Abstract
Following CD4 receptor binding to the HIV-1 envelope spike (Env), the conserved N-heptad repeat (NHR) region of gp41 forms a coiled-coil that is a precursor to the fusion reaction. Although it has been a target of drug and vaccine design, there are few monoclonal antibody (mAb) tools with which to probe the antigenicity and immunogenicity specifically of the NHR coiled-coil. Here, we have rescued HIV-1-neutralizing anti-NHR mAbs from immune phage display libraries that were prepared (i) from b9 rabbits immunized with a previously described mimetic of the NHR coiled-coil, N35CCG-N13, and (ii) from an HIV-1 infected individual. We describe a rabbit single-chain Fv fragment (scFv), 8K8, and a human Fab, DN9, which specifically recognize NHR coiled-coils that are unoccupied by peptide corresponding to the C-heptad repeat or CHR region of gp41 (e.g. C34). The epitopes of 8K8 and DN9 were found to partially overlap with that of a previously described anti-NHR mAb, IgG D5; however, 8K8 and DN9 were much more specific than D5 for unoccupied NHR trimers. The mAbs, including a whole IgG 8K8 molecule, neutralized primary HIV-1 of clades B and C in a pseudotyped virus assay with comparable, albeit relatively modest potency. Finally, a human Fab T3 and a rabbit serum (both non-neutralizing) were able to block binding of D5 and 8K8 to a gp41 NHR mimetic, respectively, but not the neutralizing activity of these mAbs. We conclude from these results that NHR coiled-coil analogs of HIV-1 gp41 elicit many Abs during natural infection and through immunization, but that due to limited accessibility to the corresponding region on fusogenic gp41 few can neutralize. Caution is therefore required in targeting the NHR for vaccine design. Nevertheless, the mAb panel may be useful as tools for elucidating access restrictions to the NHR of gp41 and in designing potential improvements to mimetics of receptor-activated Env.
Keywords: antibody neutralization, HIV-1 gp41, phage display, fusion intermediate, six-helix bundle, vaccine design, epitope exposure, virus capture
INTRODUCTION
A major goal in HIV-1 vaccine research is to develop an immunogen that can elicit neutralizing antibodies (Abs) against diverse primary isolates of HIV-1 (Klausner et al., 2003; McMichael, 2006; Srivastava, Ulmer, and Barnett, 2005; Zwick and Burton, 2007). HIV-1 neutralizing Abs target the envelope glycoprotein (Env) spikes on the surface of infectious virions, each spike consisting of a relatively labile trimer of gp120-gp41 heterodimers. Env-based immunogens typically elicit only sporadic titers of neutralizing Ab against a limited range of viral isolates (Beddows et al., 2007; Crooks et al., 2007; Grundner et al., 2005; Kim et al., 2003; Law et al., 2007; McBurney, Young, and Ross, 2007; Selvarajah et al., 2005; Yang, Wyatt, and Sodroski, 2001). Such isolate specific neutralization is often associated with Abs against gp120 epitopes that are either variable in sequence (e.g. V1, V2, V3 and V4) or exposure on trimeric Env (Pantophlet and Burton, 2006; Phogat and Wyatt, 2007). To elicit Abs of greater breadth and potency, an Env-based vaccine may need to correctly present conserved sites on Env to the immune system in a stable and immunogenic format.
Conserved sites on Env that are crucial to host receptor recognition or viral fusion are difficult targets for vaccine design. The conserved CD4 binding site on gp120 is recessed amongst more variable elements (Pantophlet and Burton, 2006), and is the target of only one described broadly neutralizing mAb, b12 (Burton et al., 1994; Zhou et al., 2007). The glycan-rich outer face of gp120 is also the target of just one described broadly neutralizing mAb, 2G12 (Calarese et al., 2003; Trkola et al., 1996). V3 and the ‘bridging sheet’ on gp120 constitute the binding site for coreceptor (CCR5 or CXCR4) (Huang et al., 2005), but Abs against these targets are typically weakly neutralizing due to limited epitope exposure on native Env, and steric and kinetic restrictions post-CD4 engagement (Binley et al., 2004; Hartley et al., 2005; Labrijn et al., 2003; Reeves et al., 2005). Gp41 is largely occluded by quaternary interactions within native Env (Chen et al., 2005; Sougrat et al., 2007; Wyatt and Sodroski, 1998; Zanetti et al., 2006; Zhu et al., 2006), although the neutralizing mAbs 2F5, Z13e1 and 4E10 against the membrane-proximal external region (MPER) in the C-terminal portion of the gp41 ectodomain (Muster et al., 1993; Nelson et al., 2007; Stiegler et al., 2001) can bind Env and block a late stage of fusion (Dimitrov et al., 2007; Zwick, 2005). Despite great effort, high neutralizing Ab titers against any of the conserved sites on Env have not yet been elicited by design (Kim et al., 2007; Law et al., 2007; Phogat and Wyatt, 2007).
Post-CD4 engagement, gp41 within Env can rearrange to adopt a transitional “pre-hairpin” state, in which the fusion peptide inserts into the host cell membrane (Eckert and Kim, 2001b; Sattentau, Zolla-Pazner, and Poignard, 1995). After several minutes and activation of Env by coreceptor, gp41 appears to collapse into energetically stable ‘six-helix bundles’ (6HBs), thus driving the fusion of virus and host cell membranes (Eckert and Kim, 2001b; Melikyan et al., 2000). Structures of pre-fusion forms of gp41 are undetermined, but X-ray and NMR structures of the 6HB core of gp41 reveal a trimer of N-heptad repeat (NHR) α-helices surrounded by three C-heptad repeat (CHR) helical peptides in an anti-parallel fashion (Caffrey et al., 1998; Chan et al., 1997; Lu, Blacklow, and Kim, 1995; Weissenhorn et al., 1997). The inner-helical NHR trimer in these 6HB structures has been used to model the receptor-activated state of gp41 and to design several mimetics of the NHR coiled-coil that can potently inhibit fusion. Examples of such NHR trimer mimetics include, NCCG-gp41 (Louis, Bewley, and Clore, 2001), N35CCG-N13 (Louis et al., 2003), IZN36 and similar mimetics (Bianchi et al., 2005; Eckert and Kim, 2001a), as well as “5-helix”, a single chain mimetic in which one CHR peptide of the 6HB is missing so as to expose a long grooved interface of the NHR coiled-coil (Root, Kay, and Kim, 2001).
The CHR peptide, C34 (Chan et al., 1997; Gustchina et al., 2005), as well as certain other fusion inhibitors, including T-20 (Wild et al., 1994), D-peptides (Eckert et al., 1999; Welch et al., 2007) and other peptidomimetics (Jiang et al., 1993; Sia and Kim, 2003) can bind to the NHR coiled-coil and potently inhibit HIV-1 entry into cells. Access of fusion inhibitors to the NHR region however appears to be limited, as CHR peptides that are tethered to cargo proteins tend to lose inhibitory potency with increasing cargo size (Hamburger et al., 2005). Recently, human mAbs have been identified from non-immune Fab phage display libraries that can bind to NHR mimetics, inhibit HIV-1-mediated fusion (Louis et al., 2005) and neutralize various isolates in a pseudotyped assay with modest potency (Gustchina et al., 2007; Miller et al., 2005). An X-ray crystal structure of one mAb, D5, in complex with the gp41-mimetic, 5-Helix, reveals the D5 Fab arm binding orthogonally to the axis of the NHR trimer and into a conserved hydrophobic pocket (Chan, Chutkowski, and Kim, 1998; Luftig et al., 2006). More recently, Fab 3674 has been identified, which is distinguished by its ability to bind tightly not only to the NHR coiled-coil, but also to a 6HB mimetic in which the NHR and CHR regions are connected by a six-residue linker (Gustchina et al., 2007). Despite the interest these fusion inhibitors and mAbs have generated about the conserved NHR region as a potential vaccine lead, the immunogenic potential of the NHR coiled-coil has not been well established.
Here, we rescued rabbit and human mAbs specifically against the NHR coiled-coil of gp41 from immune phage display libraries. One set of phage libraries was prepared from b9 rabbits (Popkov et al., 2003) that had been immunized with the NHR mimetic, N35CCG-N13 (Louis et al., 2003). N35CCG-N13 encompasses residues 546-575 of gp41, presents high-affinity binding sites for up to three CHR peptides (Louis et al., 2003), and includes pocket-forming residues implicated in D5 recognition (Luftig et al., 2006). A previous study showed that a purified fraction of polyclonal rabbit IgG, monospecific for N35CCG-N13, can block HIV-1 Env-mediated cell fusion with considerable potency (IC50 10-50 nM) (Louis et al., 2003). We also prepared a Fab phage library using bone marrow RNA of an HIV-infected individual, FDA2 (Moore et al., 1996; Parren et al., 1998; Vujcic and Quinnan, 1995). Using a unique affinity-selection strategy, we isolated rabbit scFvs (e.g. 8K8), as well as a human Fab, DN9, that can neutralize different primary isolates of HIV-1 in a pseudotyped assay. These neutralizing mAbs do not significantly recognize soluble gp41 or 6HB and thus were not elicited against immunogens structurally related to these proteins. Fab DN9 and mAb 8K8, although currently of limited potency, are nevertheless evidence that neutralizing Abs can be elicited against the NHR trimer alone. These mAbs may be useful in probing the HIV-1 fusion mechanism and pre-screening next-generation immunogens based on the gp41 NHR region.
RESULTS
Identification of anti-NHR mAbs from rabbit scFv and human Fab immune libraries
In an effort to identify neutralizing Abs against the gp41 NHR region, we explored two different approaches. In the first approach, we followed-up on a study that examined rabbit hyperimmune sera raised against the gp41-NHR mimetic, N35CCG-N13 (Louis et al., 2003). N35CCG-N13 is a soluble homotrimer corresponding to the HIV-1 gp41 NHR region residues 546-580 (i.e. SGIVQQQNNLLRAIEAQQHLLQLTVWGIKQ-CCG-RISGIVQQQNNLLRA; in which ‘CCG ’ replaces residues 576-578 for stabilization of the trimer by disulfide formation and the C-terminal residues repeat the first 13 residues). In that study, serum inhibition of HIV-1 was not readily detected, but a purified fraction of the polyclonal IgG that was specific for N35CCG-N13 could potently inhibit HIV-1 fusion (Louis et al., 2003). Thus, two b9 allotype rabbits were similarly hyperimmunized using N35CCG-N13. The b9 rabbits are well suited for scFv phage library preparation as they have a relatively low frequency of unpaired Cys80 in their kappa light chain variable regions, and can be used to generate diverse and high affinity Abs (Popkov et al., 2003). Consistent with the prior study, the resulting antisera did not neutralize the homologous HIV-1 strain, HxB2, (IC50 <1:10 serum dilution), even though the reciprocal endpoint titers against N35CCG-N13 were >105. Bone marrow and spleen were harvested from the immunized b9 rabbits, total RNA isolated, and scFv phage display libraries were prepared (Barbas et al., 2001). Immobilized N35CCG-N13 was used to affinity-select the rabbit scFv libraries, and individual clones were screened for binding activity against N35CCG-N13 and a soluble gp41 ectodomain, r-gp41HxB2. Recombinant r-gp41HxB2 corresponds to residues 541-682 of gp41, is produced in various glycosylated forms in Pichia pastoris, and is commercially available. N35CCG-N13 binders were identified from the rabbit scFv libraries, designated 13K3, R21 and 8K8 (Fig.1A). (The other antigens used in Fig.1B are discussed in a subsequent section.) Only scFv 8K8 had any neutralizing activity against HIV-1HxB2 (see below). None of the other rabbit scFvs neutralized HIV-1HxB2 (IC50s >6.6 μM; data not shown).
Figure 1.
