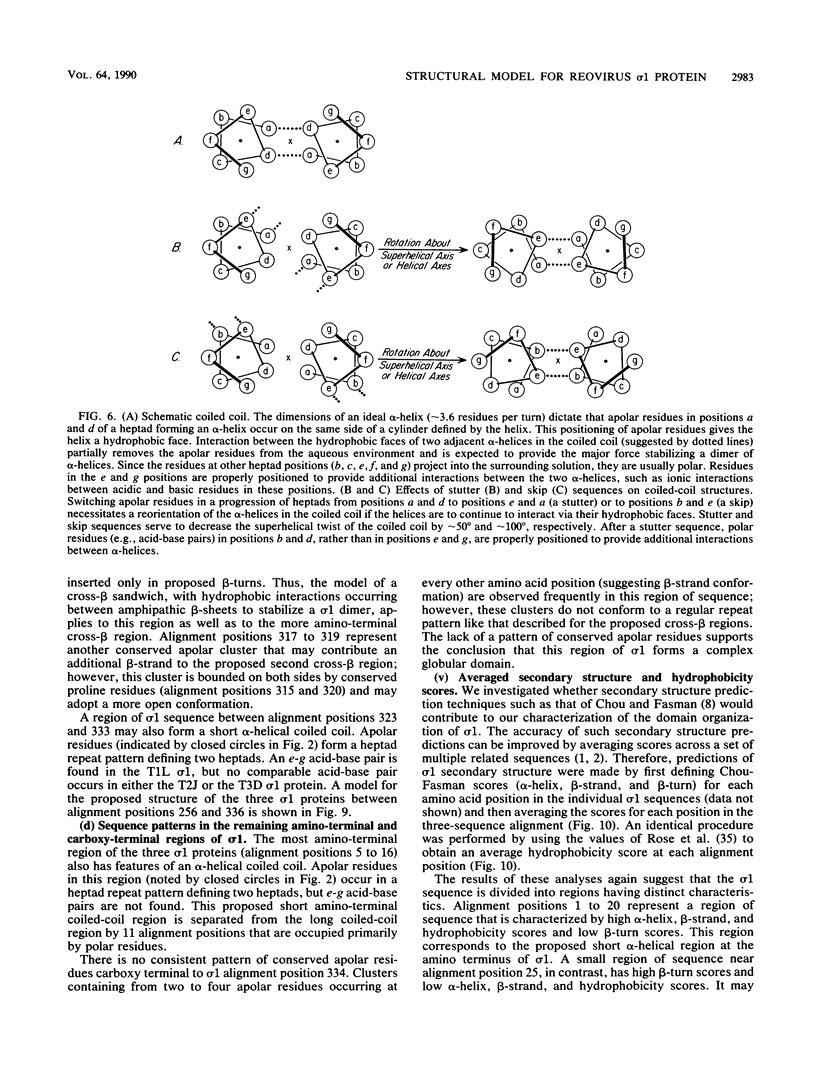
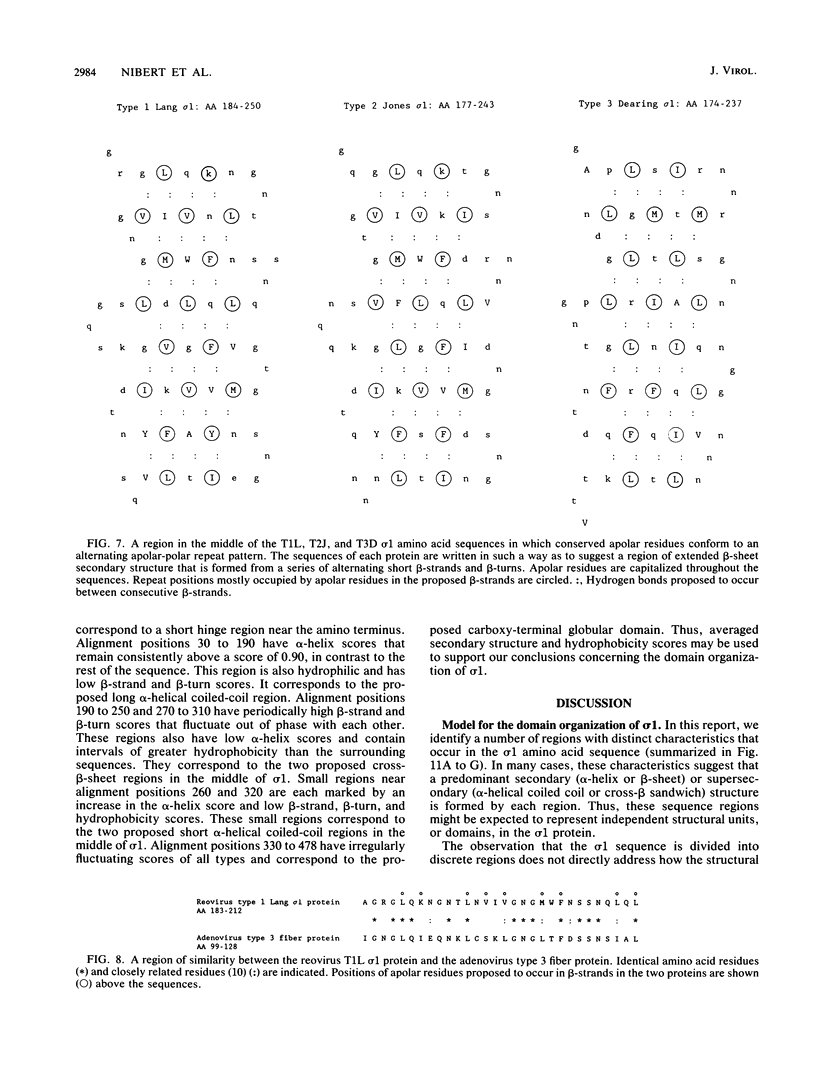
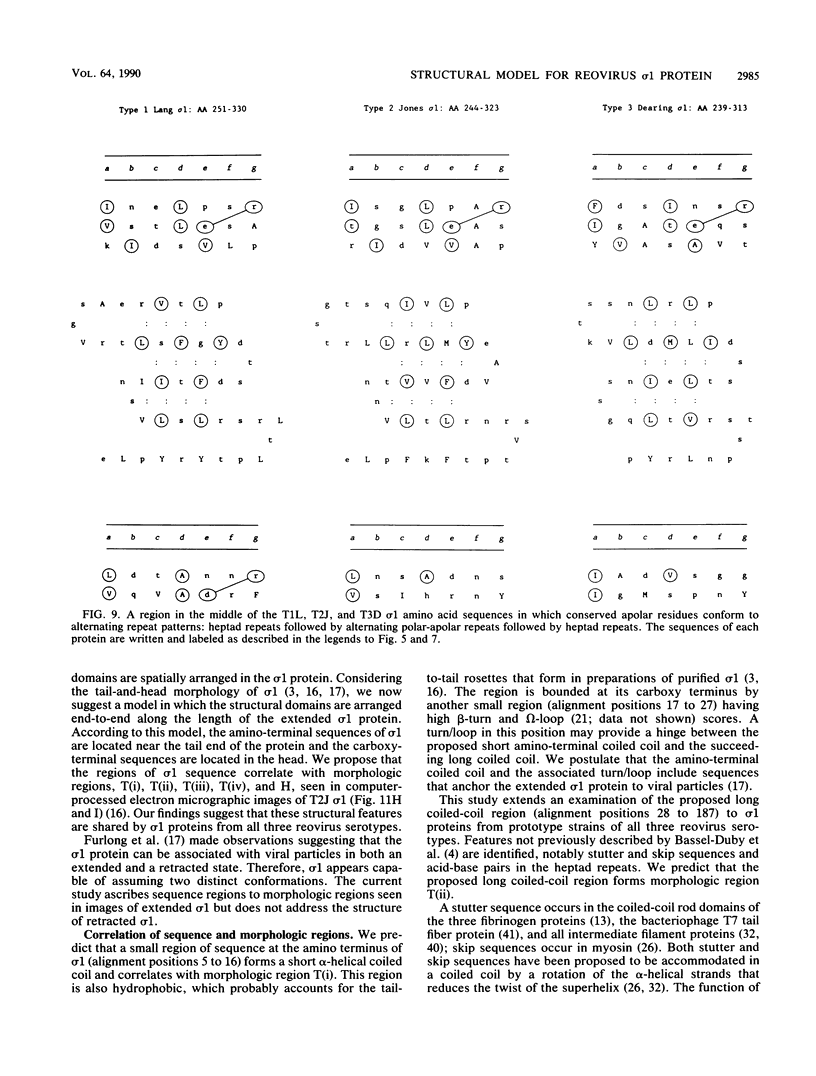
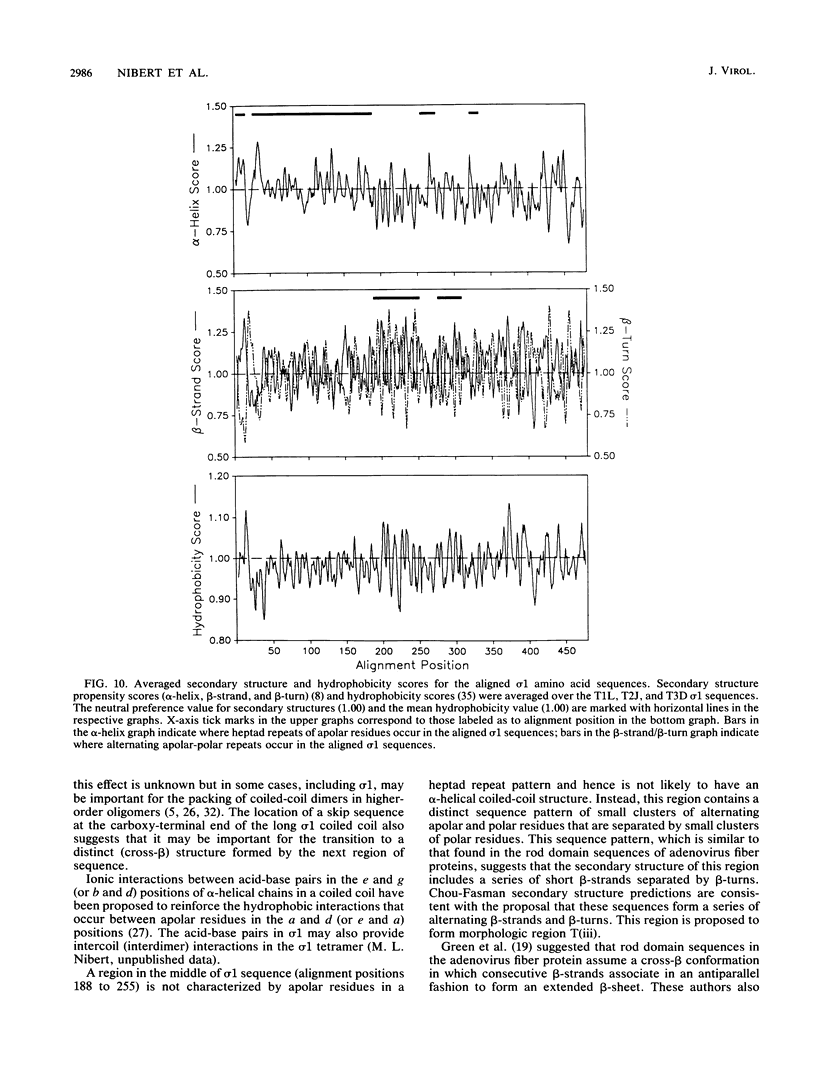
Abstract
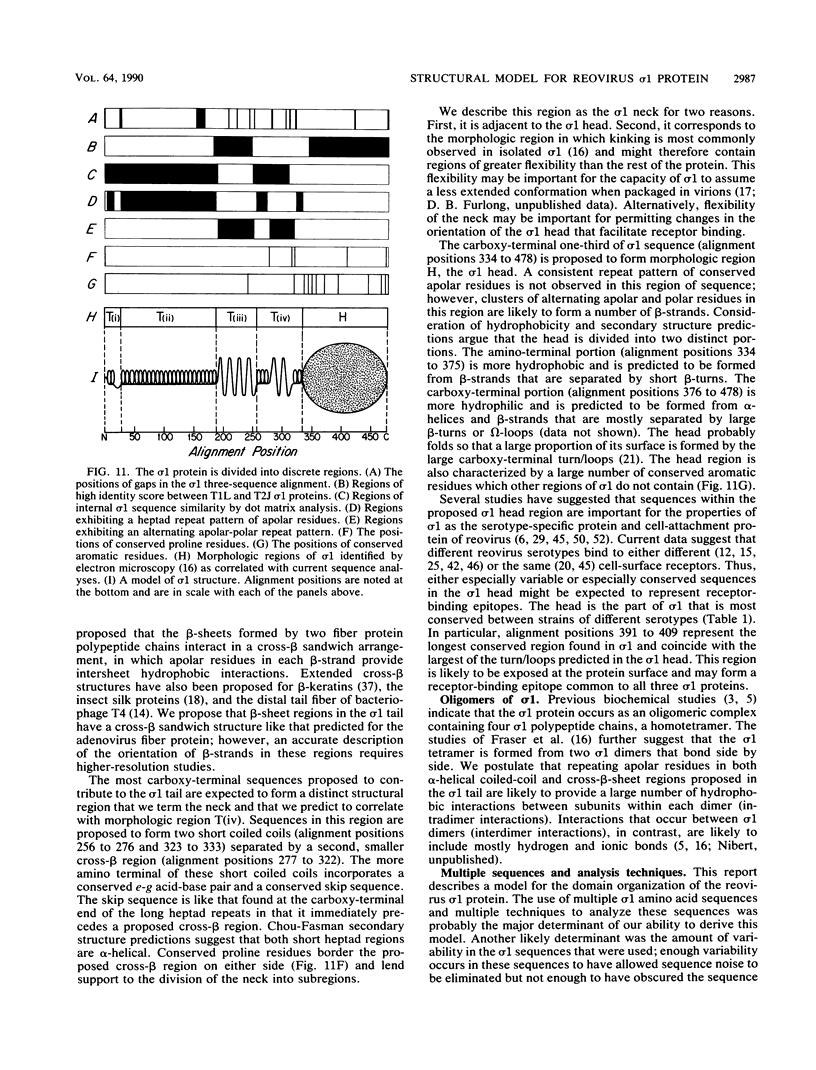
This report describes a model for the structure of the reovirus cell-attachment protein sigma 1. S1 gene nucleotide sequences were determined for prototype strains of the three serotypes of mammalian reoviruses. Deduced amino acid sequences of the S1-encoded sigma 1 proteins were then compared in order to identify conserved features of these sequences. Discrete regions in the amino-terminal two-thirds of sigma 1 sequence share characteristics with the fibrous domains of other cellular and viral proteins. Most of the amino-terminal one-third of sigma 1 sequence is predicted to form an alpha-helical coiled coil like that of myosin. The middle one-third of sigma 1 sequence appears more heterogeneous; it is predicted to form a large region of beta-sheet that is followed by a region which contains two short alpha-helical coiled coils separated by a smaller region of beta-sheet. The two beta-sheet regions are each proposed to form a cross-beta sandwich like that suggested for the rod domain of the adenovirus fiber protein (N. M. Green, N. G. Wrigley, W. C. Russell, S. R. Martin, and A. D. McLachlan, EMBO J. 2:1357-1365, 1983). The remaining carboxy-terminal one-third of sigma 1 sequence is predicted to form a structurally complex globular domain. A model is suggested in which the discrete regions of sigma 1 sequence are ascribed to morphologic regions seen in computer-processed electron micrographic images of the protein (R. D. B. Fraser, D. B. Furlong, B. L. Trus, M. L. Nibert, B. N. Fields, and A. C. Steven, J. Virol. 64:2990-3000, 1990.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P., Rao J. K. Prediction of protein structure. Methods Enzymol. 1986;130:185–207. doi: 10.1016/0076-6879(86)30012-0. [DOI] [PubMed] [Google Scholar]
