Abstract
In DNA barcoding, a short standardized DNA sequence is used to assign unknown individuals to species and aid in the discovery of new species. A fragment of the mitochondrial gene cytochrome c oxidase subunit 1 is emerging as the standard barcode region for animals. However, patterns of mitochondrial variability can be confounded by the spread of maternally transmitted bacteria that cosegregate with mitochondria. Here, we investigated the performance of barcoding in a sample comprising 12 species of the blow fly genus Protocalliphora, known to be infected with the endosymbiotic bacteria Wolbachia. We found that the barcoding approach showed very limited success: assignment of unknown individuals to species is impossible for 60% of the species, while using the technique to identify new species would underestimate the species number in the genus by 75%. This very low success of the barcoding approach is due to the non-monophyly of many of the species at the mitochondrial level. We even observed individuals from four different species with identical barcodes, which is, to our knowledge, the most extensive case of mtDNA haplotype sharing yet described. The pattern of Wolbachia infection strongly suggests that the lack of within-species monophyly results from introgressive hybridization associated with Wolbachia infection. Given that Wolbachia is known to infect between 15 and 75% of insect species, we conclude that identification at the species level based on mitochondrial sequence might not be possible for many insects. However, given that Wolbachia-associated mtDNA introgression is probably limited to very closely related species, identification at the genus level should remain possible.
Keywords: barcoding, mitochondrial introgression, insects, Wolbachia, Protocalliphora
1. Introduction
Even conservative estimates suggest that the majority of the species living on the planet are currently undescribed (e.g. Novotny et al. 2002). To achieve rapid species descriptions in the context of the current biodiversity crisis, and given the decline in the number of taxonomists, several authors have suggested the use of barcoding in taxonomy (Hebert et al. 2003a,b; Blaxter 2004; Schindel & Miller 2005). DNA barcoding is the use of a short standardized DNA sequence (in animals, a 600 bp fragment of the mitochondrial cytochrome c oxidase (COI) gene) to identify species. DNA barcoding regroups two different and relatively independent aspects: it can be used to (i) identify and assign unknown specimens to species that have previously been described and (ii) facilitate the discovery of new species.
Using a mitochondrial fragment as opposed to a nuclear one for DNA barcoding has two major advantages (Hurst & Jiggins 2005). First, because it is haploid and has highly conserved regions, the COI fragment is technically easy to amplify without cloning in a variety of species. Second, the mitochondrion has an effective population size approximately one-quarter of that of nuclear markers, and, in animals, a high evolutionary rate which therefore provides a high level of resolution. Even closely related species can usually be differentiated by using a relatively short sequence. However, these advantages are associated with a major drawback. While mitochondrial DNA was considered to be a neutral marker that reflects the history of the species, Ballard & Whitlock (2004) and Bazin et al. (2006) have recently argued that mitochondria are in fact often under strong selection and evolve under unusual evolutionary rules when compared with other genomes. Selection can act directly on the mtDNA itself, but it can also arise indirectly from disequilibrium with other maternally transmitted DNA (Hurst & Jiggins 2005).
In insects, the endosymbiotic bacteria Wolbachia are an example of such maternally transmitted DNA. These bacteria cause a number of reproductive alterations in their hosts, including induction of thelytokous parthenogenesis, feminization of genetic males, male killing and, most commonly, the induction of sperm–egg incompatibilities termed cytoplasmic incompatibility (reviewed in Werren 1997; Stouthamer et al. 1999). These reproductive phenotypes effectively increase the frequency of infected females in the host populations, often at the expense of host fitness. Thus, when a population becomes infected with Wolbachia, the bacteria will rapidly spread and the mtDNA type associated with the initial infection will hitch-hike through the population by indirect selection. Given that between 15 and 75% of insect species harbour Wolbachia (Werren et al. 1995a; West et al. 1998; Jeyaprakash & Hoy 2000; Werren & Windsor 2000), these bacteria are possibly an important cause of indirect selection on mtDNA in insects.
Wolbachia can potentially influence mtDNA variation at the intra- or interspecific level. At the intraspecific level, the influence of Wolbachia is now well documented: numerous studies have demonstrated that selection acting on Wolbachia has indirectly reduced mtDNA polymorphism in the infected population or species (e.g. Shoemaker et al. 1999, 2004; Ballard 2000a,b; Jiggins 2003; see review in Hurst & Jiggins 2005). While this means that mtDNA should not be used to make inferences about population histories in Wolbachia-infected species, this should not cause a problem for barcoding. However, Wolbachia can also affect mtDNA variation between species boundaries. In insects, at least three cases are currently described where Wolbachia infection has caused mitochondrial introgression between closely related species: between several members of the melanogaster subgroup of Drosophila (Rousset & Solignac 1995; Ballard 2000a,b), between two sister species of sub-Saharan butterflies Acraea encedon and Acraea encedana (Jiggins 2003), and between the yellow and the brown type of Eurema hecabe, two yet unnamed sibling species of Japanese butterflies (Narita et al. 2006). In such situations, barcoding is impossible because different species share an identical barcode. However, due to the relative paucity of studies where mtDNA variation and Wolbachia infection of closely related species have been investigated, it is currently not known whether such introgressions are the exception or the rule in Wolbachia-infected species.
