Abstract
Background
Osteopontin (OPN) is a potential therapeutic target in hepatocellular carcinoma (HCC) because it is a critical mediator of metastatic function. The molecular mechanisms which determine expression of OPN in HCC however, are unknown. In this study, we examine differential OPN expression in the two HCC cell lines: HepG2 and Hep3B.
Methods
OPN expression, metastatic function, OPN promoter activity , and active transcription of OPN mRNA and its decay were assessed in the two HCC cell lines using standard techniques. RNA gel-shift assays were performed to determine binding of cytoplasmic proteins to OPN mRNA.
Results
Expression of OPN cellular/secreted protein and mRNA was greater in HepG2 than Hep3B cells (p<0.01). Transient transfection of the OPN promoter construct demonstrated equivalent luciferase activities in the two cell lines; the rate of transcription was also equivalent as determined by Chromatin Immuno-precipetation assay. OPN mRNA half-life was 21 ± 1 and 3 ± 1 h in HepG2 and Hep3B respectively (p<0.02). In HepG2 and Hep3B, the nucleotide sequence of OPN and its 5’-UTR, 3’-UTR, and poly (A) tail lengths were identical. A luciferase construct coupled in-line with OPN-5’-UTR and OPN 3’-UTR presented greater expression in HepG2 (p<0.01 vs. Hep3B). Deletion of nt 10–57 of the OPN 5’-UTR restored luciferase and HA-tagged OPN protein expression in Hep3B, but not in Hep G2. RNA gel-shift assays demonstrate different patterns of protein binding to OPN 5’-UTR between the two HCC cell lines.
Conclusions
We conclude that RNA stability is a new, previously unrecognized mechanism which regulates OPN expression in HCC to convey metastatic function.
Introduction
HCC is the third leading cause of cancer-related death in the world. In the United States, the incidence of HCC has nearly doubled over the past two decades [1]. The molecular mechanisms underlying the development of HCC are not well-elucidated and have been the focus of study recently. As per the current literature, one of the biomarkers that play a crucial role in determining the oncogenic potential of various cancers is osteopontin (OPN) [2, 3].
OPN is an acidic, hydrophilic glycoprotein identified initially in the bone as a sialoprotein and later found to be expressed at basal levels in a variety of normal tissues and cell types with greater expression levels in the liver, lymph nodes, testes, and spleen [4]. OPN has been reported to be over-expressed in several neoplasms and is associated with tumorigenesis, tumor invasion, and metastasis in breast, lung, prostate, and colon cancers [5–8]. Increased levels of circulating OPN reported in many malignancies (including breast, and prostate) have been associated with poor patient prognosis [9–11] . OPN is also over-expressed in HCC [12, 13], and recent data suggest that OPN derived from neoplastic cells possesses structural and functional differences from that of OPN from untransformed cells [14, 15]. Because over-expression of OPN in cancer cells is known to be a major predictor of metastasis, regulators of OPN expression could serve as potential molecular targets for modulation of tumor phenotype. Although data that suggest that OPN expression is controlled by complex regulatory pathways at the transcriptional level in several cancers, the molecular mechanisms which determine expression of OPN in HCC are largely unknown.
In this study, we assess the differential transcriptional regulation of OPN in two HCC cell lines-HepG2 and Hep3B. Although both these tumor cell lines were obtained from patients with HCC, they express OPN mRNA and protein differentially. We hypothesized that the differential OPN mRNA expression in the two HCC cell lines occurs at the level of transcriptional and/or post-transcriptional events. Our study demonstrates that the HepG2 and Hep3B cell lines are competent equivalently in synthesizing OPN mRNA, OPN promoter activation, and possess structurally similar OPN 3’-UTR and 5’-UTR. RNA gel-shift assays demonstrate different patterns of protein binding patterns to OPN 5’-UTR between the two cell lines. In HepG2 and Hep3B cells, different repertoires of RNA binding proteins targeting OPN 5’-UTR regulate OPN mRNA stability and alter OPN expression. Our results suggest that OPN mRNA stability is the crucial factor that contributes to the differential expression observed between the two HCC cell lines.
Materials and Methods
Cell Culture
The HCC cell lines HepG2 (ATCC# HB-8065), and Hep3B (ATCC# HB-8064) obtained from American Type Tissue Collection (ATCC, Manassas, VA) were maintained as monolayers in Minimum Essential Medium (MEM, Sigma-Aldrich, St. louis, MO) plus10% fetal bovine serum at 37°C and 5% CO2. In some cases, cells were cultured with actinomycin D (20µg/ml) for 0, 4, 8, 16, 24, and 36 h at 37°C to inhibit transcription.
Western Blot analysis
Protein lysates obtained from the cell lines lysed with the buffer containing 0.8% NaCl, 0.02% KCL, 0.1%SDS, 1% Triton X-100, 0.5% Sodium deoxycholic acid, 0.144% NaH2PO4, and 0.024% KH2PO4, (pH 7.4) were separated by 12% SDS-PAGE. Transferred membranes were blocked and labeled with human monoclonal OPN primary antibody (1:1000 dilution; R&D Systems, Minneapolis, MN) or His antibody (figure 4B) for 3 h at room temperature followed by staining with HRP-conjugated secondary antibody for 1 h at room temperature. Bound peroxidase activity was detected by using Western Blot Luminol reagent as per manufacturer’s instructions (Santa Cruz Biotechnology, Santa Cruz, CA). For quantification, the bands were scanned into AlphaImager 3400 (Alpha Innotech, San Leandro, CA) and normalized by dividing the measured density of protein bands by the density of GAPDH or β-actin control bands from corresponding cell lysates.