- Argos P., Schwarz J., Schwarz J. An assessment of protein secondary structure prediction methods based on amino acid sequence. Biochim Biophys Acta. 1976 Aug 9;439(2):261–273. doi: 10.1016/0005-2795(76)90062-3. [DOI] [PubMed] [Google Scholar]
- Banerjea A. C., Brechling K. A., Ray C. A., Erikson H., Pickup D. J., Joklik W. K. High-level synthesis of biologically active reovirus protein sigma 1 in a mammalian expression vector system. Virology. 1988 Dec;167(2):601–612. [PubMed] [Google Scholar]
- Bassel-Duby R., Jayasuriya A., Chatterjee D., Sonenberg N., Maizel J. V., Jr, Fields B. N. Sequence of reovirus haemagglutinin predicts a coiled-coil structure. 1985 May 30-Jun 5Nature. 315(6018):421–423. doi: 10.1038/315421a0. [DOI] [PubMed] [Google Scholar]
- Bassel-Duby R., Nibert M. L., Homcy C. J., Fields B. N., Sawutz D. G. Evidence that the sigma 1 protein of reovirus serotype 3 is a multimer. J Virol. 1987 Jun;61(6):1834–1841. doi: 10.1128/jvi.61.6.1834-1841.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel-Duby R., Spriggs D. R., Tyler K. L., Fields B. N. Identification of attenuating mutations on the reovirus type 3 S1 double-stranded RNA segment with a rapid sequencing technique. J Virol. 1986 Oct;60(1):64–67. doi: 10.1128/jvi.60.1.64-67.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashdollar L. W., Chmelo R. A., Wiener J. R., Joklik W. K. Sequences of the S1 genes of the three serotypes of reovirus. Proc Natl Acad Sci U S A. 1985 Jan;82(1):24–28. doi: 10.1073/pnas.82.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Chroboczek J., Jacrot B. The sequence of adenovirus fiber: similarities and differences between serotypes 2 and 5. Virology. 1987 Dec;161(2):549–554. doi: 10.1016/0042-6822(87)90150-4. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O., Barker W. C., Hunt L. T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichter M. A., Weiner H. L. Infection of neuronal cell cultures with reovirus mimics in vitro patterns of neurotropism. Ann Neurol. 1984 Nov;16(5):603–610. doi: 10.1002/ana.410160512. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F., Goldbaum D. M., Doolittle L. R. Designation of sequences involved in the "coiled-coil" interdomainal connections in fibrinogen: constructions of an atomic scale model. J Mol Biol. 1978 Apr 5;120(2):311–325. doi: 10.1016/0022-2836(78)90070-0. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Goldberg E. B., Crowther R. A. The distal half of the tail fibre of bacteriophage T4. Rigidly linked domains and cross-beta structure. J Mol Biol. 1979 Jul 25;132(1):101–113. doi: 10.1016/0022-2836(79)90498-4. [DOI] [PubMed] [Google Scholar]
- Epstein R. L., Powers M. L., Rogart R. B., Weiner H. L. Binding of 125I-labeled reovirus to cell surface receptors. Virology. 1984 Feb;133(1):46–55. doi: 10.1016/0042-6822(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Fraser R. D., Furlong D. B., Trus B. L., Nibert M. L., Fields B. N., Steven A. C. Molecular structure of the cell-attachment protein of reovirus: correlation of computer-processed electron micrographs with sequence-based predictions. J Virol. 1990 Jun;64(6):2990–3000. doi: 10.1128/jvi.64.6.2990-3000.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong D. B., Nibert M. L., Fields B. N. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol. 1988 Jan;62(1):246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes A. J., Parker K. D., Atkins E. D., Beighton E. "Cross-beta" conformation in proteins. J Mol Biol. 1968 Mar 14;32(2):343–358. doi: 10.1016/0022-2836(68)90014-4. [DOI] [PubMed] [Google Scholar]