In this study, we focus on blow flies belonging to the genus Protocalliphora. Protocalliphora are the Holarctic group of species found commonly in the boreal forest and other areas. Protocalliphora are widespread, occupying virtually any habitat where nidicolous birds nest from the forests to the river valleys and from the deserts to the marshes (Sabrosky et al. 1989). Larval stages are obligate haematophagous parasites of nidicolous birds (Bennett & Whitworth 1991). Protocalliphora is the largest genus of blow flies in the Holarctic region with over 40 species described (Sabrosky et al. 1989). At least two species of Protocalliphora are known to be infected by Wolbachia, although it is not yet known whether the bacteria induce cytoplasmic incompatibility or other phenotypes in Protocalliphora (Baudry et al. 2003). To determine whether Wolbachia is involved in mtDNA introgressions, and hence the possibility of barcoding in this genus, we studied Wolbachia infection status and the nuclear and mtDNA polymorphism of 12 species of Protocalliphora.
2. Material and methods
(a) Sampling and DNA extraction
Thirty-one Protocalliphora individuals, belonging to 12 species, were included in this study (table 1). There are at least 16 other Protocalliphora species in the Nearctic, but they were not included in this study because they are rare and very difficult to sample. When available, three individuals per species were analysed. The most closely related genus to Protocalliphora is the monospecific genus Trypocalliphora (Sabrosky et al. 1989); we therefore used three Trypocalliphora braueri individuals as outgroups. However, the taxonomic status of Trypocalliphora has been debated. Rognes (1984) considered it a valid genus, while Sabrosky et al. (1989) considered it a subgenus of Protocalliphora. Whitworth (2003b) evaluated each argument and concluded that Sabrosky's conclusions were based on a misinterpretation of larval morphology and behaviour. Thus, he supported Rognes view that Trypocalliphora is a valid genus. Given the uncertainty about the status of Trypocalliphora, we also used one Lucilia sericata and one Phormia regina as outgroups.
Table 1.
Protocalliphora individuals analysed in the study. (The first four columns indicate the Protocalliphora species, the identification code of the nest where the specimen was sampled, the collection location and the bird host species for each individual, respectively. The last column shows the Wolbachia infection status (§§2 and 3). NI designates non-infected individuals.)
| species | nest label | location | bird hosta | Wolbachia infectionb |
|---|---|---|---|---|
| P. asiovora | 6852 | USA, WA | black-billed magpie | NI |
| P. bennetti | 7887 | Canada, BC | tree swallow | wA2 wB |
| P. bennetti | 7893 | Canada, BC | tree swallow | wA2 wB |
| P. bennetti | 7908 | Canada, BC | tree swallow | wA2 |
| P. deceptor | 6765 | USA, TX | Carolina chickadee | NI |
| P. deceptor | 6767 | USA, TX | Bewick's wren | NI |
| P. deceptor | 6884 | USA, OK | Bewick's wren | NI |
| P. falcozi | ari1 | France, Corsica | blue tit | wA1 wA2 |
| P. falcozi | pac13 | France, Corsica | blue tit | wA1 wA2 |
| P. falcozi | fel18 | France, Corsica | blue tit | wA1 wA2 |
| P. halli | 7884 | Canada, BC | barn swallow | wA2 wB |
| P. halli | 6998 | USA, WA | barn swallow | wA2 wB |
| P. hirundo | 6904 | USA, WA | cliff swallow | wB |
| P. hirundo | 7054-1 | USA, WA | bank swallow | wB |
| P. metallica | 6972-1 | USA, OH | Carolina wren | wA1 |
| P. occidentalis | 7887 | Canada, BC | tree swallow | wB |
| P. occidentalis | 7903 | Canada, BC | tree swallow | wB |
| P. occidentalis | 7025 | USA, AZ | western bluebird | wB |
| P. rognesi | 7054-2 | USA, WA | bank swallow | wA2 wB |
| P. rognesi | 7055 | USA, WA | bank swallow | wB |
| P. rugosa | 7887 | Canada, BC | tree swallow | wB |
| P. rugosa | 7890 | Canada, BC | tree swallow | wB |
| P. rugosa | 7893 | Canada, BC | tree swallow | wB |
| P. shannoni | 7634 | USA, OH | American robin | NI |
| P. shannoni | 7803 | USA, OH | American robin | wA1 |
| P. shannoni | 6972-4 | USA, OH | Carolina wren | wA1 |
| P. sialia | 7811 | Canada, SK | tree swallow | wA2 wB |
| P. sialia | 7220 | USA, OH | eastern bluebird | wB |
| outgroups | ||||
| T. braueri | 7851 | USA, VA | mockingbird | NI |
| T. braueri | 7903 | Canada, BC | tree swallow | NI |
| T. braueri | 7909 | Canada, BC | tree swallow | NI |
Bird host species: American robin, Turdus migratorius; bank swallow, Riparia riparia; barn swallow, Hirundo rustica; Bewick's wren, Thryomanes bewickii; black-billed magpie, Pica pica; blue tit, Parus caeruleus; Carolina chickadee, Poecile carolinensis; Carolina wren, Thryothorus ludovicianus; cliff swallow, Petrochelidon pyrrhonota; eastern bluebird, Sialia sialis; northern mockingbird, Mimus polyglottos; tree swallow, Tachycineta bicolor; western bluebird, Sialia mexicana.
Wolbachia infection status: wA1, wA2, wB refers to the three different Wolbachia strains observed in Protocalliphora (§3). wA1, wA2 belong to the A super group, and wB to the B super group.