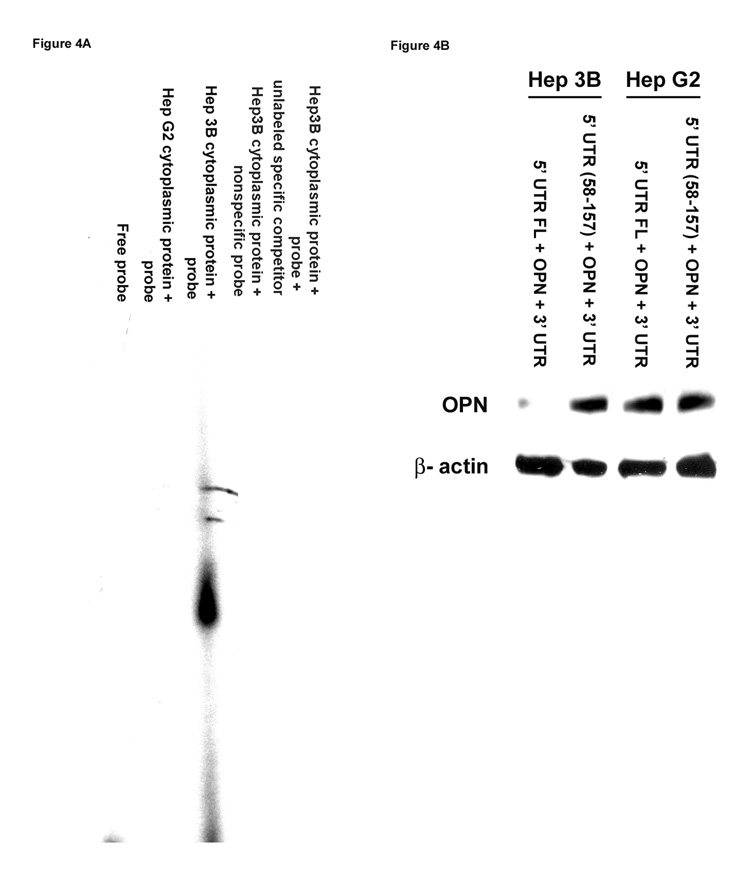
Figure 4. Determination of protein(s) binding to the 5’-UTR of the OPN promoter region using gel-shift assays.
(A) Cytoplasmic protein lysates were extracted from HepG2 and Hep3B cells and incubated with (α-32 P) ATP (2500 Ci/mmol) end-labeled synthesized 5’-UTR from the human OPN promoter sequence using T4 polynucleotide kinase followed by G-50 column purification. The final products were resolved on 6% poly-acrylamide gel electrophoresis using 0.5× Tris/borate EDTA buffer and visualized by autoradiography. Gel is representative of three experiments.
(B) Full-length OPN 5’-UTR or deletion in OPN 5’-UTR (58–157) along with OPN promoter region, OPN coding region and 3’-UTR were ligated to the pcDNA3.1 expression vector at the C-terminal followed by transient transfection of HepG2 and Hep3B cells and assayed for OPN protein expression by Western blot analysis. Blot is representative of three experiments.
Reverse transcriptase (RT)-PCR and Real-time PCR analysis
Total RNA was extracted from HepG2 and Hep3B cells using Qiagen RNeasy Mini Kit (Qiagen Inc., Valencia, CA). Reverse transcription-PCR was performed with 1µg of RNA using OPN promoter specific primers (Integrated DNA Technologies; IDT, Coralville, IA) forward: 5’-TGCTGAATGCCCATCCCGTAAATG-3’; reverse: 5’-GCTGAATGCACAACCCAGTAGCAA-3’. The following PCR parameters were used: 92 °C for 2min; 94 °C for 30sec, 54 °C for 30sec, 72 °C for 30sec for 28 cycles; 72 °C for 7min. For real-time PCR analysis, two-step RT-PCR reactions were performed using SYBR Green detection. First strand cDNA were synthesized from 1µg total RNA using the iScript Select cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA) at 48°C for 30min. The following OPN forward primer 5’-TGAATGGTGCATACAAGGCCATCC-3’, and reverse primer 5’-TTCATAACTGTCCTTCCCACGGCT-3’ were used for the quantitative PCR analysis. GAPDH was used as the endogenous control: GAPDH forward: 5’-AGCCTCAAGATCATCAGCAATGCC-3’; GAPDH reverse: 5’-TGTGGTCATGAGTCCTTCCACGAT-3’ (IDT). Real-time PCR parameters used were as follows: 95 °C for 3min; 95 °C for 30sec, 55 °C for 35sec for 40 cycles; 95 °C for 1min, and 55 °C for 10min. PCR was performed with iQ SYBR Green super mix (Bio-Rad Laboratories), using iCycler iQ Real-time PCR Detection System (Bio-Rad Laboratories). OPN was normalized to a housekeeping gene, GAPDH. the 2-delta-delta Ct formula used to calculate the results.
Si-RNA techniques
HepG2 and Hep3B cells were plated at a density of 5 × 105 cells/well in 6-well plates (Costar, Corning Inc., NY) in DMEM plus 10% FBS 24 h prior to transfection as previously described [16]. Cells were transfected with OPN SiRNA or mismatch SiRNA (Santa Cruz. Biotechnology) using Lipofectamine 2000 (Invitrogen Co., Carlsbad, CA) as per manufacturer’s instructions. Transfected cells were harvested after 48 h, and OPN protein levels were quantified by Western blot analysis in triplicate assays.
Adhesion/ Migration/ Invasion Assays
Adhesion, migration, and invasion assays were performed as described previously [17]. Briefly for adhesion assay, 1×105 cells/well were seeded in triplicates on 96-well plates pre-coated with matrigel (10µg/ml) for 2 h at 37 °C. Non-adherent cells were aspirated, and the bound cells were washed in 1× PBS and fixed with 3.7% paraformaldehyde for 10min followed by staining with 0.4% crystal violet for 10min at room temperature. After several washes in 1× PBS, 30% acetic acid was added to release the dye and assayed on a microplate reader at 590nm (Molecular Devices Co., Sunnyvale, CA). For migration and invasion assays, cells were seeded on 8 µm-pored, 12-well transwells in triplicates (1×105/well) and incubated for 24 h at 37 °C and 5% CO2. The upper chamber was coated with matrigel (0.4mg/ml) 24 h prior to seeding of cells for the invasion assay. The migrated cells were fixed for 10min in 1× PBS containing 3.7% paraformaldehyde followed by 3 washes in 1× PBS after the non-migrated cells on the top surface of the filters were scraped off with cotton swabs. The filters were stained with 0.4% crystal violet for 10min at room temperature followed by detection of dye release as described above for the adhesion assay. The results were expressed as mean +/− SEM.
Transient transfection and Luciferase activity
As described previously, a confluent monolayer of HepG2 and Hep3B cells were transfected with 4µg of human OPN promoter construct (2.1KB) and/or 50ng/ml pRL-SV40 control plasmid plus Renilla luciferase gene by Lipofection as per manufacturer’s protocol [18]. After 48 h of transient transfection, cells were processed for luciferase activity analysis using a luminometer (TD-20/20, Tuner Designs, Sunnyvale, CA) according to manufacturer’s instructions. The data represents mean-fold luminescence of the triplicates from three independent experiments.