- Green N. M., Wrigley N. G., Russell W. C., Martin S. R., McLachlan A. D. Evidence for a repeating cross-beta sheet structure in the adenovirus fibre. EMBO J. 1983;2(8):1357–1365. doi: 10.1002/j.1460-2075.1983.tb01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. W., Hayes E. C., Joklik W. K. Protein sigma 1 is the reovirus cell attachment protein. Virology. 1981 Jan 15;108(1):156–163. doi: 10.1016/0042-6822(81)90535-3. [DOI] [PubMed] [Google Scholar]
- Leszczynski J. F., Rose G. D. Loops in globular proteins: a novel category of secondary structure. Science. 1986 Nov 14;234(4778):849–855. doi: 10.1126/science.3775366. [DOI] [PubMed] [Google Scholar]
- Lim V. I. Algorithms for prediction of alpha-helical and beta-structural regions in globular proteins. J Mol Biol. 1974 Oct 5;88(4):873–894. doi: 10.1016/0022-2836(74)90405-7. [DOI] [PubMed] [Google Scholar]
- Lim W. A., Sauer R. T. Alternative packing arrangements in the hydrophobic core of lambda repressor. Nature. 1989 May 4;339(6219):31–36. doi: 10.1038/339031a0. [DOI] [PubMed] [Google Scholar]
- Maizel J. V., Jr, Lenk R. P. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maratos-Flier E., Kahn C. R., Spriggs D. R., Fields B. N. Specific plasma membrane receptors for reovirus on rat pituitary cells in culture. J Clin Invest. 1983 Aug;72(2):617–621. doi: 10.1172/JCI111010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan A. D., Karn J. Periodic features in the amino acid sequence of nematode myosin rod. J Mol Biol. 1983 Mar 15;164(4):605–626. doi: 10.1016/0022-2836(83)90053-0. [DOI] [PubMed] [Google Scholar]
- McLachlan A. D., Stewart M. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J Mol Biol. 1975 Oct 25;98(2):293–304. doi: 10.1016/s0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- Munemitsu S. M., Atwater J. A., Samuel C. E. Biosynthesis of reovirus-specified polypeptides. Molecular cDNA cloning and nucleotide sequence of the reovirus serotype 1 Lang strain bicistronic s1 mRNA which encodes the minor capsid polypeptide sigma 1a and the nonstructural polypeptide sigma 1bNS. Biochem Biophys Res Commun. 