Blow fly larvae or pupae were collected from bird nests several days after fledging of the young birds. Collections were made either directly by the authors or by naturalists, in the continental USA and Canada, except for the Protocalliphora falcozi individuals, which were collected from France. Emergent flies, when possible with their puparia, were placed into 95% ethanol. Species were then identified based on fly and pupal case morphology (Sabrosky et al. 1989; Whitworth 2002, 2003a,b). To minimize screening of siblings, only one individual per bird nest was subjected to molecular analysis. DNA from adult flies was extracted with QIAgen DNeasy kit, following the manufacturer's protocol. The lower half of the abdomen of each fly was used for DNA extraction, as it contains the reproductive tissues in which Wolbachia is predominantly found. Extracted DNA was resuspended in 100 μl elution buffer.
(b) Nuclear analysis
We first attempted to reconstruct the phylogeny of the Protocalliphora genus by using nuclear sequence data (Internal Transcribed Spacers 1 and 2), but this was unsuccessful due to a very low level of substitutions between the species, major alignment problems caused by numerous indels, and the fact that the few observed substitutions between species were almost only autapomorphies.
We therefore used the amplified fragment length polymorphism technique (AFLP; Vos et al. 1995) to analyse the nuclear structure of the Protocalliphora genus because this technique has the ability to generate a large number of informative markers with relative ease. For each individual, genomic DNA was double-digested with EcoRI and MseI. DNA fragments were ligated with EcoRI and MseI adapters, generating template DNA for polymerase chain reaction (PCR) amplification (see Baudry et al. 2003 for details). A pre-selective amplification was performed using two primers complementary to the adapters and the restriction site sequences, in the following conditions: 94°C for 1 min, 56°C for 1 min 30 s and 72°C for 2 min, for a total of 35 cycles. Next, a selective PCR was performed with primers similar to the pre-selective amplification primers but with three additional bases at the 3′-end. A total of six primer combinations was used, with the following selective bases: E-TAC and M-TAC, E-TAC and M-GAT, E-TAC and M-CTG, E-GAT and M-ATC, E-GAT and M-CTG, E-GAT and M-CAG, with the Eco primer being fluorescently labelled with 6-FAM. The PCR products were run on an ABI 3700 Capillary DNA Sequencer, thus allowing us to estimate the size of the fragments with an error less than 0.2 bp. Fragments within the size range of 50–500 bp were kept for analyses.
The character matrix of presence or absence of bands produced by the AFLP procedure was analysed with PAUP v. 4.0 (Swofford 2002). A nuclear phylogenetic tree, rooted with L. sericata and P. regina, was constructed by parsimony analysis using a heuristic search with tree bisection–reconnection. Phylogenetic reconstruction was also performed by the neighbour-joining method (Saitou & Nei 1987) using Nei & Li (1979) and Upholt (1977) distances. The reliability of the trees obtained was examined using 1000 bootstrap replicates.
(c) Mitochondrial analysis
Two conserved primer pairs C1J-2183, C1-N-2659 and C2J-3138, TKN-3772 were used to respectively amplify a 374 bp fragment of the cytochrome oxidase I gene (COI) and a 579 bp fragment of the cytochrome oxidase II gene (COII). Thermocycle conditions were as described above. The PCR products were purified and then sequenced with an ABI 377 automatic sequencer (Perkin–Elmer). All COI and COII sequences were proof read and aligned manually.
Tree reconstruction and divergence calculation performed with the COI or COII data produced almost identical results (not shown); we therefore pooled the two datasets before analysis. Using COI and COII sequences to reconstruct the mitochondrial phylogeny of the genus, we started by performing likelihood ratio tests (Huelsenbeck & Rannala 1997) to determine which model of DNA sequence evolution is the most appropriate for the COI and COII data. We used the Model test (Posada & Crandall 1998) procedure implemented in HY-PHY (www.hyphy.org) to test hierarchically the effect of unequal base frequencies, different rates between transitions and transversions, different rates between all substitutions and rate variation over nucleotide sites. The model that best fit the dataset is a general time reversible (GTR) model with rate heterogeneity among sites (gamma distribution shape parameter of 0.167). We then used this model of sequence evolution to reconstruct a phylogenetic tree, rooted with L. sericata and P. regina, by maximum likelihood analysis (heuristic tree search with tree bisection–reconnection performed with PAUP v. 4.0 (Swofford 2002). The reliability of the tree obtained was examined using 1000 bootstrap replicates. Nucleotide sequence divergences between species were calculated with MEGA v. 3.1 (Kumar et al. 2004) using the Kimura two-parameter (K2P) model, the best metric when distances are low (Nei & Kumar 2000).
(d) Wolbachia analysis
A 454 bp fragment of the wsp gene was amplified by PCR, using the general wsp primers designed by Braig et al. (1998) for Wolbachia: wsp 81F and wsp 691R. Thermocycle conditions were 95°C for 1 min, 55°C for 1 min and 72°C for 1 min 30 s, for a total of 35 cycles. In the absence of amplification, the PCR was repeated twice to confirm that the negative result was due to the absence of Wolbachia and not to a failure of the PCR procedure. The positive PCR products were purified and then sequenced with an ABI 377 automatic sequencer (Perkin–Elmer). In several cases, the sequencing results demonstrated the presence of two strains of Wolbachia in one individual. PCR products were then sequenced with primers specific for the two A groups (wA1 and wA2; Baudry et al. 2003) or the B group Wolbachia (Zhou et al. 1998).