Chromatin Immunoprecipitation (ChIP) assay
HepG2 and Hep3B cells transiently transfected with the OPN reporter plasmid construct were fixed and immunoprecipitated using the EZ-ChIP assay Kit (Upstate Biotechnology, Charlottesville, VA) according to manufacturer’s protocol. Immunoprecipitation was performed on purified chromatin using the anti-RNA pol2 antibody provided in the kit. To determine the presence of the sequence of interest, purified DNA was subjected to Real-time PCR using specific primers (IDT) for OPN promoter region as described above.
mRNA decay assay
The decay rate of OPN mRNA was measured in an actinomycin D-stimulated time-course assay. Briefly, transcription was inhibited in HepG2 and Hep3B cells by culturing in the presence of actinomycin D (20µg/ml), and total RNA was extracted at selected time points there-after (0,4,8,16,24, and 36 h). The relative abundance of OPN mRNA was determined by standard Northern blot analysis as previously described [17] using (α-32P) dCTP labeled probe (300bps) constructed based on human OPN cDNA sequence (Gene bank accession number D14813).
Assays determining the length of the Poly (A) tail
Total RNA from HepG2 and Hep3B cells was isolated using Trizol Reagent (Invitrogen Co.). RNA-3’-linker 5’-Phos/ UAA UAC GAC UCA CUA UAC GGG CGC CGC UCA UGA GUC CA-3’ (IDT) was linked to the mRNA of HepG2 and Hep3B cells using T4 RNA Ligase Kit (Promega Co., Madison WI), according to the manufacturer’s protocol. First-Strand cDNA synthesis was performed using SuperScript -RT (Invitrogen Co.) and primer 5’-GCG CCC GTA TAG TGA GTC GTA TTA-3’ followed by PCR using forward primer 5’-GCG CCT CTA GAA AAG GAG AAA AAA TAC AATT-3’, and reverse primer 5’-GCG CCC GTA TAG TGA GTC GTA TTA-3’ (IDT). PCR products were resolved on 1.2% agarose gels and visualized using ethidium-bromide staining and detection.
OPN 5’-UTR and 3’-UTR reporter vector constructs
The OPN 5’-UTR full-length fragment (5’-UTR FL (10–157)) and mutated fragment generated by the deletion in OPN 5’-UTR from nt 10 – 57 (5’-UTR (58 –157) constructs were cloned into the HindIII/NcoI sites of the luciferase containing PGL3 basic vector (Promega Co., Madison WI) using primers with restriction enzyme sites for Hind III (forward primers) or NcoI (reverse primers). The reporter vector constructs OPN 5’-UTR (10–157) and OPN 5’-UTR (58–157) were generated using forward primers 5’-CCA AGC AAG CTT GTC TGC AGC AGC ATT TAA AT-3’ and 5’- CCA AGC AAG CTT GCA GCA GGA GGC AGA GCA C-3’ respectively (IDT). Reverse primer 5’- GCA CGC CAT GGT AGT GAG TTT TCC TTG GTC G-3’ (IDT) was used in the generation of both the fragments.
For OPN 3’-UTR reporter vector constructs, OPN 3’-UTR fragments were generated by PCR using forward primer 5’-GCG CCT CTA GAA AAG GAG AAA AAA TAC AATT-3’and reverse primer 5’-GGC CGT CTA GAT TAC ATT CAA GAT AAA AGA-3’ (IDT). Luc-OPN-3’-UTR fragment was generated by restriction enzyme Xba I digest followed by insertion at a down-stream site of luciferase containing PGL3 basic vector (Promega Co.). All vector constructs were verified by DNA sequencing.
Cytoplasmic extracts and Electrophoric mobility shift assays
Cytoplasmic protein extracts obtained from HepG2 and Hep3B cells using lysis buffer containing 1× PBS, 0.1% Triton X-100, 0.5% sodium deoxycholic acid and protein cocktail inhibitors were subjected to gel-shift assay analysis as described previously [19]. Briefly, 5’-UTR from the human OPN promoter sequence was synthesized and end-labeled with (α-32 P) ATP (2500 Ci/mmol) using T4 polynucleotide kinase (Promega Co.) followed by G-50 column purification. The final products were resolved on 6% poly-acrylamide gel electrophoresis using 0.5× Tris/borate EDTA buffer and visualized by autoradiography.
Statistical Analysis
Data are expressed as mean +/− SEM. Analysis was performed using Student’s T test. Values of p<0.05 were considered significant.