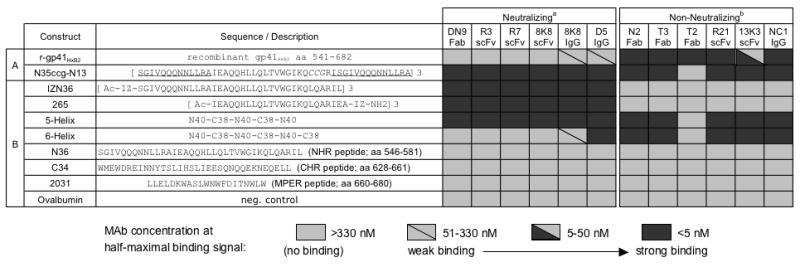
Binding by ELISA of mAbs against a panel of immobilized gp41 mimetic antigens. MAbs were titered in parallel against (A) r-gp41HxB2 and N35CCG-N13, used in the initial mAb screening process, and (B) a panel of gp41 mimetics and peptides. The mAb concentrations at half-maximal binding were determined and are represented using shaded boxes: strong binding (<5 nM, dark), moderate binding (5-50 nM, half dark), weak binding (51-330 nM, light with diagonal line), and no detectable binding (>330 nM, light). MAbs designated as a, ‘Neutralizing’, and b, ‘Non-neutralizing’ were found to neutralize HIV-1HxB2 with an IC50<100 nM, and an IC50>~4 μM in a single-round infectivity assay, respectively (see text).
In a parallel approach to identify anti-NHR specificities, we initially sourced a human Fab phage display library with a modest clonal diversity of ~107, which had been prepared previously using the bone marrow RNA extracted from an HIV-1 seropositive individual (Zwick et al., 2001). The individual, FDA2, has been shown to have relatively broad neutralizing Ab serum titers against primary HIV-1 (Moore et al., 1996; Parren et al., 1998; Vujcic and Quinnan, 1995). This human Fab library yielded several N35CCG-N13 binders, a representative clone of which, N2 Fab, bound very tightly to N35CCG-N13 as well as to r-gp41HxB2 (Fig.1A). We also found that the previously described human Fab T3 (Binley et al., 1996), as well as the murine IgG NC-1 (Dimitrov et al., 2005; Jiang, Lin, and Lu, 1998) bound similarly tightly to both N35CCG-N13 and r-gp41HxB2 (Fig.1A). A control human Fab T2, which binds to a gp41 ‘cluster I’ epitope near to the immunodominant loop, bound r-gp41HxB2 but not the other gp41 mimetics, as expected, due to the absence of the immunodominant loop region in the mimetics. The human Fabs N2, T2 and T3 did not neutralize HIV-1HxB2 (IC50s >4 μM; data not shown) (Binley et al., 1996), and NC-1 has been shown to have no detectable inhibitory activity at 1.3 μM (Binley et al., 1996; Jiang, Lin, and Lu, 1998).
Given the initial screening results, in which several non-neutralizing Abs could bind tightly to both N35CCG-N13 and recombinant r-gp41HxB2 but, in which 8K8 recognized only N35CCG-N13, we subsequently employed a “competitive selection” strategy. Thus, an excess of r-gp41HxB2 was added to the input phage prior to affinity selection against immobilized N35CCG-N13, so as to block the apparent abundance of non-neutralizing mAbs that cross-reacted with r-gp41HxB2. For these selections, a second FDA2 Fab library was prepared that has a high clonal diversity (~1010). With these modifications, two more rabbit scFvs were identified, designated R3 and R7, which have CDR H3 sequences that bear significant sequence homology with 8K8, although their light chains are unrelated (Fig.2). In addition, the larger FDA2 human Fab library (diversity ~1010) now afforded one unique Fab, designated DN9, which bound to N35CCG-N13 and not to recombinant r-gp41HxB2 (Fig.1A). The level of homology to V-region germline genes with human Fab DN9 was found to be within the range observed with the other non-neutralizing human Fabs (Fig.2). Fab DN9 has a long CDR H3 (i.e. 20 residues), as do the non-neutralizing human Fabs (i.e. CDR H3s of Fabs N2, T2 and T3 are 19, 18 and 19 residues, respectively) (Fig.2). In contrast, the rabbit scFvs bear much shorter CDR H3s, and the 8K8 family of scFvs have 7-residue H3s with high sequence homology. The H3s of the non-neutralizing rabbit scFvs R21 and 13K3 were 14 and 12 residues in length, respectively (Fig.2), which is near average for rabbit H3s (i.e. 11.6 residues) (Wu, Johnson, and Kabat, 1993).
Figure 2.

Heavy and light chain variable region amino acid sequences of human and rabbit anti-NHR mAbs used in this study. The germline genes with closest homology for each of the human heavy and light chain variable regions are also shown with %homology reported at the protein level.
Neutralization of pseudotyped HIV-1 by scFv 8K8 and Fab DN9
Because scFvs R3 and R7 share similar heavy chain sequences and binding properties with scFv 8K8 (Figs. 1 and 2) and were further found to neutralize HxB2 and a select few primary isolates with similar potency to scFv 8K8 (data not shown), we focused on scFv 8K8 and Fab DN9. We tested scFv 8K8 and Fab DN9 against a panel of HIV-1 (T-cell line adapted ‘TCLA’ strains and primary isolates) in a single-round infectivity assay using U87.CD4.(CXCR4 or CCR5) target cells (Bjorndal et al., 1997). (Fab DN9 had to be used sparingly due to poor production yields.) The IC50s of scFv 8K8 and Fab DN9 were found to be modest, typically ranging from ~50-500 nM (Table 1), and with no clear pattern of sensitivity based on the NHR sequences of the viruses chosen (sequences not shown). However, HIV-1JR-FL appeared to be completely resistant to scFv 8K8 (IC50 >5 μM). HIV-1JR-FL has a highly unusual Arg at position 564 in the NHR (<0.4% of 562 group M Envs from the Los Alamos database; 11-2-07), whereas all other viruses in the panel have the consensus His at that position. Most of the other viruses chosen from clades B, A and C were at least modestly sensitive to neutralization by the two mAbs, revealing a breadth to their neutralizing activity. Such breadth in neutralizing activity is uncommon among described anti-HIV-1 mAbs and unprecedented for a mAb elicited against an Env mimetic.
TABLE 1.
Neutralization by rabbit scFv 8K8 and human Fab DN9 of pseudotyped clade B, A and C isolates of HIV-1 in a single-round infectivity assay using U87.CD4.(CXCR4 or CCR5) target cells.
| Isolate | Clade | IC50(nM) | |
|---|---|---|---|
| 8K8 scFv | DN9 Fab | ||
| HXB2 | B | 30 | 45 |
| MN | B | 30 | 25 |
| ADA | B | 130 | 60 |
| YU2 | B | 150 | 200 |
| R2 | B | 250 | 200 |
| SF162 | B | 250 | nd |
| JR-FL | B | >5000 | nd |
| JR-CSF | B | 500 | nd |
| 92UG037.8 | A | 600 | >1000 |
| 92RW020.5 | A | 300 | 600 |
| 92BR025.9 | C | 250 | 50 |
| ZM109F.PB4 | C | 300 | 100 |
| ZM53M.PB12 | C | nd | 200 |
| SIVmac239 | >5000 | nd | |
Binding profile of the NHR mAbs to various immobilized gp41 mimetics
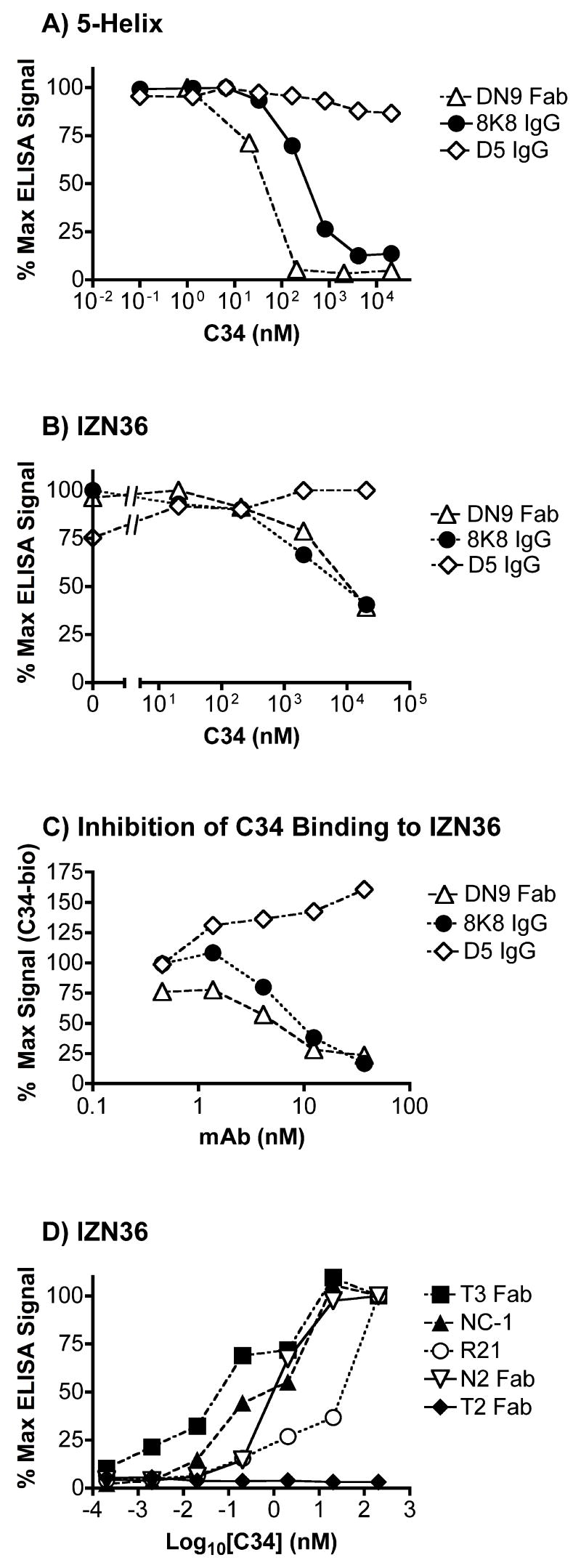
To obtain a broader perspective on the specificities of the recovered NHR mAbs, the mAbs were titered side-by-side against additional gp41 mimetics, including the trimeric NHR coiled-coil mimetic, IZN36 (Eckert and Kim, 2001a); the single-chain 6HB mimetic, 6-Helix, as well as its counterpart that lacks one outer CHR peptide domain, 5-Helix (Root, Kay, and Kim, 2001); the unconstrained linear peptides from the NHR, CHR and MPER regions, N36, C34 and 2031, respectively; and another NHR trimer mimetic that was synthesized for this study, designated 265, which bears residues 559-580 of the gp41 NHR region and therefore lacks the N-terminal 13-residue segment that is repeated twice in N35CCG-N13. Both the neutralizing and non-neutralizing NHR mAbs were found to bind tightly to 5-Helix (Fig.1B). None of the Abs recognized the linear peptides 2031, N36 and C34, indicating a specific discontinuous NHR epitope with each mAb. Interestingly, none of the non-neutralizing mAbs bound to the NHR coiled-coil mimetic IZN36 (Eckert and Kim, 2001a), 265, or the unconstrained N36 peptide, but did bind tightly to N35CCG-N13, indicating a specific conformational requirement with the non-neutralizing Abs for N35CCG-N13 recognition. The non-neutralizing murine mAb NC-1 (Jiang, Lin, and Lu, 1998) had previously been shown to recognize N35CCG-N13 (Dimitrov et al., 2005), and this was confirmed here. The results reveal that 265 is the NHR coiled-coil mimetic with the minimal segment to which only the neutralizing NHR mAbs still bound tightly (Fig.1B). A more detailed study of the binding of the NHR mAbs to 265 and related NHR mimetics is in progress (FMB, PED; manuscript in preparation).
A whole IgG molecule 8K8 was engineered using human constant IgG domains as a scaffold. The chimeric humanized IgG was found to bind very strongly to N35CCG-N13, and has a similar antigen binding profile as its scFv counterpart, as expected (Fig.1). In slight contrast to the scFv 8K8, however, a weak reactivity of IgG 8K8 with recombinant r-gp41HxB2 and 6-Helix at high concentration was observed. We speculate that this reactivity is due to an avidity effect enhancing the binding of the bivalent IgG. IgG 8K8 reactivity with 6-Helix was much weaker than that observed with IgG D5 (half-maximal binding titer was ~150-fold stronger for D5; binding curves not shown), even though both mAbs bound with similarly high titers to IZN36 and 5-Helix (Fig.1B). The scFv 8K8 and Fab DN9 did not bind significantly to r-gp41HxB2 or 6-Helix (Root, Kay, and Kim, 2001), though D5 did cross-react significantly with 6-Helix and somewhat less so with r-gp41HxB2, indicating that 8K8 and DN9 have a strong preference for NHR trimers that are unoccupied by CHR peptide (Fig.1).