3. Results and discussion
(a) Nuclear structure of the Protocalliphora genus
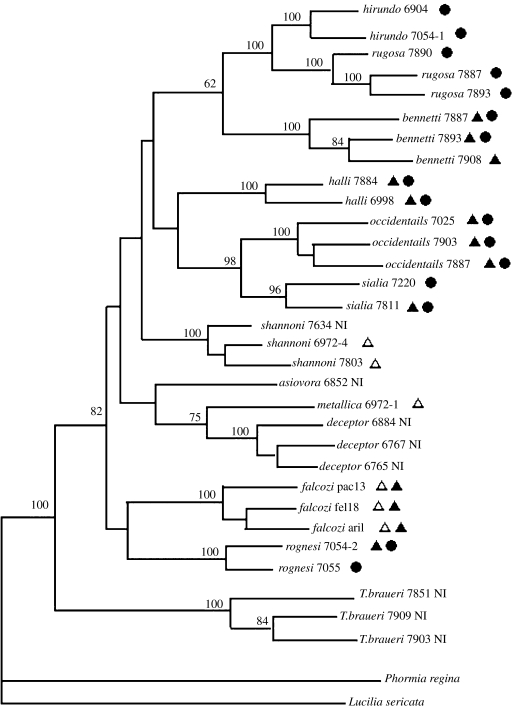
We have reconstructed the nuclear phylogeny of the genus using the AFLP technique. The six AFLP primer pairs used in this study generated a total of 1410 markers. Of these, 1391 (98.7%) were polymorphic and 897 (63.6%) were parsimony informative. The phylogenetic tree reconstructed by parsimony analysis from these data (figure 1) was almost identical to a neighbour-joining tree built with the same data (not shown).
Figure 1.
Phylogram of the Protocalliphora genus based upon AFLP data. The tree was generated by parsimony analysis using a heuristic search with tree bisection–reconnection. Bootstrap values are shown as percentage of 1000 replicates at each node only if they are 50% or greater. The Wolbachia infection status of each individual is shown on the tree. Individuals infected with wA1, wA2 or wB Wolbachia strains are respectively represented by an open triangle, a solid triangle and a circle. Non-infected individuals are symbolized by NI.
The first noticeable characteristic of the cladogram is that T. braueri occupies a well-supported position outside Protocalliphora, confirming the status of Trypocalliphora as a sister genus to Protocalliphora. Second, Protocalliphora individuals always cluster by species, with bootstrap support values of 100% except in one case where the value was 96%. This confirms that morphology correctly identifies species in the Protocalliphora genus. However, note that the multilocus approach to reconstructing the nuclear DNA phylogeny may result in the species being monophyletic even if at individual loci there are sometimes shared polymorphisms between species. Finally, although we used a very high number of characters to build the cladogram, the relationships between species remain partly unresolved, with several nodes having bootstrap values under 50%. We obtained similar results (§2) when trying to construct a phylogeny with nuclear sequences from Internal Transcribed Spacers 1 and 2 (data not shown, available from the authors upon request), suggesting that most species of the genus have diverged approximately at the same time.
(b) Mitochondrial structure of the Protocalliphora genus and barcoding
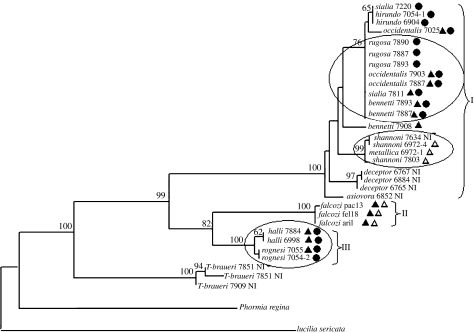
The phylogenetic tree representing the mitochondrial genetic structure of the Protocalliphora genus based upon COI and COII data is shown in figure 2. On this mitochondrial phylogeny, T. braueri also occupies a well-supported position outside Protocalliphora. However, within Protocalliphora, the mitochondrial phylogeny bears few resemblances to the nuclear one. First, in contrast to what was observed for the nuclear data, the mitochondrial haplotypes showed a limited tendency to cluster by species. In only four species (P. deceptor, P. falcozi, P. halli and P. rognesi) do all individuals group together (figure 2). Second, the relationships between species are markedly different between the two trees. For example, P. rognesi and P. halli show very closely related mitochondrial haplotypes but are only distantly related at the nuclear level.
Figure 2.
Phylogram of the Protocalliphora genus based upon COI and COII data (total of 953 bp). The tree was generated by maximum likelihood analysis using a heuristic search with tree bisection–reconnection. Bootstrap values are shown as percentage of 1000 replicates at each node only if they are 50% or greater. The Wolbachia infection status of each individual is shown on the tree. Individuals infected with wA1, wA2 or wB Wolbachia strains are respectively represented by an open triangle, a solid triangle and a circle. Non-infected individuals are symbolized by NI. Three clusters defined using 3 or 1.8% divergence as threshold values (§3) are shown on the figure. The three ellipses indicate cases where horizontal transfer of Wolbachia between species seems probable (§3).
The first objective of DNA barcoding is species identification. In practice, the sample to be identified is positioned in a previously characterized phylogeny, usually using neighbour-joining and/or parsimony analysis (Meyer & Paulay 2005). The test sample then receives the identity of its sister clade. Obviously, to be successful, this approach requires species to be monophyletic. In our sampling of Protocalliphora, reliable identification can therefore be obtained only for the four species mentioned above. Considering that our sampling comprises 10 species that are represented by more than one individual, this represents a success rate of only 40%.