Results
Differential phenotypic and functional characteristics of HepG2 and Hep3B cell lines
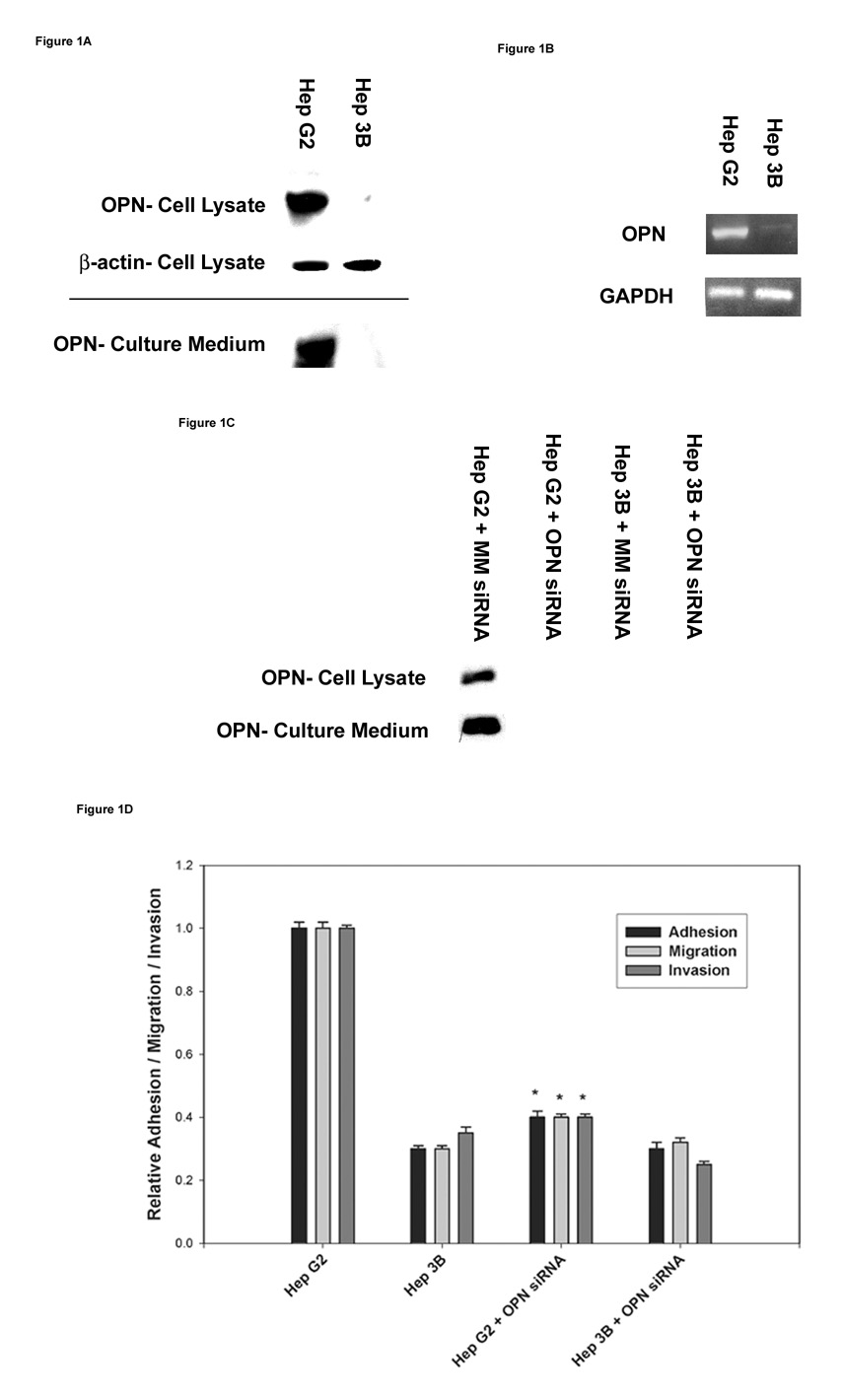
Expression of OPN protein and mRNA was determined in the two HCC cell lines. OPN protein expression was 12-fold greater in the cellular lysate and culture media (secreted protein) of HepG2 cells compared to Hep3B (p<0.01) in which OPN expression was virtually undetectable (Figure1A). OPN mRNA expression was enhanced more than 7-fold in HepG2 cells compared to Hep3B cells (p<0.01) (Figure 1B). OPN expression was suppressed specifically in the two HCC cell lines using siRNA techniques. As shown in Figure1C, OPN siRNA transfected into the HCC cell lines was able to significantly inhibit OPN protein expression with specificity as mismatch siRNA (MM siRNA) showed no effect on OPN expression. In correlation with the phenotypic characteristics, functional analysis revealed enhanced in vitro adhesion, migration and invasion behavior of HepG2 cells compared to Hep3B cells (Figure1D). Suppression of OPN expression by siRNA transfection in HepG2 cells significantly decreased in vitro adhesion, migration, and invasion properties. When MM siRNA was added to HepG2 or Hep3B, there was no change in in vitro adhesion, migration, and invasion properties. (Data not shown.) These data suggest that HepG2 and Hep3B cells possess distinct phenotypic and functional characteristics as determined by their differential OPN expression and metastatic behavior, respectively.
Figure 1. Detection of OPN mRNA and protein expression in the two HCC cell lines.
(A) Total cellular lysates and concentrated culture media from HepG2 and Hep3B cell lines were subjected to immunoblot analysis to determine OPN protein expression.
(B) RT-PCR analysis of OPN mRNA expression in HepG2 and Hep3B cell lines. GAPDH mRNA expression was used as a house keeping control gene.
(C) In vitro suppression of OPN expression by OPN siRNA transfection in the two HCC cell lines. HepG2 and Hep3B cell lines were transiently transfected with either MM siRNA (Mismatch siRNA) or OPN siRNA for 48 h. OPN protein expression was determined by western blot analysis. Blots (A–C) are representative of three separate experiments.
(D) Functional properties of the two HCC cell lines were assessed by their adhesion, migration and invasion characteristics as described in materials and methods. Correlation of OPN expression to its metastatic behavior in HepG2 and Hep3B cells was determined by specifically inhibiting OPN expression by standard OPN siRNA techniques. Data are expressed as mean ± SEM (* p< 0.05 vs. HepG2).
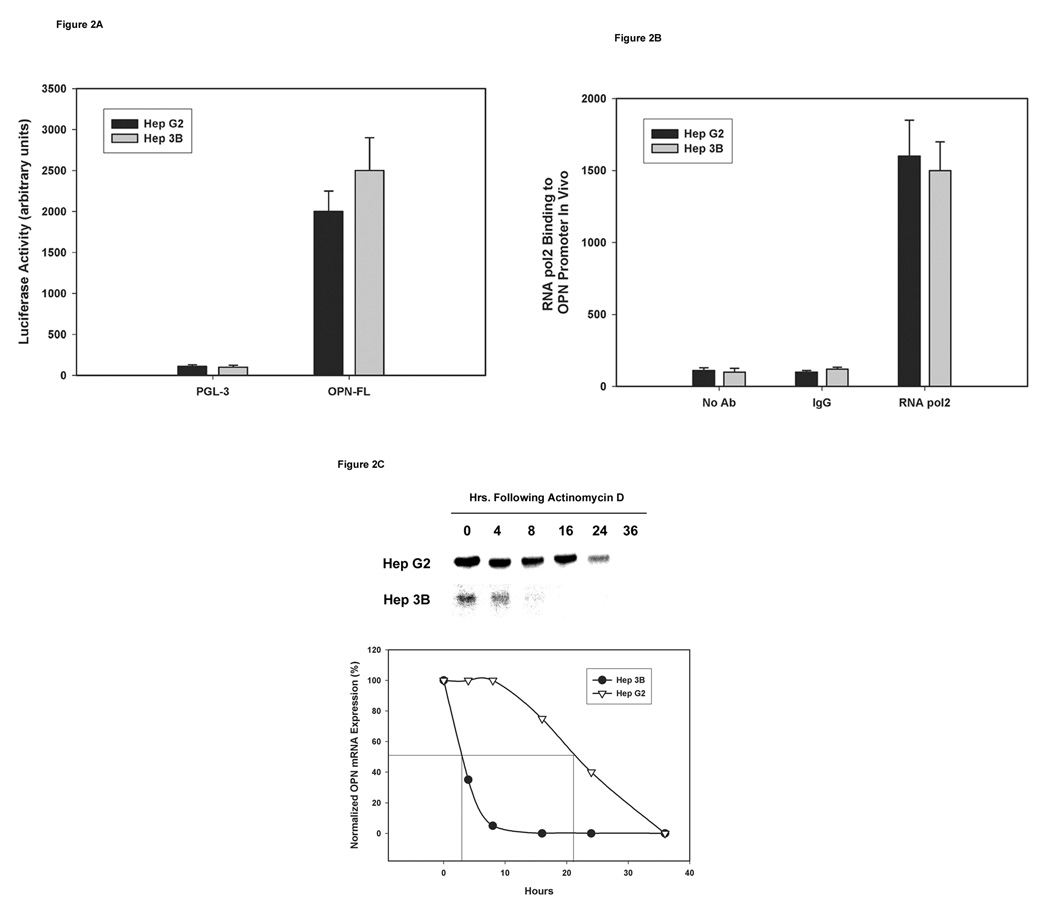
Assessment of promoter activity, transcriptional activation, and OPN mRNA half-life
To determine if there was a difference in the promoter activity between HepG2 and Hep3B cells, a luciferase expressing plasmid (PGL3) alone or in conjunction with the full-length human OPN promoter region (PGL3+OPN full-length) was transfected transiently and assayed for the luciferase activity after 48 h. Similar levels of promoter activity were observed in the two HCC cell lines (Figure 2A) suggesting intact transcriptional machinery in the two cell lines. We determined active transcription by examining RNA pol2 binding to OPN promoter region using ChIP assays. Transcriptional activity was comparable in HepG2 and Hep3B cell lines because the two HCC cell lines demonstrated similar levels of RNA pol2 binding to the OPN promoter construct as quantified by real-time PCR using specific OPN promoter primers (Figure 2B). These results suggest similar levels of active transcription of OPN mRNA in the two HCC cell lines. To assess OPN mRNA degradation, HepG2 and Hep3B cells were cultured in the medium with or without actinomycin D (20 µg/ml). At several time points after incubation (0, 4, 8, 16, 24, and 36 h), total RNA was extracted and subjected to Northern blot analysis to determine expression and quantification of OPN mRNA. Half-life of OPN mRNA in HepG2 cells was 21 ± 1 h whereas the half-life of OPN mRNA in Hep3B cells was only 3 ± 1 h (p<0.01) (Figure 2C). These data suggest that the difference in OPN mRNA expression between the HCC cell lines is neither due to the inactivation of the transcription machinery nor defects in transcriptional activation but is due to accelerated OPN mRNA degradation in Hep3B cells compared to HepG2 cells.