1986 Oct 30;140(2):508–514. doi: 10.1016/0006-291x(86)90761-8. [DOI] [PubMed] [Google Scholar]
- Nagata L., Masri S. A., Pon R. T., Lee P. W. Analysis of functional domains on reovirus cell attachment protein sigma 1 using cloned S1 gene deletion mutants. Virology. 1987 Sep;160(1):162–168. doi: 10.1016/0042-6822(87)90056-0. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Onodera T., Toniolo A., Ray U. R., Jenson A. B., Knazek R. A., Notkins A. L. Virus-induced diabetes mellitus. XX. Polyendocrinopathy and autoimmunity. J Exp Med. 1981 Jun 1;153(6):1457–1473. doi: 10.1084/jem.153.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSEN L. Serologic grouping of reoviruses by hemagglutination-inhibition. Am J Hyg. 1960 Mar;71:242–249. doi: 10.1093/oxfordjournals.aje.a120107. [DOI] [PubMed] [Google Scholar]
- RUDALL K. M. The proteins of the mammalian epidermis. Adv Protein Chem. 1952;7:253–290. doi: 10.1016/s0065-3233(08)60020-0. [DOI] [PubMed] [Google Scholar]
- Ramig R. F., Cross R. K., Fields B. N. Genome RNAs and polypeptides of reovirus serotypes 1, 2, and 3. J Virol. 1977 Jun;22(3):726–733. doi: 10.1128/jvi.22.3.726-733.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risler J. L., Delorme M. O., Delacroix H., Henaut A. Amino acid substitutions in structurally related proteins. A pattern recognition approach. Determination of a new and efficient scoring matrix. J Mol Biol. 1988 Dec 20;204(4):1019–1029. doi: 10.1016/0022-2836(88)90058-7. [DOI] [PubMed] [Google Scholar]
- Rose G. D., Geselowitz A. R., Lesser G. J., Lee R. H., Zehfus M. H. Hydrophobicity of amino acid residues in globular proteins. Science. 1985 Aug 30;229(4716):834–838. doi: 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]