Intraspecific divergence within species of Protocalliphora at the COI and COII loci ranged from 0.00 to 0.71% (table 2), with an average value of 0.18%. This value is close to the 0.25% value observed by Hebert et al. (2003a,b) for average intraspecific divergence in a large sample of Lepidoptera. Interspecific divergence shows a greater range, with values varying between 0.10 and 8.63%, and an average of 3.86% (table 2).
Table 2.
Divergence values between species at the COI and COII loci. (Within species divergence values are shown on the diagonal of the table. The three clusters defined by using the 1.8 or 3% divergence criterion (see Results and figure 2) are shown framed. Divergence values within each cluster are shown in bold. ND, not determined.)
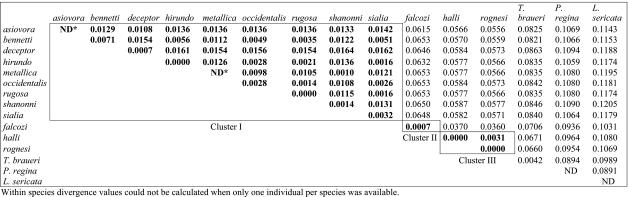 |
The second objective of DNA barcoding is to help in species discovery. The proposed method is to use a threshold value chosen to separate intraspecific from interspecific variation. An unidentified sequence differing from a known sequence by less than the threshold value will be considered to belong to this species, whereas if it differs by more than the threshold value, it will be considered to represent a new taxon. At least two methods have been proposed to choose a threshold. The first uses a fixed value considered suitable for the taxonomic group of interest. For example, it has been proposed that in insects, interspecific divergence almost always exceeds 3% (Hebert et al. 2003a,b) and this value can therefore be used as a threshold. Alternatively, Hebert et al. (2004) have proposed that a threshold of ten times the average intraspecific difference would be appropriate to screen for new animal species. In Protocalliphora, this would translate into using either 3 or 1.8% as a threshold for screening new species. In both cases, we obtained only three clusters of individuals (figure 2). Cluster II was the only one to correspond to one species (P. falcozi). Cluster III included two species, P. halli and P. rognesi, and cluster I comprised the remaining nine species. In this last cluster, the maximum divergence observed between species was only 1.64% and there was extensive haplotype sharing. Four species (P. sialia, P. occidentalis, P. rugosa and P. bennetti) showed the same haplotype. Similarly, P. sialia and P. hirundo shared one haplotype. Using barcoding to identify new species in Protocalliphora would therefore very much underestimate the species richness of the genus, as only 3 species would be recognized instead of 12 (an underestimation of 75%). Note that this major underestimation of species number could not be significantly ameliorated by using a lower threshold value because the problem is caused mostly by the non-monophyly of the species and by the overlap between intra- and interspecific divergence values. At best, using an ad hoc threshold value of 1%, three more clusters would be recognized, but still only 50% of the species would be recognized.
(c) Wolbachia infection and mitochondrial DNA variation within Protocalliphora
It would of course be interesting to determine which factors have caused the major discrepancies observed between the mitochondrial and the nuclear structure of Protocalliphora. Since the bacterium Wolbachia was known to be present in at least one species of the genus (P. sialia, Baudry et al. 2003) and because interspecific mitochondrial introgression linked to Wolbachia infection has been described in insects (Rousset & Solignac 1995; Ballard 2000a,b; Jiggins 2003; Narita et al. 2006), Wolbachia seemed a possible candidate. We therefore determined the Wolbachia infection status of each Protocalliphora individual using a fragment of the wsp gene (table 1; figures 1 and 2).
Among the 12 species of our sample, only two, P. asiovora and P. deceptor, showed no Wolbachia-infected individuals. The Protocalliphora genus therefore seems to present a very high level of Wolbachia infection, with more than 80% of the species harbouring the bacteria. Moreover, it is important to emphasize that the two species where we found no evidence of Wolbachia may not be infection free. The small sample size per species (one to three individuals) means that we are unlikely to detect infections within a species unless they are present at a very high frequency.
The results of the sequencing showed that only three Wolbachia strains were present in the Protocalliphora genus. Two bacteria belong to Wolbachia-A group (Werren et al. 1995b), hereafter called wA1 and wA2, and are observed in three and six of the species, respectively. The third one is a B group Wolbachia (Werren et al. 1995b), hereafter called wB, which is present in seven species. Note that these identifications are based on the wsp gene only and that the strains identity could be checked using the more powerful MLST method developed by Baldo et al. (2006). The same three strains were previously observed in P. sialia (Werren & Bartos 2001; Baudry et al. 2003). Several individuals are infected by two of the three strains. Finally, even with the small number of individuals analysed per species, we observed an intraspecific polymorphism of infection in four of the species.
There are two general explanations for the extensive sharing of Wolbachia strains and mitochondrial haplotypes among Protocalliphora species: (i) maintenance of an ancestral mitochondrial and infection polymorphism that existed prior to divergence of the infected species or (ii) movement of Wolbachia and their associated mitochondrial haplotype between species by interspecific hybridization after their speciation. In the first case, there should be a correlation between the nuclear phylogeny and the mitochondrial one, as well as with the Wolbachia infection status, i.e. closely related species at the nuclear level should have a higher tendency to share mitochondrial haplotype and Wolbachia strains. On the contrary, if mitochondrial introgression associated with Wolbachia infection did occur, we should observe cases where species not closely related at the nuclear level share very similar mitochondrial haplotype and Wolbachia strains.