IgGs 8K8, D5 and Fab DN9 do not recognize Env in typical binding assay formats
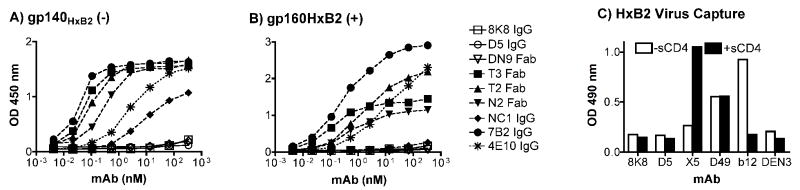
Given the relatively specific binding requirements of 8K8 and DN9 for a trimeric configuration of the NHR region, these mAbs may not bind to common soluble forms of Env that are often used in mAb binding assays. To test this notion, we assayed the ability of IgG 8K8 and Fab DN9 to bind to soluble gp140HxB2(-) in which the furin-cleavage site had been knocked out, as well as to cleavage-competent gp160HxB2(+) that had been detergent-liberated from infectious pseudotyped HIV-1HxB2 virions and captured using immobilized lectin from galanthus nivalis. Neither gp140HxB2(-) or detergent-dissociated gp160HxB2(+) were recognized by 8K8, DN9 or D5, whereas these two Env species were bound tightly by other gp41 mAbs, including Fab T2, Fab T3, Fab N2, 4E10, 7B2 (against the immunodominant loop; (Moore et al., 2006) (Fig.3A and 3B), as well as by 2F5 and D50 (a murine IgG against an immunodominant CHR ‘cluster II’ epitope on gp41; (Earl et al., 1997) (data not shown). NC-1 recognized the gp140HxB2(-) but not gp160HxB2(+); the structural basis for this discrepancy is not clear. The above results indicate that the epitopes of 8K8, DN9 and D5 are occluded with soluble preparations of gp140HxB2(-) and gp160HxB2(+).
Figure 3.
IgG 8K8, Fab DN9 and/or IgG D5 do not recognize HIV-1 Env in typical binding assay formats. Anti-gp41 mAbs were used to probe (A) soluble gp140HxB2(-) in which the furin-cleavage site is knocked out, and (B) cleavage-competent gp160HxB2(+) freshly liberated from infectious HIV-1HxB2 virions using detergent. In both cases, soluble Env was captured in microtiter wells using immobilized lectin from galanthus nivalis, prior to probing the wells with the various mAbs. In addition to the mAbs already described in Fig.1, IgGs 7B2 and 4E10, against the immunodominant loop and MPER of gp41, respectively, were used in the comparison. (C) Immobilized IgG 8K8 and D5 were used in a standard virus capture assay in the presence or absence of 20 μg/ml soluble CD4 (sCD4), and captured virus quantitated using an in-house p24 ELISA using the irrelevant IgG DEN3 as a negative control, and IgGs X5, D49 and b12 as positive control mAbs against the coreceptor binding site on gp120, the immunodominant loop on gp41, and the CD4 binding site on gp120, respectively.
In models of HIV-1 Env fusion, the NHR trimer is not yet formed in the native untriggered form of Env and is available only for a relatively short period of time post receptor engagement. We attempted to capture infectious HIV-1HxB2 whole virions using immobilized IgGs 8K8 and D5 in a standard virus capture assay that measures the p24 content of the captured virions (Nyambi et al., 2000; Poignard et al., 2003), with the expectation that no virus will be captured due to the absence of the NHR trimer epitopes on the virion surface. Indeed, the levels of captured HIV-1HxB2 virions (p24) using immobilized IgGs 8K8 and D5 were no different than that using a negative (irrelevant) control mAb, IgG DEN3, despite efficient virion capture using IgG b12 (against the CD4 binding site of gp120) and IgG D49 (against the immunodominant loop of gp41) (Fig.3C, open bars). The addition of soluble CD4 (sCD4) did not induce capture of HIV-1HxB2 virions by 8K8 and D5, despite significantly enhancing virion capture by IgG X5 (against the coreceptor binding site of gp120) and diminishing capture by IgG b12 (Fig.3C, closed bars). These data show that 8K8 and D5 cannot capture HIV-1 virions in the presence or absence of soluble CD4, indicating that accessibility to their epitopes on virion-associated Env is at least somewhat restricted.
IgGs 8K8, D5 and Fab DN9 bind to overlapping but distinct epitopes on NHR coiled-coil mimetics
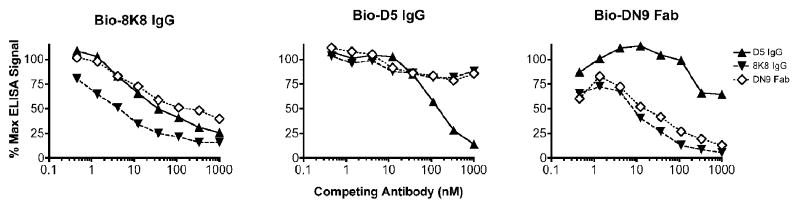
To further explore the binding properties of 8K8, D5 and Fab DN9 we performed competition experiments. We found that D5 was able to block IgG 8K8 binding to 5-Helix (unidirectionally) though not as efficiently as 8K8 inhibited itself (Fig.4, first two panels). Fab DN9 inhibited and was inhibited by 8K8, but it did not block D5 binding to 5-Helix. IgG 8K8 efficiently blocked Fab DN9 binding to 5-Helix, but did not block D5 (Fig.4, right panel). These mAb competition experiments show that the specificities of 8K8 and DN9 overlap somewhat, but are clearly distinct from each other and from D5. Next, we wished to determine the effect of free C34 peptide on mAb recognition of 5-Helix and IZN36. Figures 5A and 5B show that at high concentrations, C34 efficiently blocked recognition of immobilized 5-Helix and IZN36 by Fab DN9 and IgG 8K8, but not by IgG D5. In reverse format, recognition of IZN36 by biotinylated C34 could be blocked using IgG 8K8 and Fab DN9, but not using D5 (Fig.5C). We conclude that the binding sites of 8K8 and DN9 overlap more with that of C34 than does the D5 epitope. We further observed that low nanomolar concentrations of C34 are sufficient to induce recognition of IZN36 by the non-neutralizing mAbs Fab T3, Fab N2, IgG NC-1 and scFv R21 (Fig.5D). These non-neutralizing mAbs had no observable reactivity with C34 alone (Fig.1). The neutralizing mAbs 8K8, DN9 and D5 were therefore unique among the panel of mAbs in their ability to recognize IZN36 in the absence of C34.
Figure 4.
Competition ELISA using IgGs D5 and 8K8, and Fab DN9 against their biotinylated (Bio) counterparts for recognition of immobilized 5-Helix. Bio-8K8 IgG (left panel), Bio-D5 IgG (middle panel), and Bio-DN9 Fab (right panel) at concentrations previously determined to generate non-saturating binding signals were added to wells coated with immobilized 5-Helix that were pretreated with various concentrations of competing mAb. Results are plotted as a percentage of the maximal ELISA binding signal of the Bio-mAb in the absence of competitor.
Figure 5.
The effect of C34 on recognition of NHR mimetics 5-Helix and IZN36 by mAbs in ELISA. (A) C34 efficiently blocks recognition of 5-Helix by IgG 8K8 and Fab DN9, but does not inhibit IgG D5. (B) C34 efficiently blocks recognition of IZN36 by IgG 8K8 and Fab DN9, but does not inhibit IgG D5. (C) IgG 8K8 and Fab DN9, but not IgG D5, can block binding of biotinylated (Bio)-C34 to immobilized IZN36. (D) C34 induces recognition of IZN36 by the non-neutralizing anti-gp41 Abs, Fab T3, Fab N2, murine mAb NC-1, and rabbit scFv R21 at subnanomolar concentrations.
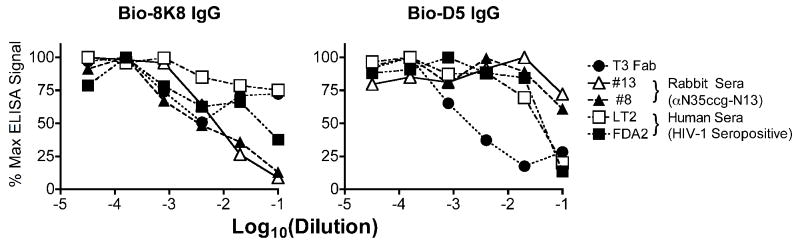
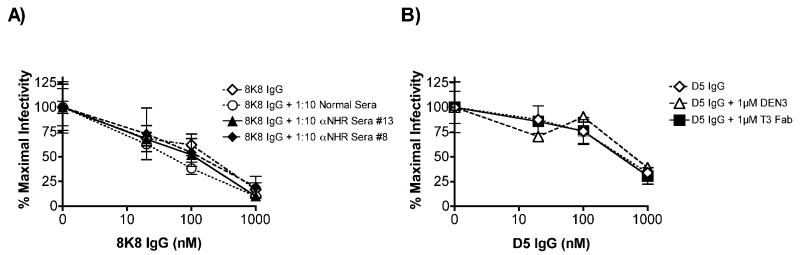
To further distinguish between the epitopes of the neutralizing IgGs 8K8 and D5 and those of other NHR Abs, competition experiments were performed using 5-Helix as the target antigen and with various competitors including, mAbs, rabbit sera against N35CCG-N13, and the neutralizing sera of two HIV-1 seropositive individuals, FDA2 (Moore et al., 1996; Parren et al., 1998; Vujcic and Quinnan, 1995) and LT2 (Dhillon et al., 2007). Of the completely non-neutralizing mAbs, Fab T3 was able to efficiently block D5 but not 8K8 binding to immobilized 5-Helix (Fig.6). In contrast, the non-neutralizing polyclonal rabbit sera could only block 8K8 binding to 5-Helix. Thus, the epitopes of 8K8 and D5 on 5-Helix overlap with epitopes of separate and distinct non-neutralizing anti-NHR Abs. Finally, the FDA2 serum (diluted 1:10) was able to block both IgGs 8K8 and D5, whereas the LT2 serum only blocked D5, indicating a specific difference between the NHR Ab subspecificities in the sera of these two HIV-1 seropositive individuals (Fig.6).
Figure 6.
Binding of IgGs 8K8 and D5 to immobilized 5-Helix can be blocked using non-neutralizing anti-NHR sera or mAb in an ELISA. Biotinylated (Bio)-8K8 IgG (left panel) is inhibited by non-neutralizing αNHR Sera #13 and #8 (drawn from N35CCG-N13-immunized rabbits) and, to a lesser degree, by a neutralizing serum from patient FDA2. Bio-D5 IgG (right panel) is inhibited by non-neutralizing anti-NHR Fab T3, and by neutralizing sera from patients LT2 and FDA2. At the highest concentration, the competing sera are added 1:10, and Fab T3 and the negative control mAb DEN3 are each added at 1 μM, and a 5-fold dilution series is performed.
Neutralization by IgGs 8K8 and D5 is not affected by non-neutralizing anti-NHR Abs
Because the non-neutralizing sera against N35CCG-N13 block 8K8 binding to 5-Helix, one might argue that a preponderance of non-neutralizing Abs in the sera may similarly interfere with the neutralizing activity of 8K8 and related Abs in the same serum. However, the same concentrations of these rabbit sera, and of Fab T3, which efficiently block the binding of 8K8 and D5 to 5-Helix, respectively, have no effect on the neutralization potency of these mAbs against HIV-1HxB2 (Fig.7). These data suggest that the non-neutralizing NHR Abs simply do not recognize the NHR trimer on the authentic gp41 fusion intermediate, even though they bind tightly to overlapping epitopes on NHR peptidomimetics.
Figure 7.