Figure 2. Determination of transcription activity and OPN mRNA half-life in the two HCC cell lines.
(A) HepG2 and Hep3B cells were transfected with firefly luciferase reporter plasmid vector PGL3 or PGL3 plus OPN full-length region along with 50ng/ml SV-40 expressing renilla luciferase for 48 h. Firefly luciferase activity was standardized against Renilla luciferase activity and was expressed as the mean fold luminescence for three independent experiments.
(B) Binding of RNA pol2 to the OPN promoter region was assessed using ChIP assays in HepG2 and Hep3B cells followed by quantification using real-time PCR analysis; data are expressed as mean ± SEM of three independent experiments.
(C) OPN mRNA half-life was determined in the two HCC cell lines in the presence of actinomycin D (20µg/ml) by Northern blot analysis after 0, 4, 8, 16, 24, and 36 h of incubation. Bands on the blots were quantified and presented in a graphical form. Blot is representative of three experiments. Graph combines data from the three experiments.
Contribution of the UTR’s to the differential regulation of OPN mRNA in HepG2 and Hep3B cells
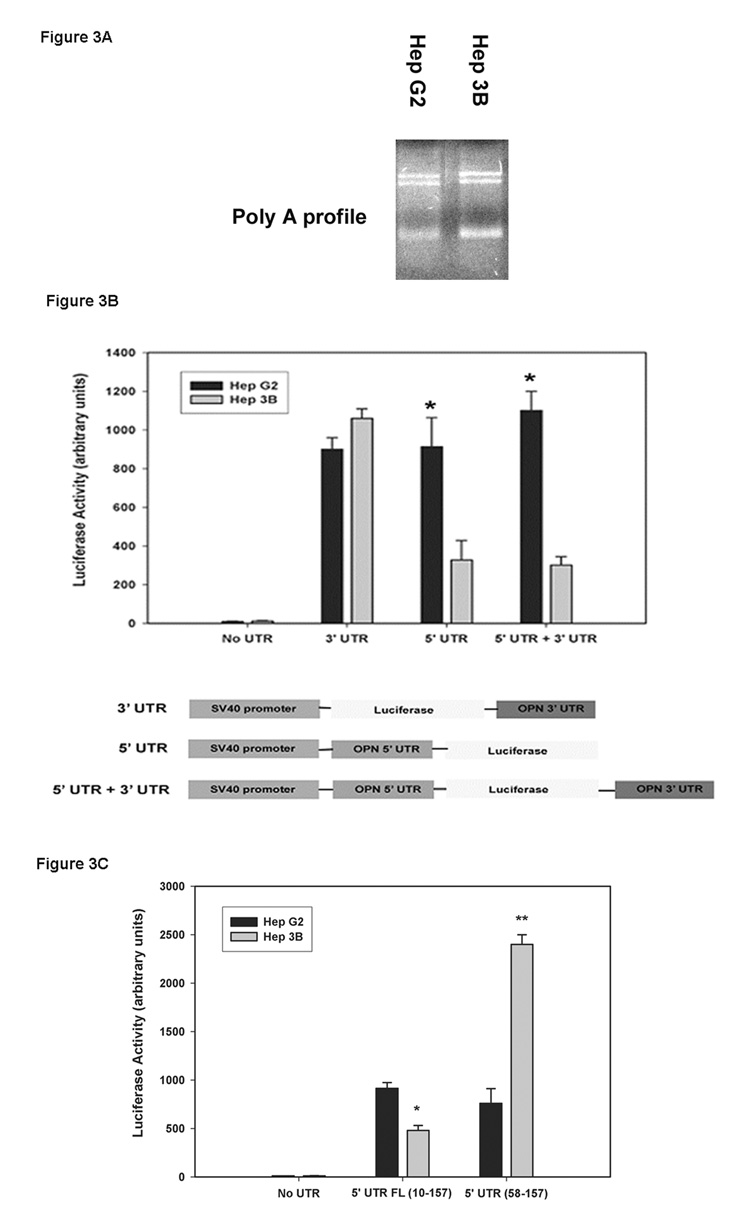
Because mRNA half-life can be regulated by the poly (A) tail, we sought to determine if there was a difference in poly (A) tail length contributing to the differential OPN stability between the two cell lines [20]. The poly (A) tail length in both HepG2 and Hep3B cells had same size distribution (Figure 3A). To determine if the post- transcription modifications in OPN 3’-, 5’- or both- UTR’s affect OPN mRNA stability in Hep3B cells, the effect of UTR fragments on steady state levels of mRNA was tested using a complete luciferase reporter system. Briefly, 3’-UTR and/or 5’-UTR segments were cloned separately into PGL3 vector with the SV40 promoter followed by transient transfection and assayed for transcription activity by luciferase reporter system. Luciferase acts as a surrogate for OPN in this setting. The data show that presence of OPN 3’- UTR had no effect on luciferase activity in both the HCC cell lines, but the presence of OPN 5’-UTR in Hep3B cells significantly decreased luciferase expression compared to HepG2 cells (Figure 3B). Expression of luciferase in the No-UTR group was subtracted out to normalize the data. To further evaluate the region of the 5’-UTR that could be affecting the transcriptional activity in Hep3B cells, a 5’-UTR mutant construct with deletion of fragment nt 10–57 was created and subjected to the similar experimental setting as described in Figure 3B. In HepG2, the deletion of nt 10–57 (5’-UTR (58–157)) in the OPN 5’-UTR had no effect on luciferase activity compared to full-length 5’-UTR (1–157). In contrast, the OPN 5’-UTR deletion (58 –157) construct in Hep3B cells significantly increased luciferase expression (Figure 3C). The augmented luciferase expression in Hep3B was 2.5 fold greater than HepG2 cells. Our data suggest that the potential cause for differential OPN mRNA expression between HepG2 and Hep3B cells lies in the nt 10–57 portion of the 5’-UTR, whereas the poly (A) tail and 3’-UTR have minimal or no effect.
Figure 3. Assessment of post-transcription modifications in the OPN 5’-UTR and 3’-UTR in HepG2 and Hep3B cells.
(A) Poly (A) tail profile of HepG2 and Hep3B OPN mRNA was determined by using oligo (dT) linker and T4 ligase followed by RT-PCR. PCR products were resolved on 1.2% agarose gel and visualized using ethidium-bromide staining and detection.
(B) Schematic representation of OPN 3’-UTR and/or 5’-UTR cloned into luciferase containing PGL-3 plasmid vector with SV-40 promoter. Luciferase activity was determined in HepG2 and Hep3B cells transiently transfected with the above constructs (n=3). Data are expressed as mean ± SEM. (* p<0.05 vs. Hep3B).