In the Protocalliphora mitochondrial phylogeny, we observe three such cases (indicated by ellipses in figure 2). First, the four species that share an identical haplotype (P. sialia, P. occidentalis, P. rugosa and P. bennetti, see above) all harbour wB Wolbachia (and three of them also wA2). However, while P. sialia and P. occidentalis on one hand, and P. rugosa and P. bennetti on the other, are closely related at the nuclear level, the two pairs are not (figure 1). Second, P. metallica and P. shannoni show almost identical mtDNA haplotypes (1 bp difference), are both infected by the wA1 Wolbachia strain, but are only distantly related at the nuclear level. Similarly, P. halli and P. rognesi show very similar mtDNA haplotypes (3 bp difference) and are both infected by the wA2 and wB Wolbachia strain, but are not closely related at the nuclear level. In these three cases, a parsimonious explanation for the observed pattern is that interspecific transfer of mtDNA and Wolbachia strain did occur. Of course, these two explanations are not mutually exclusive; while we believe that the observed patterns strongly suggest that interspecific mitochondrial and Wolbachia transfers did take place in at least three cases, maintenance of ancestral polymorphism in other cases is also possible.
Finally, it is interesting to note that, of the two species that show mitochondrial monophyly with a strong bootstrap support (P. deceptor and P. falcozi), the first is not infected by Wolbachia, while P. falcozi is the only species of our sample not from North America. This suggests that mitochondrial monophyly is observed in this genus only when mitochondrial introgression associated with Wolbachia transfer cannot occur, either because Wolbachia is absent or owing to geographical isolation.
(d) Barcoding in insects
In insects, three cases were already described where Wolbachia infection indirectly caused an interspecific mtDNA introgression, and our study adds a fourth one. In all four cases, the mtDNA introgression occurred between very closely related species, and was not accompanied by detectable nuclear introgression. The most probable explanation for this is that Wolbachia strains and associated mitochondrial haplotypes have been occasionally transferred from species to species by rare hybridization events. The rarity of these events, and the fact that the interspecific hybrids probably have a low fitness, would make the nuclear gene flow associated with these hybridizations negligible. In contrast, the selective advantage of Wolbachia results in its increase in frequency, and the infection and associated mtDNA haplotype spreads into the new species (Hurst & Jiggins 2005). It should be noted that interspecific mtDNA introgression associated with Wolbachia infections precludes identification at the species level based on COI barcoding. However, these introgressions are restricted to species that can hybridize, even if rarely, and therefore to very closely related species. This means that barcoding at a higher taxonomic rank, for example the genus, should remain possible in Wolbachia-infected species.
Although still controversial (e.g. Ebach & Holdrege 2005; Will et al. 2005; Rubinoff 2006), the scientific benefits expected of DNA barcoding include accelerating assignment of specimens to species that have been previously described and facilitating discovery of new species (Meyer & Paulay 2005; Savolainen et al. 2005; Lefebure et al. 2006). To produce accurate results, these two aspects of barcoding have different requirements. In comprehensively studied groups, assignment of a specimen to the correct species only requires species to be monophyletic at the mitochondrial level. Accurate species discovery also necessitate species to be monophyletic but, additionally, there should be an absence of overlap between intra- and interspecific variations, i.e. a barcoding ‘gap’ (Meyer & Paulay 2005).
Numerous studies have investigated the performance of the two aspects of barcoding. Most studies published to date suggest that barcoding achieves high accuracy in the task of assigning specimen to known species (e.g. Hebert et al. 2004; Janzen et al. 2005; Ward et al. 2005; Hajibabaei et al. 2006; Smith et al. 2006), implying that species are usually monophyletic at the mitochondrial level (but see Funk & Omland 2003). In contrast, there is a disagreement regarding the performance of barcoding for the discovery of new species. Earlier studies suggested a low error rate but they usually undersampled intraspecific variation (because very few individuals were sampled by species) and interspecific divergence (because closely related species were not always sampled; Meyer & Paulay 2005). A recent study by Meyer & Paulay (2005) provides the first examination of barcoding performance in a comprehensively sampled, diverse group (cypraeid marine gastropods, or cowries). They found that due to a substantial overlap between intra- and interspecific variations, discovery of new species using barcoding would lead to an unacceptable error rate. In contrast, our study on the Protocalliphora genus showed a very high error rate for both aspects of barcoding, specimen identification and species discovery. Studies on the performance of barcoding in comprehensively sampled insects groups are needed to determine whether or not the Protocalliphora case is an exception in insects.