Neutralization of HIV-1HxB2 by IgGs 8K8 and D5 in the presence or absence of non- neutralizing anti-NHR sera or mAb. Virus was pre-incubated for 1 hr with incremental concentrations of IgG 8K8 or IgG D5 in the presence or absence of sera, Fab T3, or negative control IgG DEN3 (constant concentration of 1:10 for the sera and 1 μM for the mAbs, corresponding to the highest concentrations used in Fig.6) and then added to U87.CD4.CXCR4 cells. Luciferase activity was measured after 72 h. (A) IgG 8K8 neutralization in the presence of normal sera (negative control), αNHR sera #13, or αNHR sera #8. (B) IgG D5 neutralization in the presence of IgG DEN3 (negative control) or Fab T3.
Neutralization of HIV-1 (clades B and C) using IgGs 8K8, D5 and 4E10
The neutralizing activity of IgG 8K8 was compared with IgGs D5 and 4E10 against a panel of pseudotyped HIV-1, including TCLA and primary isolates from clades B and C. As expected, all of the HIV-1 isolates are sensitive to neutralization by the broadly neutralizing mAb 4E10, which targets a highly conserved epitope on the MPER of gp41 (IC50s typically ranging from ~3-30 nM), and all except HIV-1JR-FL are at least modestly sensitive to IgG 8K8 (IC50s typically ~200-1000 nM; Table 2). For the eleven HIV-1 isolates that were tested also against D5, seven of these were found to be slightly more sensitive to D5 than to IgG 8K8 (IC50s within a few fold of each other), the exceptions being HIV-1ADA and HIV-1YU2 that were more sensitive to 8K8, and HIV-1JR-FL, which was completely resistant to IgG 8K8 (Table 2). As a whole, these data show that IgG 8K8 has cross-clade HIV-1 neutralizing activity in a single-round infectivity assay, which approximates to the breadth and potency of IgG D5, but is typically 1 or 2 orders of magnitude less potent than IgG 4E10, depending on the isolate.
TABLE 2.
Neutralization of pseudotyped clade B and clade C primary isolates of HIV-1 by IgGs 8K8, D5 and 4E10 in a single-round infectivity assay using U87.CD4.(CXCR4 or CCR5) target cells.
| Isolate | Clade | IC50 (nM) | ||
|---|---|---|---|---|
| 8K8 IgG | D5 IgG | 4E10 IgG | ||
| HXB2 (TCLA) | B | 40 | 30 | <1 |
| ADA | B | 150 | 170 | 3 |
| YU2 | B | 100 | 270 | 4 |
| SF162 | B | 470 | 200 | 3 |
| JR-FL | B | >3300 | 2600 | 30 |
| JR-CSF | B | 800 | 200 | 20 |
| ZM197M.PB7 | C | 1200 | 1000 | 8 |
| Du172 | C | 500 | 200 | 10 |
| CAP45.2.00.G3 | C | 50 | nd | 15 |
| CAP210.2.00.E8 | C | 500 | 500 | 8 |
| ZM214M.PL15 | C | 1000 | 800 | 10 |
| ZM109F.PB4 | C | 400 | 400 | 4 |
| ZM53M.PB12 | C | 1100 | nd | 330 |
| SIVmac239 | >3300 | >3300 | >3300 | |
NHR Ala substitutions that confer resistance or hypersensitivity to neutralization by IgG 8K8
We also performed neutralization assays using IgGs 8K8 and D5 against a series of four mutants of pseudotyped HIV-1LAI, each engineered to bear a single Ala substitution in the NHR region. The fusion inhibitor drug T-20 (enfuvirtide) was also included as a positive control in the comparison. Two of the mutants, H564A and L565A, were chosen based upon the resistant isolate HIV-1JR-FL, which has two uncommon residues at these positions (i.e. R564 and M565). The other two mutants, L568A and K574A, reportedly confer resistance to neutralization by IgG D5 (Miller et al., 2005). As suspected, the H564A substitution made HIV-1LAI resistant specifically to 8K8 (>30-fold increase in IC50), indicating that H564 is critical for 8K8 recognition of the gp41 NHR region. The L565A substitution however made the virus ~10-20-fold more sensitive to 8K8 and D5 as well as T20 (Table 3). The L568A and K574A mutants, which are reportedly 19-fold and 11-fold more resistant to D5 (Miller et al., 2005), were found in our hands to be about >10-fold and 2-fold more resistant to D5, respectively (Table 3). Interestingly, neutralization sensitivity to 8K8 was less affected by these D5 resistance mutations (2-fold change or less). The L565A, L568A and K574A mutants, but not H564A, were slightly more sensitive to T-20, perhaps due to slower fusion kinetics with the former three Ala mutants, as the former mutants, but not the latter, were less infectious than wt HIV-1LAI, and similar mutations reportedly can affect both fusion kinetics and T20 sensitivity (Miller et al., 2005; Reeves et al., 2002). In general, the neutralization sensitivity profiles of 8K8, D5 and T-20 against the four NHR Ala mutant viruses were distinct, suggesting a difference in the binding mechanisms for each Ab/inhibitor.
TABLE 3.
Relative neutralization sensitivities of NHR Ala-mutants of pseudotyped HIV-1LAI to IgG 8K8, T20 and IgG D5 in the single-round infectivity assay using U87.CD4.CXCR4 target cells.
| HIV-1LAI clone | Infectivitya | Fold-resistance [(IC50mutant)/(IC50wt)] | ||
|---|---|---|---|---|
| 8K8 | T20 | D5 | ||
| LAI (wild type) | ++ | 1 | 1 | 1 |
| H564A | ++ | >30 | 1 | 1 |
| L565A | + | 0.05 | 0.1 | 0.05 |
| L568A | + | 0.5 | 0.3 | >10, 19b |
| K574A | + | 1 | 0.2 | 2, 11b |
Infectivity of Ala mutant pseudoviruses relative to parental HIV-1LAI as measured by relative light units (RLU) produced in target cells by viral supernatants. ++, infectivity similar to that of wild-type HIV-1LAI; +, infectivity between≈ 6-20% that of wild-type.
D5 data taken from ref.(Miller et al., 2005) for comparison (performed using the closely-related strain HxB2)
IgGs 8K8 and D5 do not cross react with cardiolipin
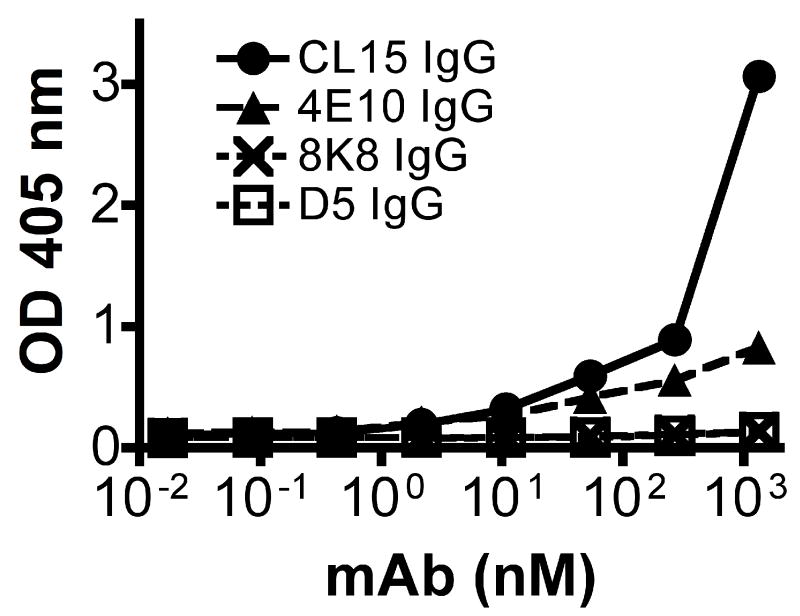
A recent study has suggested that the difficulty in eliciting broadly neutralizing Abs against HIV-1 may be due to down regulation of cognate B-cells that are autoreactive to host antigens, such as cardiolipin (Haynes et al., 2005; Scherer et al., 2007), although this mechanism is controversial (Scherer et al., 2007; Vcelar et al., 2007). We considered whether the apparently low serum titers of 8K8-like antibodies in rabbits that had been immunized with N35CCG-N13 could be due to cross-reactivity of such Abs with cardiolipin. In an ELISA that has been used in a multicenter study for determining cardiolipin cross-reactivity, we observed no binding to cardiolipin with IgGs 8K8 and D5, under conditions in which binding was clearly observed with the previously described cardiolipin cross-reactive mAb CL15 (Hwang et al., 2001), and to a lesser extent 4E10 (Fig.8). Thus, cardiolipin crossreactivity is not an explanation for the poor titers of the 8K8 specificity in the sera of N35CCG-N13-immunized rabbits, nor is it a general phenotype for neutralizing mAbs against the NHR region of gp41.
Figure 8.
MAb binding to cardiolipin using a standardized ELISA format (Harris, Pierangeli, and Birch, 1994). Assayed mAbs include the gp41 IgGs 8K8, D5, 4E10 and a described cardiolipin crossreactive mAb CL15 (Hwang et al., 2001).
DISCUSSION
This study provides mAb evidence of HIV-1 neutralizing specificities generated (i) in animals by immunization using a gp41 NHR mimetic protein, and (ii) in natural HIV-1 infection in humans. The successful identification of neutralizing Abs against the NHR region depended on the use of the stable, helical NHR gp41-mimetic, N35CCG-N13 (Louis et al., 2003). For example, the selection of the human Fab DN9 appeared to require an initial selection round using N35CCG-N13, followed by the competitive selection strategy involving high concentrations of recombinant r-gp41HxB2. In contrast, Fab DN9 was not identified in parallel selections using IZN36, against which Fab DN9 binds very tightly (our unpublished observations). We presume that co-selection of non-neutralizing specificities in the first round of affinity-selection was beneficial for subsequent enrichment of the rare neutralizing Fab DN9 in later competitive selection rounds.
The antigen-binding and mAb-competition experiments using 8K8, DN9 and D5 strongly suggest that the epitopes of 8K8 and DN9 are more closely related to each other than to that of D5. Although D5 has a reported preference to bind NHR mimetics in the absence of CHR peptide (Luftig et al., 2006; Miller et al., 2005), our experiments indicate significant cross-reactivity of D5 with immobilized 6-Helix. Similarly, C34 competes efficiently with 8K8 and DN9 binding to immobilized NHR mimetics (e.g. 5-Helix and IZN36), but not with D5 in our experiments. Recently, a novel Ab, Fab 3764, has been shown to bind to the NHR region with approximately equally high affinity whether free (cf. N35CCG-N13) or interacting with CHR peptide in the form of a 6HB (cf. the minimal stable ectodomain core of gp41 and NCCG-gp41) (Gustchina et al., 2007). Given our observation that C34 also induces non-neutralizing epitopes on IZN36 at very low concentrations, determining the minimal requirements of neutralization activity for NHR mAbs will be required before mimetics of the NHR region that include the CHR region can be considered as vaccine candidates.
The affinity of the NHR region for CHR peptide has been largely attributed to the N-terminal portion of the NHR whereas the affinity of the NHR for itself has been ascribed to the C-terminal portion of the NHR that includes the hydrophobic pocket (Chang and Hsu, 2007; Dwyer et al., 2003). Given the strong competition between 8K8/DN9 and C34, these mAbs might bind N-terminal to Fab 3764, which is unaffected by N-C interactions. We would further argue that DN9 and 8K8 contact two adjacent NHR helices simultaneously, similar to C34. In support of this notion, Western blot analysis revealed that IgG 8K8 recognized only NHR trimer constructs and not the 6HB, and therefore behaved like a ‘class C’ Ab, as defined in an earlier study ((Louis et al., 2005); JML, CAB, GMC, unpublished observations). More precise structural details of the 8K8 and DN9 epitopes can be determined using crystallography to shed light on the observed differences in the functional epitopes amongst our mAbs and Fab 3674 and D5.