(C) Luciferase activity was performed on the HCC cell lines transiently transfected with full-length 5’-UTR as shown in Figure 3B or deletion of nt 10–57 fragment in the 5’-UTR (58–157) construct (n=3). Data are expressed as mean +/− SEM. *p values <0.05 were considered significant.
Binding of a cytoplasmic protein(s) to the 5’-UTR of OPN promoter in Hep3B cells
To determine whether cytoplasmic proteins extracted from HepG2 and Hep3B cells bind to the nt 10–57 fragment of the OPN 5’-UTR promoter region, RNA gel-shift assays were performed using an oligo-nucleotide sequence that corresponds to the segment of interest. Binding of cytoplasmic proteins to the nt 10–57 segment of the OPN promoter 5’-UTR was observed only with the lysates extracted from Hep3B cells and not from HepG2 cells (Figure 4A). Binding was ablated with the addition of 20-fold excess unlabeled probe with the labeled probe. No binding was found in the presence of labeled nonspecific probe. We evaluated the effect of deletion in OPN 5’-UTR on OPN protein expression. 5’-UTR full-length or deletion in 5’-UTR (58–157) segment along with OPN promoter region, OPN coding region and 3’-UTR were cloned into the pcDNA3.1/His expression vector followed by transient transfection and assessed for OPN protein expression by Western blots using OPN antibody. The data in Figure 4B show that the deletion of the 5’-UTR (58–157) had no effect on OPN expression in HepG2 cells but augmented OPN protein expression in Hep3B cells, suggesting that the nt 10–57 fragment of the OPN 5’-UTR regulates OPN mRNA stability and protein expression in Hep3B cells.
Discussion
Osteopontin is a sialic-acid rich, secreted glycoprotein expressed at low levels in a variety of human tissues but with higher expression in the liver, lymph nodes, testes, and spleen [4]. Increased OPN levels in the neoplasms compared to their corresponding normal tissues have been detected in several malignancies, including lung, kidney, liver, breast, gastric, ovarian, and prostate tissues [21, 22]. Recent data suggest a direct correlation between OPN over-expression and tumor metastasis for several malignancies [23, 24]. OPN is over-expressed in HCC and may ultimately become a marker for cancer metastasis [23]. Although the functional properties of OPN are well-studied, the molecular mechanisms underlying the enhanced regulation of OPN expression in malignancy (including HCC) have not been elucidated.
Our data demonstrated increased expression of OPN mRNA and protein in HepG2 but not Hep3B cells. Consistent with other reports that correlate OPN expression to metastatic behavior [16, 17], over-expression of OPN in HepG2 but not Hep3B also correlated with enhanced in vitro correlates of metastasis. To further evaluate the observed difference, we examined the transcriptional and post-transcriptional regulation of OPN mRNA expression in the two HCC cell lines.
In eukaryotic cells, initiation of messenger RNA transcription is carried-out by binding of RNA pol2 to the promoter region along with general transcription factors (TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) and accessory factors such as transcription activators and co-activators. The eukaryotic promoters are made of a combination of DNA sequences consisting of core promoter elements which bind RNA pol2, proximal promoter elements which bind transcriptional activators, and distal promoter elements which bind factors that modulate RNA pol2 activity [25, 26]. Promoter target sequences cannot be recognized directly by RNA pol2. Reports suggest that general transcription factors, mainly TFIID, guide binding and positioning of RNA pol2 to the TATA consensus sequence located 25 bp upstream of the transcription start site [27]. In this study, we show equivalent promoter activity in HepG2 and Hep3B cell lines (figure 2A) and similar rate of OPN mRNA synthesis by ChIP assay by isolating RNA pol2 bound DNA complex followed by RT-PCR using specific primers to further assess OPN mRNA expression.