References
- Baldo L, et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 2006;72:7098–7110. doi: 10.1128/AEM.00731-06. doi:10.1128/AEM.00731-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard J.W. Comparative genomics of mitochondrial DNA in Drosophila simulans. J. Mol. Evol. 2000a;51:64–75. doi: 10.1007/s002390010067. [DOI] [PubMed] [Google Scholar]
- Ballard J.W. When one is not enough: introgression of mitochondrial DNA in Drosophila. Mol. Biol. Evol. 2000b;17:1126–1130. doi: 10.1093/oxfordjournals.molbev.a026394. [DOI] [PubMed] [Google Scholar]
- Ballard J.W, Whitlock M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. doi:10.1046/j.1365-294X.2003.02063.x [DOI] [PubMed] [Google Scholar]
- Baudry E, Bartos J, Emerson K, Whitworth T, Werren J.H. Wolbachia and genetic variability in the birdnest blowfly Protocalliphora sialia. Mol. Ecol. 2003;12:1843–1854. doi: 10.1046/j.1365-294x.2003.01855.x. doi:10.1046/j.1365-294X.2003.01855.x [DOI] [PubMed] [Google Scholar]
- Bazin E, Glemin S, Galtier N. Population size does not influence mitochondrial genetic diversity in animals. Science. 2006;312:570–572. doi: 10.1126/science.1122033. doi:10.1126/science.1122033 [DOI] [PubMed] [Google Scholar]
- Bennett G, Whitworth T. Studies on the life history of some species of Protocalliphora (Diptera: Calliphoridae) Can. J. Zool. 1991;69:2048–2058. [Google Scholar]
- Blaxter M.L. The promise of a DNA taxonomy. Phil. Trans. R. Soc. B. 2004;359:669–679. doi: 10.1098/rstb.2003.1447. doi:10.1098/rstb.2003.1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig H.R, Zhou W, Dobson S.L, O'Neill S.L. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J. Bacteriol. 1998;180:2373–2378. doi: 10.1128/jb.180.9.2373-2378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebach M.C, Holdrege C. DNA barcoding is no substitute for taxonomy. Nature. 2005;434:697. doi: 10.1038/434697b. doi:10.1038/434697b [DOI] [PubMed] [Google Scholar]
- Funk D, Omland K. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2003;34:397–423. doi:10.1146/annurev.ecolsys.34.011802.132421 [Google Scholar]
- Hajibabaei M, Janzen D.H, Burns J.M, Hallwachs W, Hebert P.D. DNA barcodes distinguish species of tropical Lepidoptera. Proc. Natl Acad. Sci. USA. 2006;103:968–971. doi: 10.1073/pnas.0510466103. doi:10.1073/pnas.0510466103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D, Cywinska A, Ball S.L, deWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B. 2003a;270:313–321. doi: 10.1098/rspb.2002.2218. doi:10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D, Ratnasingham S, deWaard J.R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B. 2003b;270(Suppl. 1):S96–S99. doi: 10.1098/rsbl.2003.0025. doi:10.1098/rsbl.2003.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D, Stoeckle M.Y, Zemlak T.S, Francis C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004;2:e312. doi: 10.1371/journal.pbio.0020312. doi:10.1371/journal.pbio.0020312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Rannala B. Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science. 1997;276:227–232. doi: 10.1126/science.276.5310.227. doi:10.1126/science.276.5310.227 [DOI] [PubMed] [Google Scholar]
- Hurst G.D, Jiggins F.M. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc. R. Soc. B. 2005;272:1525–1534. doi: 10.1098/rspb.2005.3056. doi:10.1098/rspb.2005.3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen D.H, Hajibabaei M, Burns J.M, Hallwachs W, Remigio E, Hebert P.D. Wedding biodiversity inventory of a large and complex Lepidoptera fauna with DNA barcoding. Phil. Trans. R. Soc. B. 2005;360:1835–1845. doi: 10.1098/rstb.2005.1715. doi:10.1098/rstb.2005.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaprakash A, Hoy M.A. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol. Biol. 2000;9:393–405. doi: 10.1046/j.1365-2583.2000.00203.x. doi:10.1046/j.1365-2583.2000.00203.x [DOI] [PubMed] [Google Scholar]
- Jiggins F.M. Male-killing Wolbachia and mitochondrial DNA: selective sweeps, hybrid introgression and parasite population dynamics. Genetics. 2003;164:5–12. doi: 10.1093/genetics/164.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. doi:10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- Lefebure T, Douady C.J, Gouy M, Gibert J. Relationship between morphological taxonomy and molecular divergence within Crustacea: proposal of a molecular threshold to help species delimitation. Mol. Phylogenet. Evol. 2006;40:435–447. doi: 10.1016/j.ympev.2006.03.014. doi:10.1016/j.ympev.2006.03.014 [DOI] [PubMed] [Google Scholar]
- Meyer C.P, Paulay G. DNA barcoding: error rates based on comprehensive sampling. PLoS Biol. 2005;3:e422. doi: 10.1371/journal.pbio.0030422. doi:10.1371/journal.pbio.0030422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita S, Nomura M, Kato Y, Fukatsu T. Genetic structure of sibling butterfly species affected by Wolbachia infection sweep: evolutionary and biogeographical implications. Mol. Ecol. 2006;15:1095–1108. doi: 10.1111/j.1365-294X.2006.02857.x. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Oxford University Press; Oxford, UK: 2000. Molecular evolution and phylogenetics. [Google Scholar]