HIV-1 natural infection elicits many non-neutralizing mAbs against various epitope clusters on gp41, making it difficult to identify and distinguish these from neutralizing ones. Non-neutralizing anti-gp41 mAbs have been mapped to the disulfide loop region, the NHR region proximal to this loop, and the CHR region adjacent to the MPER (Binley et al., 1996; Earl et al., 1997; Opalka et al., 2004; Xu et al., 1991). The binding of many of these mAbs to gp41 has been shown to depend on the coiled-coil association of the NHR and CHR regions (Chen et al., 2000; Gorny and Zolla-Pazner, 2000; Jiang, Lin, and Lu, 1998; Xu et al., 1991). We also identified non-neutralizing mAbs (rabbit and human) against the NHR trimer mimetic, N35CCG-N13. The epitopes of DN9 and 8K8 are clearly distinct from the non-neutralizing epitopes described above, as DN9 and 8K8 do not bind tightly to soluble r-gp41HxB2, gp140, or gp160, as many non-neutralizing mAbs do. We also found that immobilized IgGs 8K8 and D5 are unable to capture infectious HIV-1 virions in the presence or absence of soluble CD4, in contrast to that observed with many non-neutralizing mAbs to gp41 that bind to gp41 from which gp120 has been shed, or to gp41 within non-functional gp41-gp120 heterodimers (Burrer et al., 2005; Moore et al., 2006; Nyambi et al., 2000; Poignard et al., 2003). The fact that 8K8 binding to 5-Helix is blocked by both DN9 and D5, the latter of which binds to the hydrophobic pocket on the NHR coiled-coil (Luftig et al., 2006), together with the observation that all three mAbs bind to the relatively short NHR trimer mimetic 265, links these neutralizing anti-NHR specificities to a relatively restricted region of the NHR near to the hydrophobic pocket. Although the non-neutralizing Fab T3 can block D5 from binding to 5-Helix, Fab T3 likely lacks neutralizing activity because it cannot bind to fusogenic NHR trimers. Indeed, we found that Fab T3 only recognizes IZN36 if it is pre-complexed with C34, which would in turn, mimic the 6HB (post fusion) form of gp41. Fab T3 has further been shown unable to bind to native Env trimers (i.e. [gp120-gp41]3) in a Blue-native PAGE gel-mobility shift assay (Moore et al., 2006). Further clarification of the precise epitope of Fab T3 may be useful in determining the exact features of the NHR coiled-coil that are accessible to Ab solely on fusogenic gp41, as the T3 epitope is apparently accessible on N35CCG-N13 but not on IZN36. With respect to DN9, it would seem unlikely that the antigen that elicited this specificity in vivo would have been the bona fide fusogenic ‘pre-hairpin’ gp41, due to its apparent limited stability and exposure, but may have been an antigenically related form of gp41, which might have originated on the surface of virus, infected cells, or as soluble debris.
Several possible explanations may account for the inability of many anti-NHR Abs to neutralize HIV-1. (i) Part(s) of the NHR region that are distal to the hydrophobic pocket may be occluded by intermolecular interactions within functional Env, by membrane, and/or by host cell receptors. (ii) The angle of approach of the Abs to their epitope, rather than the precise epitope surface they recognize, may preclude access of anti-NHR Abs to the fusion intermediate. Indeed, post CD4-engagement, steric restrictions to the NHR have been demonstrated previously (Hamburger et al., 2005). (iii) Subtle differences in secondary, tertiary or quaternary structure may exist between the authentic fusion-intermediate and the cognate regions on gp41 NHR mimetics. (iv) Kinetic restrictions may limit the period in which Abs can bind to the fusion intermediate, especially those with slow on-rates and fully reversible binding mechanisms (Dimitrov et al., 2005; Steger and Root, 2006).
The neutralization potency of D5 has been reported to be roughly equivalent, on a molar scale, among its whole IgG, Fab and single-chain Fv (scFv) counterparts (Miller et al., 2005). In contrast, a bivalent construct of Fab 3674 was typically, though not always, more potent than its monovalent counterpart (Gustchina et al., 2007). Other mAbs have been reported to recognize conformational epitopes on gp41 (gp140), in which there does not appear to be a clear and consistent effect on neutralization potency between the Fab and IgG formats (Choudhry et al., 2007; Zhang et al., 2006). Here, we observed that the neutralization potency of scFv 8K8 and its IgG counterpart were, in the main, roughly comparable, though the scFv was occasionally more potent. This finding is not unexpected given the study by Hamburger et al., which demonstrated steric blocks surrounding the NHR coiled-coil that become apparent when C34 is fused with increasingly bulky fusion partners (Hamburger et al., 2005). Fab DN9 is poorly produced in E. coli, which limited its use in assays. Hence, a whole IgG DN9 is under construction, which is expected to produce greater Ab yields and facilitate future structure-function studies with this mAb.
The neutralization potencies of IgGs 8K8 and D5 were broadly similar, although D5 was able to neutralize HIV-1JR-FL, which 8K8 could not. The result with HIV-1JR-FL may be explained by the rare R564 residue in JR-FL, which replaces the more commonly observed H564 that is so critical for 8K8 neutralization as shown by the Ala-scan analysis. We note that the resistance of HIV-1JR-FL to NHR-targeted anti-HIV-1 mAbs and inhibitors has been previously reported, and may relate to the kinetics of fusion, another structural feature of the NHR region in JR-FL, or some combination of these factors (Gustchina et al., 2007; Miller et al., 2005; Welch et al., 2007). An Ala-scan study involving soluble NHR coiled-coils is in progress to determine the relative contributions of different side-chains to anti-NHR mAb recognition (manuscript in preparation, FMB, PED). It is noteworthy that the IC50s with IgG D5 in our study are several-fold lower than those reported in an earlier study using D5 (Miller et al., 2005). The differences in assay sensitivity may be due, at least in part, to differences in coreceptor density on target cells that reportedly can affect the neutralization potency of mAbs and inhibitors, particularly those that may be sensitive to fusion kinetics (Choudhry et al., 2006; Ketas et al., 2007; Reeves et al., 2005). Nevertheless, in both the Merck D5 study and in our own, the MPER mAb controls (i.e. 2F5 and 4E10, respectively) were found to be more potent than the anti-NHR mAbs by ~1-2 orders of magnitude and sometimes more. As we found in a prior study involving weaker neutralization with resistant isolates of HIV-1 (Zwick et al., 2005), the shape of neutralization curves with IgGs D5 and 8K8 can make IC50 determinations more difficult than with more potent mAbs.
There is no doubt that the moderate potency of the described anti-NHR mAbs, being about 1 to 2 orders of magnitude weaker than IgG 4E10, is a major concern with respect to any approach to vaccine design involving this region. Nevertheless, 8K8 and DN9 show modest but relatively broad neutralization that includes isolates from clades B and C, which is an uncommon phenotype. Moreover, the described NHR mAbs may not reflect the potency of all Abs in their class. More potent anti-NHR Abs might exist given that a monospecific fraction of rabbit polyclonal IgG against the NHR mimetic, N35CCG-N13, inhibited fusion more potently than 2G12 (Louis et al., 2003). The present mAb competition data would seem to suggest that the weak neutralization in the serum of the hyper-immunized rabbit is unlikely to be a function of non-neutralizing (or enhancing) Abs counteracting the activity of neutralizing Abs. Rather, our results and the results of Louis et al. (Louis et al., 2003) would suggest that the poor neutralizing activity in the serum is mainly due to relatively low titers of Abs that can recognize the gp41 fusion intermediate.
Finely elucidating the steric and kinetic access restrictions to the NHR region will presumably be beneficial to drug and vaccine design. If the steric restrictions derive from Env itself, these may be opportune surfaces against which other drugs or more potent neutralizing Abs may be sought. Immunogen design must consider the presentation of the NHR region with the permissible geometry of Ab approach, and the close apposition of the target cell membrane prior to fusion. The parameters of binding kinetics can be addressed in part using mutagenesis and affinity optimization of existing mAbs (Steger and Root, 2006). With respect to drug design, the NHR mimetics themselves, while extremely potent (Bianchi et al., 2005; Louis, Bewley, and Clore, 2001; Louis et al., 2003), are very likely to be highly immunogenic in vivo, which may significantly diminish their potential as therapeutics (we found, for example, that non-neutralizing sera against N35CCG-N13 can block the anti-HIV-1 activity of N35CCG-N13; RJ, MBZ, unpublished observations). Thus, the effects of immunogenicity of NHR mimetics on vaccine design and therapeutic design are reciprocal in nature and must be dealt with accordingly. Therapeutically, a small molecule inhibitor of the NHR region would therefore be preferable, providing that sufficient potency and breadth of activity can be reached in vivo.
It is unclear in the present rabbit study whether the apparent immunodominance of non- neutralizing Abs against N35CCG-N13 may have been exacerbated by clonotypic dominance of pre-existing B-cell pools. The absence of detectable cardiolipin reactivity with 8K8 suggests that autoreactivity, at least to cardiolipin (Haynes et al., 2005; Scherer et al., 2007), cannot explain the weak titers of 8K8-like Abs in the rabbits. However, the heavy chain variable regions of the neutralizing scFvs (i.e. 8K8, R3, R7) were clearly clonally related and encoded by a rarely expressed VH gene (x32), whereas those of non-neutralizing scFvs were encoded by the usually expressed VH1a1 (13K3) or VH1a2 (R21). Importantly, our data suggest that if the NHR mimetic does not accurately mimic the fusogenic target on HIV-1, then non-neutralizing anti-NHR Abs may inhibit the elicitation of potential neutralizing Abs to adjacent epitopes. MAbs against both neutralizing and non-neutralizing epitopes on existing NHR mimetics may therefore be useful in pre-screening second generation immunogens in vitro, prior to initiating immunization experiments. In addition, three-dimensional structural information of the mAb-mimetic complexes may be used to better predict the accessibility to the NHR on fusogenic gp41 for prospective next-generation therapeutics.
MATERIALS AND METHODS
Materials
The following reagents were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (ARRRP) with contributors in square brackets: pNL4-3.Luc.R-E- (Connor et al., 1995) [N. Landau], U87.CD4 (CCR5+ or CXCR4+) cells (Bjorndal et al., 1997) [H. Deng and D. Littman], Env complementation plasmids [F. Gao, B. Hahn, L. Morris, D. Montefiori, S.A. Karim, G. Ramjee, Y. Li, J.F. Salazar-Gonzalez, K. Mlisana, E. Hunter and C. Derdeyn], N36 and C34 peptides [DAIDS, NIAID]. The gp160MN-encoding plasmid was kindly provided by J. Moore and S. Beddows (Cornell). All other Env-complementation plasmids have been described: HxB2, YU2, ADA, JR-FL and JR-CSF (Sullivan et al., 1998; Zwick et al., 2003); R2 [G. Quinnan] (Quinnan et al., 1999; Zwick et al., 2003); pCAGGS SF162 gp160 [L. Stamatatos and C. Cheng-Mayer] (Cheng-Mayer et al., 1997; Stamatatos, Lim, and Cheng-Mayer, 2000). The gp41 NHR peptidomimetic and fusion inhibitor IZN36 (Eckert and Kim, 2001a) was kindly provided by D. Eckert (Merck). The plasmids encoding 5-Helix and 6-Helix (Root, Kay, and Kim, 2001) were generously provided by M. Root (TJU); the cognate proteins were produced in E. coli, and purified using Ni-NTA chromatography (Qiagen), according to a described method (Root, Kay, and Kim, 2001), and found to be >95% pure by SDS-PAGE. Human IgG 4E10 (Buchacher et al., 1994) was procured from H. Katinger through the IAVI NAC (R. Pantophlet). IgG D5 (Miller et al., 2005) was a generous gift from M. Miller (Merck). Human Fabs T2 and T3 (Binley et al., 1996) were produced in E. coli and purified using Protein G chromatography. Murine IgG NC-1 (Jiang, Lin, and Lu, 1998) was a kind gift from S. Jiang (NYBC). Anti-cardiolipin antibody CL15 (Hwang et al., 2001) was a generous gift from P. Chen (UCLA) via J.K. Scott. Recombinant r- gp41HxB2 (aa 541-682) was purchased from Viral Therapeutics (Ithaca, NY). Serum from the HIV-1 seropositive individuals, FDA2 (Moore et al., 1996; Parren et al., 1998; Vujcic and Quinnan, 1995) (gift from J. Tavel), and LT2 (Dhillon et al., 2007), were from blood drawn on 2-9-05 and in 2005, respectively.