- Signäs C., Akusjärvi G., Pettersson U. Adenovirus 3 fiber polypeptide gene: implications for the structure of the fiber protein. J Virol. 1985 Feb;53(2):672–678. doi: 10.1128/jvi.53.2.672-678.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs D. R., Bronson R. T., Fields B. N. Hemagglutinin variants of reovirus type 3 have altered central nervous system tropism. Science. 1983 Apr 29;220(4596):505–507. doi: 10.1126/science.6301010. [DOI] [PubMed] [Google Scholar]
- Steinert P. M., Steven A. C., Roop D. R. The molecular biology of intermediate filaments. Cell. 1985 Sep;42(2):411–420. doi: 10.1016/0092-8674(85)90098-4. [DOI] [PubMed] [Google Scholar]
- Steven A. C., Trus B. L., Maizel J. V., Unser M., Parry D. A., Wall J. S., Hainfeld J. F., Studier F. W. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol. 1988 Mar 20;200(2):351–365. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- Tardieu M., Weiner H. L. Viral receptors on isolated murine and human ependymal cells. Science. 1982 Jan 22;215(4531):419–421. doi: 10.1126/science.6276976. [DOI] [PubMed] [Google Scholar]
- Tyler K. L., McPhee D. A., Fields B. N. Distinct pathways of viral spread in the host determined by reovirus S1 gene segment. Science. 1986 Aug 15;233(4765):770–774. doi: 10.1126/science.3016895. [DOI] [PubMed] [Google Scholar]
- Valentine R. C., Pereira H. G. Antigens and structure of the adenovirus. J Mol Biol. 1965 Aug;13(1):13–20. doi: 10.1016/s0022-2836(65)80076-6. [DOI] [PubMed] [Google Scholar]
- Verdin E. M., King G. L., Maratos-Flier E. Characterization of a common high-affinity receptor for reovirus serotypes 1 and 3 on endothelial cells. J Virol. 1989 Mar;63(3):1318–1325. doi: 10.1128/jvi.63.3.1318-1325.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner H. L., Ault K. A., Fields B. N. Interaction of reovirus with cell surface receptors. I. Murine and human lymphocytes have a receptor for the hemagglutinin of reovirus type 3. J Immunol. 1980 May;124(5):2143–2148. [PubMed] [Google Scholar]
