar]
- Taft R.J., Pheasant M., Mattick J.S., Pheasant M., Mattick J.S., Mattick J.S. The relationship between non–protein-coding DNA and eukaryotic complexity. BioEssays. 2007;29:288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- Tsukahara Y., Lian Z., Zhang X., Whitney C., Kluger Y., Tuck D., Yamaga S., Nakayama Y., Weissman S.M., Newburger P.E., Lian Z., Zhang X., Whitney C., Kluger Y., Tuck D., Yamaga S., Nakayama Y., Weissman S.M., Newburger P.E., Zhang X., Whitney C., Kluger Y., Tuck D., Yamaga S., Nakayama Y., Weissman S.M., Newburger P.E., Whitney C., Kluger Y., Tuck D., Yamaga S., Nakayama Y., Weissman S.M., Newburger P.E., Kluger Y., Tuck D., Yamaga S., Nakayama Y., Weissman S.M., Newburger P.E., Tuck D., Yamaga S., Nakayama Y., Weissman S.M., Newburger P.E., Yamaga S., Nakayama Y., Weissman S.M., Newburger P.E., Nakayama Y., Weissman S.M., Newburger P.E., Weissman S.M., Newburger P.E., Newburger P.E. Gene expression in human neutrophils during activation and priming by bacterial lipopolysaccharide. J. Cell. Biochem. 2003;89:848–861. doi: 10.1002/jcb.10526. [DOI] [PubMed] [Google Scholar]
- Venkataraman K., Brown K.M., Gilmartin G.M., Brown K.M., Gilmartin G.M., Gilmartin G.M. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes & Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C.L., Ng P., Chiu K.P., Wong C.H., Ang C.C., Lipovich L., Liu E.T., Ruan Y., Ng P., Chiu K.P., Wong C.H., Ang C.C., Lipovich L., Liu E.T., Ruan Y., Chiu K.P., Wong C.H., Ang C.C., Lipovich L., Liu E.T., Ruan Y., Wong C.H., Ang C.C., Lipovich L., Liu E.T., Ruan Y., Ang C.C., Lipovich L., Liu E.T., Ruan Y., Lipovich L., Liu E.T., Ruan Y., Liu E.T., Ruan Y., Ruan Y. 5′ Long serial analysis of gene expression (LongSAGE) and 3′ LongSAGE for transcriptome characterization and genome annotation. Proc. Natl. Acad. Sci. 2004;101:11701–11706. doi: 10.1073/pnas.0403514101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S., Zaret K., Proudfoot N.J., Zaret K., Proudfoot N.J., Proudfoot N.J. Transcriptional termination sequences in the mouse serum albumin gene. RNA. 2006;12:655–665. doi: 10.1261/rna.2232406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang P., Fang X., Yin W., Barkess G., Li Q., Fang X., Yin W., Barkess G., Li Q., Yin W., Barkess G., Li Q., Barkess G., Li Q., Li Q. Non-coding transcripts far upstream of the epsilon-globin gene are distinctly expressed in human primary tissues and erythroleukemia cell lines. Biochem. Biophys. Res. Commun. 2006;344:623–630. doi: 10.1016/j.bbrc.2006.03.189. [DOI] [PubMed] [Google Scholar]
- Yang P.K., Kuroda M.I., Kuroda M.I. Noncoding RNAs and intranuclear positioning in monoallelic gene expression. Cell. 2007;128:777–786. doi: 10.1016/j.cell.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Zhang Z.D., Rozowsky J., Lam H.Y., Du J., Snyder M., Gerstein M., Rozowsky J., Lam H.Y., Du J., Snyder M., Gerstein M., Lam H.Y., Du J., Snyder M., Gerstein M., Du J., Snyder M., Gerstein M., Snyder M., Gerstein M., Gerstein M. Tilescope: Online analysis pipeline for high-density tiling microarray data. Genome Biol. 2007;8:R81. doi: 10.1186/gb-2007-8-5-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Kim A., Song S.H., Dean A., Kim A., Song S.H., Dean A., Song S.H., Dean A., Dean A. Enhancer blocking by chicken beta-globin 5′-HS4: Role of enhancer strength and insulator nucleosome depletion. J. Biol. Chem. 2006;281:30573–30580. doi: 10.1074/jbc.M606803200. [DOI] [PubMed] [Google Scholar]