Several modifications in the mRNA, such as 5’-capping, 3’end-cleavage/ polyadenylation, and RNA splicing, occur during and after its synthesis. The two major UTR modifications of mRNA that contribute directly to its stability are 5’-capping and 3’-polyadenylation. After the synthesis of about 25–30 nucleotides, 7-methylguanosine is added to the 5’-end of the newly synthesized RNA by a dimeric capping enzyme that binds RNA pol2 at the phosphorylated carboxyl-terminal tail domain [28]. Formation of this 5’-5’ m7 GTP containing cap in the 5’-end is involved in a variety of functions, including mRNA processing and export to the cytoplasm [29–31]. Capping is essential for initiation of translation in the cytoplasm and is also known to enhance mRNA stability [32]. In mammalian cells, capping helps protect mRNA from 5’-3’ exonuclease degradation [33, 34] . The other post-transcriptional modification is the 3’-end cleavage/polyadenylation. The mRNA synthesis is terminated at AAUAAA consensus sequence cleaved by an endonuclease generating a 3’-end to which 200–250 adenine residues are added by poly (A) polymerase. Binding of poly (A) binding protein (PABP) to the poly (A) tail is crucial in maintaining mRNA stability by protecting against exonuclease degradation [35, 36]. Recent data correlate longer poly (A) tails to enhanced stability because they are protected against nuclease digestion [37]. In addition, the translational efficiency of mRNA is associated directly with the changes in the length of the poly (A) tail during maturation. The nucleotide sequence of the OPN 5’- and 3’- UTR are the same in HepG2 and Hep3B cell lines (data not shown). Importantly, there was also no difference in the length of the poly (A) tails between the two HCC cell lines.
We then examined 5’-UTR and 3’-UTR regulated expression of a surrogate target gene, luciferase, in the two cell lines. Our results suggest that proteins bound to the nt 10–57 region of the OPN 5’-UTR in Hep3B cells contribute to the decreased stability of OPN mRNA when compared to HepG2 cells. Deletion of this region results in enhanced stability and expression of OPN mRNA. This OPN 5’-UTR-bound protein(s) has not yet been identified but has a molecular weight of approximately 54 Kd. Several nucleus-cap binding proteins have been reported to influence transport of mRNA to the cytoplasm [38], and polyadenylation at the 3’-end [39], and to effect pre-mRNA splicing [40]. In the cytoplasm, the complex of translation initiation factors (eIF4 protein) bind mRNA that helps binding of ribosomes and initiates translation. The eIF4 complex interacts physically with poly (A) binding protein/s (PABP) to initiate cap-dependent translation [41, 42]. Several data suggest an association between cap-bound mRNA degradative enzymes (such as DAN, a deadenylase) and the poly (A) tail that influences directly the stability of mRNA by stimulating deadenylation [43]. Therefore, the competition between transcription initiation factors and mRNA degrading enzymes may be involved in determining mRNA stability and further protein translation.
In conclusion, our results indicate that different repertoires of RNA binding proteins target the OPN 5’-UTR to regulate OPN mRNA stability and thus, alter OPN expression. These proteins are yet to be identified and are the subject of ongoing investigation. We further conclude that RNA stability is one mechanism which may regulate OPN expression in HCC cell lines to convey metastatic function.
Acknowledgments
This work was supported by NIH grants DK070642 (PCK), GM65113 (PCK), and AI44629 (PCK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Rittling SR, Chambers AF. Role of osteopontin in tumour progression. Br J Cancer. 2004;90:1877–1881. doi: 10.1038/sj.bjc.6601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber GF. The metastasis gene osteopontin: a candidate target for cancer therapy. Biochim Biophys Acta. 2001;1552:61–85. doi: 10.1016/s0304-419x(01)00037-3. [DOI] [PubMed] [Google Scholar]
- 4.Coppola D, Szabo M, Boulware D, Muraca P, Alsarraj M, Chambers AF, et al. Correlation of osteopontin protein expression and pathological stage across a wide variety of tumor histologies. Clin Cancer Res. 2004;10:184–190. doi: 10.1158/1078-0432.ccr-1405-2. [DOI] [PubMed] [Google Scholar]
- 5.Chambers AF, Wilson SM, Kerkvliet N, O'Malley FP, Harris JF, Casson AG. Osteopontin expression in lung cancer. Lung cancer (Amsterdam, Netherlands) 1996;15:311–323. doi: 10.1016/0169-5002(95)00595-1. [DOI] [PubMed] [Google Scholar]
- 6.Oates AJ, Barraclough R, Rudland PS. The role of osteopontin in tumorigenesis and metastasis. Invasion Metas. 1997;17:1–15. [PubMed] [Google Scholar]
- 7.Thalmann GN, Sikes RA, Devoll RE, Kiefer JA, Markwalder R, Klima I, et al. Osteopontin: possible role in prostate cancer progression. Clin Cancer Res. 1999;5:2271–2277. [PubMed] [Google Scholar]
- 8.Tuck AB, O'Malley FP, Singhal H, Tonkin KS, Harris JF, Bautista D, et al. Osteopontin and p53 expression are associated with tumor progression in a case of synchronous, bilateral, invasive mammary carcinomas. Arch Pathol Lab Med. 1997;121:578–584. [PubMed] [Google Scholar]
- 9.Fedarko NS, Jain A, Karadag A, Van Eman MR, Fisher LW. Elevated serum bone sialoprotein and osteopontin in colon, breast, prostate, and lung cancer. Clin Cancer Res. 2001;7:4060–4066. [PubMed] [Google Scholar]
- 10.Kim JH, Skates SJ, Uede T, Wong KK, Schorge JO, Feltmate CM, et al. Osteopontin as a potential diagnostic biomarker for ovarian cancer. JAMA. 2002;287:1671–1679. doi: 10.1001/jama.287.13.1671. [DOI] [PubMed] [Google Scholar]
- 11.Singhal H, Bautista DS, Tonkin KS, O'Malley FP, Tuck AB, Chambers AF, et al. Elevated plasma osteopontin in metastatic breast cancer associated with increased tumor burden and decreased survival. Clin Cancer Res. 1997;3:605–611. [PubMed] [Google Scholar]