- Weiner H. L., Drayna D., Averill D. R., Jr, Fields B. N. Molecular basis of reovirus virulence: role of the S1 gene. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5744–5748. doi: 10.1073/pnas.74.12.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner H. L., Fields B. N. Neutralization of reovirus: the gene responsible for the neutralization antigen. J Exp Med. 1977 Nov 1;146(5):1305–1310. doi: 10.1084/jem.146.5.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner H. L., Ramig R. F., Mustoe T. A., Fields B. N. Identification of the gene coding for the hemagglutinin of reovirus. Virology. 1978 May 15;86(2):581–584. doi: 10.1016/0042-6822(78)90099-5. [DOI] [PubMed] [Google Scholar]
- Williams W. V., Guy H. R., Rubin D. H., Robey F., Myers J. N., Kieber-Emmons T., Weiner D. B., Greene M. I. Sequences of the cell-attachment sites of reovirus type 3 and its anti-idiotypic/antireceptor antibody: modeling of their three-dimensional structures. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6488–6492. doi: 10.1073/pnas.85.17.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J. L., Kauffman R. S., Finberg R., Dambrauskas R., Fields B. N., Trier J. S. Determinants of reovirus interaction with the intestinal M cells and absorptive cells of murine intestine. Gastroenterology. 1983 Aug;85(2):291–300. [PubMed] [Google Scholar]
- Yeung M. C., Lim D., Duncan R., Shahrabadi M. S., Cashdollar L. W., Lee P. W. The cell attachment proteins of type 1 and type 3 reovirus are differentially susceptible to trypsin and chymotrypsin. Virology. 1989 May;170(1):62–70. doi: 10.1016/0042-6822(89)90352-8. [DOI] [PubMed] [Google Scholar]