- Nei M, Li W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl Acad. Sci. USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. doi:10.1073/pnas.76.10.5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny V, Basset Y, Miller S.E, Weiblen G.D, Bremer B, Cizek L, Drozd P. Low host specificity of herbivorous insects in a tropical forest. Nature. 2002;416:841–844. doi: 10.1038/416841a. doi:10.1038/416841a [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rognes K. Revision of the bird-parasitic blowfly genus Trypocalliphora-Peus 1960 (Diptera: Calliphoridae) Entomol. Scand. 1984;15:371–382. [Google Scholar]
- Rousset F, Solignac M. Evolution of single and double Wolbachia symbioses during speciation in the Drosophila simulans complex. Proc. Natl Acad. Sci. USA. 1995;92:6389–6393. doi: 10.1073/pnas.92.14.6389. doi:10.1073/pnas.92.14.6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinoff D. Utility of mitochondrial DNA barcodes in species conservation. Conserv. Biol. 2006;20:1026–1033. doi: 10.1111/j.1523-1739.2006.00372.x. doi:10.1111/j.1523-1739.2006.00542.x [DOI] [PubMed] [Google Scholar]
- Sabrosky C.W, Bennett G.F, Whitworth T.L. Smithsonian Institution Press; Washington, DC: 1989. Bird blowflies (Protocalliphora) in North America (Diptera: Calliphoridae), with notes on Palearctic species. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Savolainen V, Cowan R.S, Vogler A.P, Roderick G.K, Lane R. Towards writing the encyclopedia of life: an introduction to DNA barcoding. Phil. Trans. R. Soc. B. 2005;360:1805–1811. doi: 10.1098/rstb.2005.1730. doi:10.1098/rstb.2005.1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindel D.E, Miller S.E. DNA barcoding a useful tool for taxonomists. Nature. 2005;435:17. doi: 10.1038/435017b. doi:10.1038/435017b [DOI] [PubMed] [Google Scholar]
- Shoemaker D, Katju V, Jaenike J. Wolbachia and the evolution of reproductive isolation between Drosophila recens and Drosophila subquinaria. Evolution. 1999;53:1157–1164. doi: 10.1111/j.1558-5646.1999.tb04529.x. doi:10.2307/2640819 [DOI] [PubMed] [Google Scholar]
- Shoemaker D.D, Dyer K.A, Ahrens M, McAbee K, Jaenike J. Decreased diversity but increased substitution rate in host mtDNA as a consequence of Wolbachia endosymbiont infection. Genetics. 2004;168:2049–2058. doi: 10.1534/genetics.104.030890. doi:10.1534/genetics.104.030890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.A, Woodley N.E, Janzen D.H, Hallwachs W, Hebert P.D. DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae) Proc. Natl Acad. Sci. USA. 2006;103:3657–3662. doi: 10.1073/pnas.0511318103. doi:10.1073/pnas.0511318103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouthamer R, Breeuwer J.A, Hurst G.D. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 1999;53:71–102. doi: 10.1146/annurev.micro.53.1.71. doi:10.1146/annurev.micro.53.1.71 [DOI] [PubMed] [Google Scholar]
- Swofford, D. L. 2002 PAUP*: phylogenetic analysis using parsimony (*and other methods). Sunderland, MA: Sinauer Associates.
- Upholt W.B. Estimation of DNA sequence divergence from comparison of restriction endonuclease digests. Nucl. Acids Res. 1977;4:1257–1265. doi: 10.1093/nar/4.5.1257. doi:10.1093/nar/4.5.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. doi:10.1093/nar/23.21.4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward R.D, Zemlak T.S, Innes B.H, Last P.R, Hebert P.D. DNA barcoding Australia's fish species. Phil. Trans. R. Soc. B. 2005;360:1847–1857. doi: 10.1098/rstb.2005.1716. doi:10.1098/rstb.2005.1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J.H. Biology of Wolbachia. Annu. Rev. Entomol. 1997;42:587–609. doi: 10.1146/annurev.ento.42.1.587. doi:10.1146/annurev.ento.42.1.587 [DOI] [PubMed] [Google Scholar]
- Werren J.H, Bartos J.D. Recombination in Wolbachia. Curr. Biol. 2001;11:431–435. doi: 10.1016/s0960-9822(01)00101-4. doi:10.1016/S0960-9822(01)00101-4 [DOI] [PubMed] [Google Scholar]
- Werren J.H, Windsor D.M. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc. R. Soc. B. 2000;267:1277–1285. doi: 10.1098/rspb.2000.1139. doi:10.1098/rspb.2000.1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J.H, Windsor D, Guo L. Distribution of Wolbachia among neotropical arthropods. Proc. R. Soc. B. 1995a;262:197–204. doi:10.1098/rspb.1995.0196 [Google Scholar]
- Werren J.H, Zhang W, Guo L.R. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc. R. Soc. B. 1995b;261:55–63. doi: 10.1098/rspb.1995.0117. doi:10.1098/rspb.1995.0117 [DOI] [PubMed] [Google Scholar]
- West S.A, Cook J.M, Werren J.H, Godfray H.C. Wolbachia in two insect host–parasitoid communities. Mol. Ecol. 1998;7:1457–1465. doi: 10.1046/j.1365-294x.1998.00467.x. doi:10.1046/j.1365-294x.1998.00467.x [DOI] [PubMed] [Google Scholar]
- Whitworth T. Two new species of North American Protocalliphora (Diptera: Calliphoridae) from bird nests. Proc. Entomol. Soc. Wash. 2002;104:801–811. [Google Scholar]
- Whitworth T. A new species of North American Protocalliphora (Diptera: Calliphoridae) from bird nests. Proc. Entomol. Soc. Wash. 2003a;105:664–673. [Google Scholar]
- Whitworth T. A key to the puparia of 27 species of North American Protocalliphora Hough (Diptera: Calliphoridae) from bird nests and two new puparial descriptions. Proc. Entomol. Soc. Wash. 2003b;105:995–1033. [Google Scholar]
- Will K.W, Mishler B.D, Wheeler Q.D. The perils of DNA barcoding and the need for integrative taxonomy. Syst. Biol. 2005;54:844–851. doi: 10.1080/10635150500354878. doi:10.1080/10635150500354878 [DOI] [PubMed] [Google Scholar]
- Zhou W, Rousset F, O'Neil S. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. R. Soc. B. 1998;265:509–515. doi: 10.1098/rspb.1998.0324. doi:10.1098/rspb.1998.0324 [DOI] [PMC free article] [PubMed] [Google Scholar]