- 12.Budhu AS, Zipser B, Forgues M, Ye QH, Sun Z, Wang XW. The molecular signature of metastases of human hepatocellular carcinoma. Oncology. 2005;69:23–27. doi: 10.1159/000086628. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, Ye QH, Ren N, Zhao L, Wang YF, Wu X, et al. The prognostic significance of preoperative plasma levels of osteopontin in patients with hepatocellular carcinoma. J Cancer Res Clin Onc. 2006;132:709–717. doi: 10.1007/s00432-006-0119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kon S, Maeda M, Segawa T, Hagiwara Y, Horikoshi Y, Chikuma S, et al. Antibodies to different peptides in osteopontin reveal complexities in the various secreted forms. J Cell Biochem. 2000;77:487–498. [PubMed] [Google Scholar]
- 15.Crawford HC, Matrisian LM, Liaw L. Distinct roles of osteopontin in host defense activity and tumor survival during squamous cell carcinoma progression in vivo. Cancer Res. 1998;58:5206–5215. [PubMed] [Google Scholar]
- 16.Wai PY, Mi Z, Guo H, Sarraf-Yazdi S, Gao C, Wei J, et al. Osteopontin silencing by small interfering RNA suppresses in vitro and in vivo CT26 murine colon adenocarcinoma metastasis. Carcinogenesis. 2005;26:741–751. doi: 10.1093/carcin/bgi027. [DOI] [PubMed] [Google Scholar]
- 17.Mi Z, Guo H, Wai PY, Gao C, Wei J, Kuo PC. Differential osteopontin expression in phenotypically distinct subclones of murine breast cancer cells mediates metastatic behavior. J Biol Chem. 2004;279:46659–46667. doi: 10.1074/jbc.M407952200. [DOI] [PubMed] [Google Scholar]
- 18.Guo H, Marroquin CE, Wai PY, Kuo PC. Nitric oxide-dependent osteopontin expression induces metastatic behavior in HepG2 cells. Dig Dis Sci. 2005;50:1288–1298. doi: 10.1007/s10620-005-2775-6. [DOI] [PubMed] [Google Scholar]
- 19.Gao C, Guo H, Mi Z, Wai PY, Kuo PC. Transcriptional regulatory functions of heterogeneous nuclear ribonucleoprotein-U and -A/B in endotoxin-mediated macrophage expression of osteopontin. J Immunol. 2005;175:523–530. doi: 10.4049/jimmunol.175.1.523. [DOI] [PubMed] [Google Scholar]
- 20.Kuraishi T, Sun Y, Aoki F, Imakawa K, Sakai S. The poly(A) tail length of casein mRNA in the lactating mammary gland changes depending upon the accumulation and removal of milk. Biochem J. 2000;347:579–583. doi: 10.1042/0264-6021:3470579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furger KA, Menon RK, Tuck AB, Bramwell VH, Chambers AF. The functional and clinical roles of osteopontin in cancer and metastasis. Curr Mol Med. 2001;1:621–632. doi: 10.2174/1566524013363339. [DOI] [PubMed] [Google Scholar]
- 22.Tuck AB, Hota C, Chambers AF. Osteopontin(OPN)-induced increase in human mammary epithelial cell invasiveness is urokinase (uPA)-dependent. Breast Cancer Res Treat. 2001;70:197–204. doi: 10.1023/a:1013095329825. [DOI] [PubMed] [Google Scholar]
- 23.Agrawal D, Chen T, Irby R, Quackenbush J, Chambers AF, Szabo M, et al. Osteopontin identified as lead marker of colon cancer progression, using pooled sample expression profiling. J Natl Cancer Inst. 2002;94:513–521. doi: 10.1093/jnci/94.7.513. [DOI] [PubMed] [Google Scholar]
- 24.Yeatman TJ, Chambers AF. Osteopontin and colon cancer progression. Clin Exp Metastasis. 2003;20:85–90. doi: 10.1023/a:1022502805474. [DOI] [PubMed] [Google Scholar]
- 25.Zawel L, Reinberg D. Initiation of transcription by RNA polymerase II: a multi-step process. Prog Nucleic Acid Res Mol Biol. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]
- 26.Roeder RG. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem Sci. 1991;16:402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- 27.Nikolov DB, Burley SK. RNA polymerase II transcription initiation: a structural view. Proc Natl Acad Sci U S A. 1997;94:15–22. doi: 10.1073/pnas.94.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muthukrishnan S, Both GW, Furuichi Y, Shatkin AJ. 5'-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33–37. doi: 10.1038/255033a0. [DOI] [PubMed] [Google Scholar]
- 29.Konarska MM, Padgett RA, Sharp PA. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 30.Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj IW. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edery I, Sonenberg N. Cap-dependent RNA splicing in a HeLa nuclear extract. Proc Natl Acad Sci U S A. 1985;82:7590–7594. doi: 10.1073/pnas.82.22.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furuichi Y, LaFiandra A, Shatkin AJ. 5'-Terminal structure and mRNA stability. Nature. 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 33.Walther TN, Wittop Koning TH, Schumperli D, Muller B. A 5'-3' exonuclease activity involved in forming the 3' products of histone pre-mRNA processing in vitro. RNA. 1998;4:1034–1046. doi: 10.1017/s1355838298971771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sachs AB. Messenger RNA degradation in eukaryotes. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 35.Ford LP, Bagga PS, Wilusz J. The poly(A) tail inhibits the assembly of a 3'-to-5' exonuclease in an in vitro RNA stability system. Mol Cell Biol. 1997;17:398–406. doi: 10.1128/mcb.17.1.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coller JM, Gray NK, Wickens MP. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 1998;12:3226–3235. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagaoka K, Takahara K, Tanaka K, Yoshida H, Steinman RM, Saitoh S, et al. Association of SIGNR1 with TLR4-MD-2 enhances signal transduction by recognition of LPS in gram-negative bacteria. Int Immunol. 2005;17:827–836. doi: 10.1093/intimm/dxh264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 199;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 39.Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM. Participation of the nuclear cap binding complex in pre-mRNA 3' processing. Proc Natl Acad Sci USA. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 41.Tarun SZ, Jr, Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao M, Fritz DT, Ford LP, Wilusz J. Interaction between a poly(A)-specific ribonuclease and the 5' cap influences mRNA deadenylation rates in vitro. Mol Cell. 2000;5:479–488. doi: 10.1016/s1097-2765(00)80442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]