Immunization of b9 rabbits
An immunization procedure similar to that of Louis et al was followed (Louis et al., 2003). Briefly, two b9 allotype-defined rabbits, (ZZ135-5 and 2ZZ8-5 both heterozygous VH1a1/a2 and homozygous b9k; “13” and “8”), housed at the NIAID rabbit facility (Spring Valley Laboratories, Woodbine, MD), were immunized using 250 μg N35CCG-N13 in complete Freunds adjuvant, and boosted 4 times every 3 weeks with 200 μg N35CCG-N13 in incomplete Freunds adjuvant. The rabbits were sacrificed 5 days after the final boost in which the serum ELISA endpoint titers against immobilized N35CCG-N13 were >1:105, and the spleens and bone marrow harvested. The tissues were immediately placed in TRI reagent (Sigma) and frozen at -80°C.
Preparation of rabbit scFv and human Fab phage libraries
Rabbit libraries
Rabbit single-chain (sc) Fv (VL-VH) libraries were prepared according to a previously described method (Barbas et al., 2001). Briefly, total RNA was prepared separately from the bone marrow and spleen of the two b9 rabbits using the TRI reagent (Sigma). First-strand cDNA was prepared from the total RNA using Superscript III (Invitrogen), and the cDNA used in PCR reactions using Expand High Fidelity polymerase (Roche) and Ab specific primers to amplify Vκ, Vλ and VH genes. Vκ and Vλ PCR products were linked to VH PCR product using splicing overlap extension PCR employing the long linker (‘LL’, 18 amino acids) and short linker (‘SL’, 7 amino acids), respectively. Lambda and kappa libraries for each of the two rabbits were subcloned separately into pComb3X (Barbas et al., 2001), though the 8λ library was discarded due to poor ligation efficiency. Library sizes were between 2-5 × 107 for the 13κ, 13λ and 8κ libraries.
Human Fab FDA2 library
A human Fab (kappa) library was constructed using the bone marrow of an HIV-1 seropositive individual, essentially as described previously (Wild et al., 2003). Briefly, mRNA purified from the bone marrow total RNA was extracted using Oligotex mRNA kit (Qiagen), followed by first strand cDNA synthesis using SuperScript II RT First Strand cDNA Synthesis Kit and oligo (dT) (Invitrogen). Kappa light chains and IgG Fd fragments were amplified, as previously described (Wild et al., 2003). Kappa light chains, followed by the IgG Fd fragments were sequentially cloned into the PAX243 hGK vector. The final FDA IgGκ library size was 1.2 × 1010.
Affinity-selection of scFvs and Fabs from phage libraries
The scFv rabbit and human FDA2 Fab libraries were each minimally subjected to a single round of affinity selection against immobilized N35CCG-N13 in a 96-well plate (Corning) using established methods (Barbas et al., 2001). The output scFv phage were pooled for subsequent rounds. In an initial selection, four rounds of selection against N35CCG-N13 were used to identify scFv 8K8. In follow-up selections, an in-solution competition strategy was used to mask non-neutralizing gp41 epitopes. Thus, rabbit scRvs R3 and R7, as well as a human Fab DN9 were isolated using a single round against N35CCG-N13, followed by four rounds against N35CCG-N13 in the presence of an excess (20 μg/ml) of soluble recombinant r-gp41HxB2 (Viral Therapeutics). Phagemid DNA was prepared from the enriched phage pools and used to transform the non-amber suppressing E. coli strain, Top10 (Invitrogen), for production of soluble scFv and Fab. Individual colonies were picked and screened for reactivity in the crude bacterial culture supernatants against N35CCG-N13 and r-gp41HxB2, and for neutralization of HIV-1HxB2 pseudotyped virus in the single-round infectivity assay. Purification of scFvs and Fabs was achieved using Ni-NTA (Qiagen) and Protein G chromatography, respectively, as previously described (Barbas et al., 2001).
Construction and production of chimeric rabbit-human IgG1 8K8
Phagemid DNA encoding 8K8 (scFv) was used as template for amplification of variable heavy (PCR 1) and light (PCR 2) chain regions, using VH4 / VHrev and VK1 / VKrev primers, respectively (Barbas et al., 2001). Human CH1 (PCR 3; primers VThe humanized Fd and kappa chain DNA segments were sequentially Hfor and HG33rev) and CK (PCR 4; RCSF39 and leadB) were appended to the rabbit variable regions using splicing overlap extension (SOE) PCR and the respective outer primers (SOE 1). subcloned as XbaI-SacI and HindIII-EcoRI fragments, respectively, into a mammalian IgG expression vector, pDR12 (Burton et al., 1994). The pDR12-8K8 DNA was verified by DNA sequencing, and then used with the FuGENE6 transfection reagent (Roche) to transfect Chinese hamster ovary (CHO-K1) cells. Stable transfected cells were expanded using methionine sulfoximine (MSX; Sigma) selection and subjected to limiting dilution. The clone with the highest production of IgG1 8K8, as determined using enzyme-linked immunosorbent assay (ELISA) with cell culture supernatant, was chosen for scale-up in the CellCube System (Corning) and purified by affinity chromatography with protein A (Pharmacia). Purified IgG1 8K8 was concentrated, dialyzed against PBS and sterile filtered (0.2 μm). Ab purity (>95%) and concentration was determined using SDS-PAGE, and using absorbance at 280 nm.
ELISAs
(i) Direct ELISA
Microwells (96-well, half-diameter, high protein binding; Corning) were coated overnight at 4°C with 0.2 μg 5-Helix in 50 μl of PBS. The wells were washed twice using PBS containing 0.05% Tween 20, and blocked with 4% NFDM (non-fat dry milk) in PBS for 1 h at 37°C. Fifty microliters of mAb, diluted in 0.4% NFDM and 0.02% Tween 20, were added to the wells and incubated for 2 h at 37°C. Following incubation, the wells were washed five times and a goat-anti-human (Fab’)2-horseradish peroxidase conjugate (1:1000; Pierce; for human IgGs and Fabs) or rat anti-HA high affinity- horse radish peroxidase conjugate (1:1000; Roche; for the HA-tagged rabbit scFvs) was added to the wells and incubated for 1h. The wells were washed five times and then developed by adding 50 μl of tetramethylbenzidine (TMB) solution (Pierce) according to the manufacturer’s instructions. Wells containing TMB solution were stopped after 10 to 20 min before the maximal signals reached an optical density of 1.8, by adding 50 μl of H2SO4 (0.2 M), and the optical density at 450 nm was read on a microplate reader (Molecular Devices).
(ii) Competition ELISA: mAb inhibition of biotinylated mAbs
Microwells were coated with antigen and blocked as above. Twenty-five microliters of competitor mAb or sera, diluted in PBS containing 0.4% NFDM and 0.02% Tween 20, were added to the wells immediately followed by an equal volume of biotinylated Ab (final concentrations of biotinylated Abs were as follows: for 8K8, 10 ng/ml; for D5, 142 ng/ml; for DN9, 50 ng/ml). Following a 2-h incubation at 37°C, the wells were washed five times, and a streptavidin-horseradish peroxidase conjugate (1:1,000; Jackson ImmunoResearch) was added to the wells and incubated for 1 h. The wells were washed, developed and then read on a microplate reader as described above. Each Ab competitor was tested in at least two separate experiments with similar results.
(iii) Competition ELISA: inhibition of mAbs using C34
Microwells were coated with 5-Helix or IZN36 and blocked as above. Twenty-five microliters of C34, diluted in PBS containing 0.4% NFDM and 0.02% Tween 20 at incremental concentrations, were added to the wells immediately followed by an equal volume of Ab. (Final concentrations of Abs for 5-Helix and IZN36, respectively, were as follows: for 8K8, 80 ng/ml (both antigens); for D5, 200 and 400 ng/ml; for DN9, 2000 and 80 ng/ml). Following a 2-h incubation at 37°C, the wells were washed five times, and a goat anti-human Fab’2-HRP conjugate (1:1000; Jackson ImmunoResearch) was added to the wells and incubated for 1 h. The wells were washed, developed and then read, as described above.
(iv) Competition ELISA: inhibition of biotinylated C34 using mAbs
Microwells were coated with IZN36 and blocked, as above. The wells were washed twice, and 25 μl of competitor mAb diluted in PBS containing 0.4% NFDM and 0.02% Tween 20 were added. In a separate tube, biotinylated C34 (40 nM final concentration) and a streptavidin-peroxidase conjugate (1:1000 final concentration; Jackson ImmunoResearch) were pre-complexed for 1 h at 37°C. Following a brief (15 min) pre-incubation with mAb competitor in the IZN36-containing wells, the pre-complexed bio-C34-streptavidin-peroxidase mixture was added directly to the wells and the mixture incubated for a further 1 hr at 37°C. The wells were washed, developed and then read, as described above.
(v) Gp140HxB2(-) and gp160HxB2(+) sandwich ELISA
To generate gp140HxB2(-), a substitution at the furin cleavage site (i.e. ‘REKR’->’REKS’) was introduced using Quikchange mutagenesis (Stratagene) into the Env complementation vector (pSVIIIexe7pA-HxB2 (Sullivan et al., 1998)). Microwells were coated with 0.25 μg Galanthus nivalis lectin (GNL) and blocked, as above. Wells were washed twice and 50 μl were added of cell culture supernatant containing either uncleaved gp140HxB2(-), or infectious HIV-1HxB2 pseudotyped virus (10-fold concentrated) that was treated with 1% Empigen to dissociate Env, and the plates incubated for 3 h at 37°C. Wells were washed five times and 50 μl of mAb, diluted in PBS containing 0.4% NFDM and 0.05% Tween 20, were added to wells and incubated 2 h at 37°C. Wells were washed five more times and 50 μl goat-anti-human or anti-mouse (Fab’)2-HRP conjugate (1:1000, Jackson) was added and incubated for 45 min at room temperature. Wells were washed five times and developed with TMB solution and read on a microplate reader as described above.
(vi) Cardiolipin ELIS
Cardiolipin ELISAs were performed as previously described (Harris, Pierangeli, and Birch, 1994), using Protocol A (Nelson et al., 2007)
HIV-1 virion capture assay
Pseudotyped HIV-1HxB2 were produced as described below for the neutralization assays. For virus capture, ELISA microtiter wells were coated overnight at 4°C with 5 μg/ml capture mAb in PBS, similar to methods previously described (Moore et al., 2006; Nyambi et al., 2000). The wells were blocked with 4% NFDM in PBS for 1 h at 37°C and then 50 μl of virus was added and incubated for 3 h at 37°C. The wells were washed six times with PBS and captured virus was lysed with 50 μl 1% Empigen in PBS. Lysed virions were added to microwells previously coated with sheep anti-p24 (D7320; Aalto Bioreagents) and incubated for 3 h at room temperature. Wells were washed 5 times and 50 μl of sheep-anti-p24 alkaline phosphatase conjugate (Cliniqa) was added for 1 h at room temperature. Wells were developed using the AMPAK amplification kit (Argene) and read at 490 nm.
HIV-1 neutralization assays
Pseudotyped HIV-1, competent for a single round of infection, were generated by co-transfection of 293T cells using the luciferase reporter plasmid pNL4-3.Luc.R-E- (Connor et al., 1995), an Env complementation vector, and the PEI transfection reagent (Kirschner et al., 2006), as described previously (Zwick et al., 2003). In the case of the LAI Env complementation vector, the plasmid pcDNA3.1/V5-His-TOPO was used to subclone LAI env (PCR amplified using pLAI-2 (Peden, Emerman, and Montagnier, 1991) as template, according to a previously described method (Li et al., 2005)). HIV-1LAI Ala-mutants were generated using Quikchange mutagenesis (Stratagene), and verified by DNA sequencing.
The pseudotyped viruses were assayed for neutralization using U87.CD4.CCR5 cells (or U87.CD4.CXCR4 cells for HxB2, LAI and MN viruses) as target cells (Bjorndal et al., 1997). Abs were added to HIV-1 in 100 μl DMEM (Gibco) containing 10% fetal calf serum, and the virus-Ab mixtures incubated for 1 h at 37°C prior to transferring (1:1 by vol) to the target cells (final vol. 200 μl). In the case of the mAb-serum co-incubation experiments, IgGs 8K8 and D5 were diluted in the presence of a fixed, excess concentration of polyclonal sera (1:10 final dilution) or mAbs (1 μM final concentration), respectively. Following a 72-h incubation at 37°C, the cells were lysed, luciferase reagent (Promega) added, and the luminescence in relative light units (RLUs) was measured using an Orion microplate luminometer (Berthold Detection Systems). The extent of virus neutralization was determined as a percentage reduction of viral infectivity against that of an Ab-free control. All experiments were performed in triplicate.
Acknowledgments
We gratefully thank Barbara Newman, Sarah Church, Meng Wang, Kenix Vo, Linh Tran and Ann Hessell for valuable technical assistance, and Mikhail Popkov and Christoph Rader for technical advice. We thank Michael Miller (IgG D5), Debra Eckert and Peter Kim (IZN peptides), Michael Root (5-Helix and 6-Helix), and Ralph Pantophlet and the IAVI NAC (4E10) for key reagents, and especially thank ‘FDA2’ for the generous donation of a bone marrow sample. We acknowledge support from an AmFAR fellowship and from fellowship #F06-SRI-260 of the California HIV/AIDS Research Program (FMB), the AIDS Targeted Antiviral Program of the Office of the Director of the NIH (CAB, GMC), the Intramural Research Programs of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases (GMC), NIAID (RGM), the Neutralizing Antibody Consortium of the International AIDS Vaccine Initiative, and the NIH, AI 058725 (MBZ), AI 33292 (DRB), GM-46192 (IAW), RO1 AI58763, P01 GM048870 (PED).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barbas CF, 3rd, Burton DR, Scott JK, Silverman GJ. Phage Display: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- Beddows S, Franti M, Dey AK, Kirschner M, Iyer SP, Fisch DC, Ketas T, Yuste E, Desrosiers RC, Klasse PJ, Maddon PJ, Olson WC, Moore JP. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology. 2007;360(2):329–340. doi: 10.1016/j.virol.2006.10.032. [DOI] [PubMed] [Google Scholar]
- Bianchi E, Finotto M, Ingallinella P, Hrin R, Carella AV, Hou XS, Schleif WA, Miller MD, Geleziunas R, Pessi A. Covalent stabilization of coiled coils of the HIV gp41 N region yields extremely potent and broad inhibitors of viral infection. Proc Natl Acad Sci U S A. 2005;102(36):12903–12908. doi: 10.1073/pnas.0502449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Ditzel HJ, Barbas CF, 3rd, Sullivan N, Sodroski J, Parren PW, Burton DR. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res Hum Retroviruses. 1996;12(10):911–924. doi: 10.1089/aid.1996.12.911. [DOI] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorndal A, Deng H, Jansson M, Fiore JR, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman DR, Fenyo EM. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71(10):7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10(4):359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- Burrer R, Haessig-Einius S, Aubertin AM, Moog C. Neutralizing as well as non-neutralizing polyclonal immunoglobulin (Ig)G from infected patients capture HIV-1 via antibodies directed against the principal immunodominant domain of gp41. Virology. 2005;333(1):102–113. doi: 10.1016/j.virol.2004.12.034. [DOI] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266(5187):1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Caffrey M, Cai M, Kaufman J, Stahl SJ, Wingfield PT, Covell DG, Gronenborn AM, Clore GM. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 1998;17(16):4572–84. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300(5628):2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- Chan DC, Chutkowski CT, Kim PS. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc Natl Acad Sci U S A. 1998;95(26):15613–7. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89(2):263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- Chang DK, Hsu CS. Biophysical evidence of two docking sites of the carboxyl heptad repeat region within the amino heptad repeat region of gp41 of human immunodeficiency virus type 1. Antiviral Res. 2007;74(1):51–8. doi: 10.1016/j.antiviral.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433(7028):834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- Chen CH, Greenberg ML, Bolognesi DP, Matthews TJ. Monoclonal antibodies that bind to the core of fusion-active glycoprotein 41. AIDS Res Hum Retroviruses. 2000;16(18):2037–2041. doi: 10.1089/088922200750054765. [DOI] [PubMed] [Google Scholar]
- Cheng-Mayer C, Liu R, Landau NR, Stamatatos L. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J Virol. 1997;71(2):1657–61. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry V, Zhang MY, Harris I, Sidorov IA, Vu B, Dimitrov AS, Fouts T, Dimitrov DS. Increased efficacy of HIV-1 neutralization by antibodies at low CCR5 surface concentration. Biochem Biophys Res Commun. 2006;348(3):1107–15. doi: 10.1016/j.bbrc.2006.07.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry V, Zhang MY, Sidorov IA, Louis JM, Harris I, Dimitrov AS, Bouma P, Cham F, Choudhary A, Rybak SM, Fouts T, Montefiori DC, Broder CC, Quinnan GV, Jr, Dimitrov DS. Cross-reactive HIV-1 neutralizing monoclonal antibodies selected by screening of an immune human phage library against an envelope glycoprotein (gp140) isolated from a patient (R2) with broadly HIV-1 neutralizing antibodies. Virology. 2007;363(1):79–90. doi: 10.1016/j.virol.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206(2):935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- Crooks ET, Moore PL, Franti M, Cayanan CS, Zhu P, Jiang P, deVries R, Wiley C, Zharkikh I, Schülke N, Roux KH, Montefiori DC, Burton DR, Binley JM. A Comparative Immunogenicity Study of HIV-1 Virus-Like Particles Bearing Various Forms of Envelope Proteins, Particles Bearing no Envelope and soluble monomeric gp120. Virology. 2007;366(2):245–62. doi: 10.1016/j.virol.2007.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon AK, Donners H, Pantophlet R, Johnson WE, Decker JM, Shaw GM, Lee FH, Richman DD, Doms RW, Vanham G, Burton DR. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from HIV-1 infected donors. J Virol. 2007;81:6548–62. doi: 10.1128/JVI.02749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov AS, Jacobs A, Finnegan CM, Stiegler G, Katinger H, Blumenthal R. Exposure of the membrane-proximal external region of HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion. Biochemistry. 2007;46(5):1398–1401. doi: 10.1021/bi062245f. [DOI] [PubMed] [Google Scholar]
- Dimitrov AS, Louis JM, Bewley CA, Clore GM, Blumenthal R. Conformational changes in HIV-1 gp41 in the course of HIV-1 envelope glycoprotein-mediated fusion and inactivation. Biochemistry. 2005;44(37):12471–12479. doi: 10.1021/bi051092d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer JJ, Hasan A, Wilson KL, White JM, Matthews TJ, Delmedico MK. The hydrophobic pocket contributes to the structural stability of the N-terminal coiled coil of HIV gp41 but is not required for six-helix bundle formation. Biochemistry. 2003;42(17):4945–53. doi: 10.1021/bi027283n. [DOI] [PubMed] [Google Scholar]
- Earl PL, Broder CC, Doms RW, Moss B. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J Virol. 1997;71(4):2674–2684. doi: 10.1128/jvi.71.4.2674-2684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert DM, Kim PS. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc Natl Acad Sci U S A. 2001a;98(20):11187–11192. doi: 10.1073/pnas.201392898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001b;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- Eckert DM, Malashkevich VN, Hong LH, Carr PA, Kim PS. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell. 1999;99(1):103–115. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Zolla-Pazner S. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J Virol. 2000;74(13):6186–6192. doi: 10.1128/jvi.74.13.6186-6192.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundner C, Li Y, Louder M, Mascola J, Yang X, Sodroski J, Wyatt R. Analysis of the neutralizing antibody response elicited in rabbits by repeated inoculation with trimeric HIV-1 envelope glycoproteins. Virology. 2005;331(1):33–46. doi: 10.1016/j.virol.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Gustchina E, Hummer G, Bewley CA, Clore GM. Differential inhibition of HIV-1 and SIV envelope-mediated cell fusion by C34 peptides derived from the C-terminal heptad repeat of gp41 from diverse strains of HIV-1, HIV-2, and SIV. J Med Chem. 2005;48(8):3036–3044. doi: 10.1021/jm049026h. [DOI] [PubMed] [Google Scholar]
- Gustchina E, Louis JM, Lam SN, Bewley CA, Clore GM. A Monoclonal Fab Derived from a Human Nonimmune Phage Library Reveals a New Epitope on gp41 and Neutralizes Diverse Human Immunodeficiency Virus Type 1 Strains. J Virol. 2007;81(23):12946–53. doi: 10.1128/JVI.01260-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger AE, Kim S, Welch BD, Kay MS. Steric accessibility of the HIV-1 gp41 N-trimer region. J Biol Chem. 2005;280(13):12567–12572. doi: 10.1074/jbc.M412770200. [DOI] [PubMed] [Google Scholar]
- Harris EN, Pierangeli S, Birch D. Anticardiolipin wet workshop report. Fifth International Symposium on antiphospholipid antibodies. Am J Clin Pathol. 1994;101(5):616–624. doi: 10.1093/ajcp/101.5.616. [DOI] [PubMed] [Google Scholar]
- Hartley O, Klasse PJ, Sattentau Q, Moore JP. V3: HIV-1’s switch-hitter. AIDS Res Hum Retroviruses. 2005;21:171–189. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- Haynes BF, Fleming J, St Clair WE, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, Ortel TL, Liao HX, Alam MS. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang KK, Grossman JM, Visvanathan S, Chukwuocha RU, Woods VL, Jr, Le DT, Hahn BH, Chen PP. Identification of anti-thrombin antibodies in the antiphospholipid syndrome that interfere with the inactivation of thrombin by antithrombin. J Immunol. 2001;167(12):7192–7198. doi: 10.4049/jimmunol.167.12.7192. [DOI] [PubMed] [Google Scholar]
- Jiang S, Lin K, Lu M. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1998;72(12):10213–10217. doi: 10.1128/jvi.72.12.10213-10217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Lin K, Strick N, Neurath AR. HIV-1 inhibition by a peptide. Nature. 1993;365(6442):113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- Ketas TJ, Kuhmann SE, Palmer A, Zurita J, He W, Ahuja SK, Klasse PJ, Moore JP. Cell surface expression of CCR5 and other host factors influence the inhibition of HIV-1 infection of human lymphocytes by CCR5 ligands. Virology. 2007;364(2):281–90. doi: 10.1016/j.virol.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Qiao Z, Yu J, Montefiori D, Reinherz EL. Immunogenicity of recombinant human immunodeficiency virus type 1-like particles expressing gp41 derivatives in a pre-fusion state. Vaccine. 2007;25(27):5102–5114. doi: 10.1016/j.vaccine.2006.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YB, Han DP, Cao C, Cho MW. Immunogenicity and ability of variable loop-deleted human immunodeficiency virus type 1 envelope glycoproteins to elicit neutralizing antibodies. Virology. 2003;305(1):124–137. doi: 10.1006/viro.2002.1727. [DOI] [PubMed] [Google Scholar]
- Kirschner M, Monrose V, Paluch M, Techodamrongsin N, Rethwilm A, Moore JP. The production of cleaved, trimeric human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein vaccine antigens and infectious pseudoviruses using linear polyethylenimine as a transfection reagent. Protein Expr Purif. 2006;48(1):61–8. doi: 10.1016/j.pep.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Klausner RD, Fauci AS, Corey L, Nabel GJ, Gayle H, Berkley S, Haynes BF, Baltimore D, Collins C, Douglas RG, Esparza J, Francis DP, Ganguly NK, Gerberding JL, Johnston MI, Kazatchkine MD, McMichael AJ, Makgoba MW, Pantaleo G, Piot P, Shao Y, Tramont E, Varmus H, Wasserheit JN. Medicine. The need for a global HIV vaccine enterprise. Science. 2003;300(5628):2036–2039. doi: 10.1126/science.1086916. [DOI] [PubMed] [Google Scholar]
- Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang CC, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol. 2003;77(19):10557–10565. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M, Cardoso RM, Wilson IA, Burton DR. Antigenic and Immunogenic Study of Membrane-Proximal External Region-Grafted gp120 Antigens by a DNA Prime-Protein Boost Immunization Strategy. J Virol. 2007;81(8):4272–85. doi: 10.1128/JVI.02536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79(16):10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis JM, Bewley CA, Clore GM. Design and properties of N(CCG)-gp41, a chimeric gp41 molecule with nanomolar HIV fusion inhibitory activity. J Biol Chem. 2001;276(31):29485–9. doi: 10.1074/jbc.C100317200. [DOI] [PubMed] [Google Scholar]
- Louis JM, Bewley CA, Gustchina E, Aniana A, Clore GM. Characterization and HIV-1 fusion inhibitory properties of monoclonal Fabs obtained from a human non-immune phage library selected against diverse epitopes of the ectodomain of HIV-1 gp41. J Mol Biol. 2005;353(5):945–51. doi: 10.1016/j.jmb.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Louis JM, Nesheiwat I, Chang L, Clore GM, Bewley CA. Covalent trimers of the internal N-terminal trimeric coiled-coil of gp41 and antibodies directed against them are potent inhibitors of HIV envelope-mediated cell fusion. J Biol Chem. 2003;278(22):20278–20285. doi: 10.1074/jbc.M301627200. [DOI] [PubMed] [Google Scholar]
- Lu M, Blacklow SC, Kim PS. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2(12):1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- Luftig MA, Mattu M, Di Giovine P, Geleziunas R, Hrin R, Barbato G, Bianchi E, Miller MD, Pessi A, Carfi A. Structural basis for HIV-1 neutralization by a gp41 fusion intermediate-directed antibody. Nat Struct Biol. 2006;13(8):740–747. doi: 10.1038/nsmb1127. [DOI] [PubMed] [Google Scholar]
- McBurney SP, Young KR, Ross TM. Membrane embedded HIV-1 envelope on the surface of a virus-like particle elicits broader immune responses than soluble envelopes. Virology. 2007;358(2):334–346. doi: 10.1016/j.virol.2006.08.032. [DOI] [PubMed] [Google Scholar]
- McMichael AJ. HIV vaccines. Annu Rev Immunol. 2006;24:227–255. doi: 10.1146/annurev.immunol.24.021605.090605. [DOI] [PubMed] [Google Scholar]
- Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol. 2000;151(2):413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MD, Geleziunas R, Bianchi E, Lennard S, Hrin R, Zhang H, Lu M, An Z, Ingallinella P, Finotto M, Mattu M, Finnefrock AC, Bramhill D, Cook J, Eckert DM, Hampton R, Patel M, Jarantow S, Joyce J, Ciliberto G, Cortese R, Lu P, Strohl W, Schleif W, McElhaugh M, Lane S, Lloyd C, Lowe D, Osbourn J, Vaughan T, Emini E, Barbato G, Kim PS, Hazuda DJ, Shiver JW, Pessi A. A human monoclonal antibody neutralizes diverse HIV-1 isolates by binding a critical gp41 epitope. Proc Natl Acad Sci U S A. 2005;102(41):14759–14764. doi: 10.1073/pnas.0506927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Cao Y, Leu J, Qin L, Korber B, Ho DD. Inter- and intraclade neutralization of human immunodeficiency virus type 1: genetic clades do not correspond to neutralization serotypes but partially correspond to gp120 antigenic serotypes. J Virol. 1996;70(1):427–444. doi: 10.1128/jvi.70.1.427-444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, Grise H, Corcoran P, Zwick MB, Franti M, Morris L, Roux KH, Burton DR, Binley JM. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol. 2006;80(5):2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67(11):6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JD, Brunel FM, Jensen R, Crooks ET, Cardoso RM, Wang M, Hessell A, Wilson IA, Binley JM, Dawson PE, Burton DR, Zwick MB. An Affinity-Enhanced Neutralizing Antibody against the Membrane-Proximal External Region of Human Immunodeficiency Virus Type 1 gp41 Recognizes an Epitope between Those of 2F5 and 4E10. J Virol. 2007;81(8):4033–4043. doi: 10.1128/JVI.02588-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyambi PN, Mbah HA, Burda S, Williams C, Gorny MK, Nadas A, Zolla-Pazner S. Conserved and exposed epitopes on intact, native, primary human immunodeficiency virus type 1 virions of group M. J Virol. 2000;74(15):7096–7107. doi: 10.1128/jvi.74.15.7096-7107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalka D, Pessi A, Bianchi E, Ciliberto G, Schleif W, McElhaugh M, Danzeisen R, Geleziunas R, Miller M, Eckert DM, Bramhill D, Joyce J, Cook J, Magilton W, Shiver J, Emini E, Esser MT. Analysis of the HIV-1 gp41 specific immune response using a multiplexed antibody detection assay. J Immunol Methods. 2004;287(12):49–65. doi: 10.1016/j.jim.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Pantophlet R, Burton DR. GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- Parren PW, Wang M, Trkola A, Binley JM, Purtscher M, Katinger H, Moore JP, Burton DR. Antibody neutralization-resistant primary isolates of human immunodeficiency virus type 1. J Virol. 1998;72(12):10270–10274. doi: 10.1128/jvi.72.12.10270-10274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185(2):661–72. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- Phogat S, Wyatt R. Rational modifications of HIV-1 envelope glycoproteins for immunogen design. Curr Pharm Des. 2007;13(2):213–227. doi: 10.2174/138161207779313632. [DOI] [PubMed] [Google Scholar]
- Poignard P, Moulard M, Golez E, Vivona V, Franti M, Venturini S, Wang M, Parren PW, Burton DR. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J Virol. 2003;77(1):353–365. doi: 10.1128/JVI.77.1.353-365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popkov M, Mage RG, Alexander CB, Thundivalappil S, Barbas CF, 3rd, Rader C. Rabbit immune repertoires as sources for therapeutic monoclonal antibodies: the impact of kappa allotype-correlated variation in cysteine content on antibody libraries selected by phage display. J Mol Biol. 2003;325(2):325–335. doi: 10.1016/s0022-2836(02)01232-9. [DOI] [PubMed] [Google Scholar]
- Quinnan GV, Jr, Zhang PF, Fu DW, Dong M, Alter HJ. Expression and characterization of HIV type 1 envelope protein associated with a broadly reactive neutralizing antibody response. AIDS Res Hum Retroviruses. 1999;15(6):561–570. doi: 10.1089/088922299311088. [DOI] [PubMed] [Google Scholar]
- Reeves JD, Gallo SA, Ahmad N, Miamidian JL, Harvey PE, Sharron M, Pohlmann S, Sfakianos JN, Derdeyn CA, Blumenthal R, Hunter E, Doms RW. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc Natl Acad Sci U S A. 2002;99(25):16249–16254. doi: 10.1073/pnas.252469399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves JD, Lee FH, Miamidian JL, Jabara CB, Juntilla MM, Doms RW. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J Virol. 2005;79(8):4991–4999. doi: 10.1128/JVI.79.8.4991-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root MJ, Kay MS, Kim PS. Protein design of an HIV-1 entry inhibitor. Science. 2001;291(5505):884–888. doi: 10.1126/science.1057453. [DOI] [PubMed] [Google Scholar]
- Sattentau QJ, Zolla-Pazner S, Poignard P. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology. 1995;206(1):713–717. doi: 10.1016/s0042-6822(95)80094-8. [DOI] [PubMed] [Google Scholar]
- Scherer EM, Zwick MB, Teyton L, Burton DR. Difficulties in eliciting broadly neutralizing anti-HIV antibodies are not explained by cardiolipin autoreactivity. AIDS. 2007;21:2131–9. doi: 10.1097/QAD.0b013e3282a4a632. [DOI] [PubMed] [Google Scholar]
- Selvarajah S, Puffer B, Pantophlet R, Law M, Doms RW, Burton DR. Comparing antigenicity and immunogenicity of engineered gp120. J Virol. 2005;79(19):12148–12163. doi: 10.1128/JVI.79.19.12148-12163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia SK, Kim PS. Protein grafting of an HIV-1-inhibiting epitope. Proc Natl Acad Sci U S A. 2003;100(17):9756–9761. doi: 10.1073/pnas.1733910100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sougrat R, Bartesaghi A, Lifson JD, Bennett AE, Bess JW, Zabransky DJ, Subramaniam S. Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry. PLoS Pathog. 2007;3(5):e63. doi: 10.1371/journal.ppat.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava IK, Ulmer JB, Barnett SW. Role of neutralizing antibodies in protective immunity against HIV. Hum Vaccin. 2005;1(2):45–60. doi: 10.4161/hv.1.2.1764. [DOI] [PubMed] [Google Scholar]
- Stamatatos L, Lim M, Cheng-Mayer C. Generation and structural analysis of soluble oligomeric gp140 envelope proteins derived from neutralization-resistant and neutralization-susceptible primary HIV type 1 isolates. AIDS Res Hum Retroviruses. 2000;16(10):981–94. doi: 10.1089/08892220050058407. [DOI] [PubMed] [Google Scholar]
- Steger HK, Root MJ. Kinetic dependence to HIV-1 entry inhibition. J Biol Chem. 2006;281(35):25813–21. doi: 10.1074/jbc.M601457200. [DOI] [PubMed] [Google Scholar]
- Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, Katinger H. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17(18):1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. CD4-Induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998;72(6):4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70(2):1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vcelar B, Stiegler G, Wolf HM, Muntean W, Leschnik B, Mehandru S, Markowitz M, Armbruster C, Kunert R, Eibl MM, Katinger H. Reassessment of autoreactivity of the broadly neutralizing HIV antibodies 4E10 and 2F5 and retrospective analysis of clinical safety data. AIDS. 2007;21(16):2161–70. doi: 10.1097/QAD.0b013e328285da15. [DOI] [PubMed] [Google Scholar]
- Vujcic LK, Quinnan GV., Jr Preparation and characterization of human HIV type 1 neutralizing reference sera. AIDS Res Hum Retroviruses. 1995;11(7):783–787. doi: 10.1089/aid.1995.11.783. [DOI] [PubMed] [Google Scholar]
- Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387(6631):426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- Welch BD, VanDemark AP, Heroux A, Hill CP, Kay MS. Potent D-peptide inhibitors of HIV-1 entry. Proc Natl Acad Sci U S A. 2007;104(43):16828–33. doi: 10.1073/pnas.0708109104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci U S A. 1994;91(21):9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild MA, Xin H, Maruyama T, Nolan MJ, Calveley PM, Malone JD, Wallace MR, Bowdish KS. Human antibodies from immunized donors are protective against anthrax toxin in vivo. Nat Biotechnol. 2003;21(11):1305–6. doi: 10.1038/nbt891. [DOI] [PubMed] [Google Scholar]
- Wu TT, Johnson G, Kabat EA. Length distribution of CDRH3 in antibodies. Proteins. 1993;16(1):1–7. doi: 10.1002/prot.340160102. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280(5371):1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Xu JY, Gorny MK, Palker T, Karwowska S, Zolla-Pazner S. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J Virol. 1991;65(9):4832–4838. doi: 10.1128/jvi.65.9.4832-4838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wyatt R, Sodroski J. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J Virol. 2001;75(3):1165–1171. doi: 10.1128/JVI.75.3.1165-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. Cryo-Electron Tomographic Structure of an Immunodeficiency Virus Envelope Complex In Situ. PLoS Pathog. 2006;2(8):e83. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MY, Choudhry V, Sidorov IA, Tenev V, Vu BK, Choudhary A, Lu H, Stiegler GM, Katinger HW, Jiang S, Broder CC, Dimitrov DS. Selection of a novel gp41-specific HIV-1 neutralizing human antibody by competitive antigen panning. J Immunol Methods. 2006;317(12):21–30. doi: 10.1016/j.jim.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445(7129):732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- Zwick M, Burton DR. HIV-1 neutralization: mechanisms and relevance to vaccine design. Curr HIV Res. 2007;5(6):608–624. doi: 10.2174/157016207782418443. [DOI] [PubMed] [Google Scholar]
- Zwick MB. The membrane-proximal external region of HIV-1 gp41: a vaccine target worth exploring. AIDS. 2005;19(16):1725–1737. doi: 10.1097/01.aids.0000189850.83322.41. [DOI] [PubMed] [Google Scholar]
- Zwick MB, Jensen R, Church S, Wang M, Stiegler G, Kunert R, Katinger H, Burton DR. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J Virol. 2005;79:1252–1261. doi: 10.1128/JVI.79.2.1252-1261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick MB, Kelleher R, Jensen R, Labrijn AF, Wang M, Quinnan GV, Jr, Parren PW, Burton DR. A novel human antibody against human immunodeficiency virus type 1 gp120 is V1, V2, and V3 loop dependent and helps delimit the epitope of the broadly neutralizing antibody immunoglobulin G1 b12. J Virol. 2003;77(12):6965–6978. doi: 10.1128/JVI.77.12.6965-6978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75(22):10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]