Abstract
PDZK1 is a four-PDZ domain-containing scaffold protein that, via its first PDZ domain (PDZ1), binds to the C terminus of the high density lipoprotein (HDL) receptor scavenger receptor, class B, type I (SR-BI). Abolishing PDZK1 expression in PDZK1 knock-out (KO) mice leads to a post-transcriptional, tissue-specific decrease in SR-BI protein level and an increase in total plasma cholesterol carried in abnormally large HDL particles. Here we show that, although hepatic overexpression of PDZK1 restored normal SR-BI protein abundance and function in PDZK1 KO mice, hepatic overexpression of only the PDZ1 domain was not sufficient to restore normal SR-BI function. In wild-type mice, overexpression of the PDZ1 domain overcame the activity of the endogenous hepatic PDZK1, resulting in a 75% reduction in hepatic SR-BI protein levels and intracellular mislocalization of the remaining SR-BI. As a consequence, the plasma lipoproteins in PDZ1 transgenic mice resembled those in PDZK1 KO mice (hypercholesterolemia due to large HDL). These results indicate that the PDZ1 domain can control the abundance and localization, and therefore the function, of hepatic SR-BI and that structural features of PDZK1 in addition to its SR-BI-binding PDZ1 domain are required for normal hepatic SR-BI regulation.
The high density lipoprotein (HDL)3 receptor SR-BI (scavenger receptor, class B, type I) plays a key role in lipoprotein metabolism (1) by mediating the cellular uptake of cholesteryl esters from HDL and other lipoproteins into cells (2–7) via a mechanism called selective lipid uptake (3, 8–10). SR-BI also mediates bidirectional flux of unesterified cholesterol between cells and lipoproteins (11–14).
Most in vivo analyses of the physiological function of SR-BI have been conducted in mice. SR-BI is highly expressed in the liver and steroidogenic tissues (adrenal gland, ovary, and testis), which exhibit the highest levels of HDL cholesterol uptake (3). Experimental manipulations of murine SR-BI expression (e.g. hepatic overexpression, homozygous null gene knock-out) can lead to changes in total plasma cholesterol levels, the ratio of plasma unesterified to total cholesterol, HDL structure, biliary cholesterol secretion, the amounts of cholesterol stored in steroidogenic tissues, and susceptibility to atherosclerosis (15–27). Homozygous null SR-BI knock-out mice exhibit abnormally elevated (∼2.2-fold) plasma cholesterol in large HDL particles with a unesterified cholesterol/total cholesterol ratio roughly double that of wild-type (WT) mice (19, 52). This dyslipidemia is associated with female infertility and defects in the maturation and/or structure as well as reductions in the survival times of red blood cells (21, 28, 29, 30).
To date, only one intracellular protein has been shown to interact directly with and influence the function of SR-BI (31). This protein, PDZK1, regulates SR-BI expression in a tissue-specific, post-transcriptional manner (32). PDZK1 is a scaffold protein containing four PDZ protein interaction domains (Fig. 1A, top), which bind to the carboxyl termini of a number of membrane transporter proteins, including ion channels (e.g. CFTR) and cell surface receptors (33, 34). The activity of PDZK1 can be modulated by phosphorylation near its C terminus (35). Its most N-terminal PDZ domain, PDZ1, binds to the carboxyl terminus of SR-BI (31, 36) but apparently not to a minor, alternatively spliced form of SR-BI called SR-BII, whose 39-residue C-terminal sequence (encoded by an alternatively spliced exon) differs from that of SR-BI (32, 37–41). Unlike the four C-terminal residues in murine SR-BI (EAKL), those in SR-BII (SAMA) are not expected to bind to PDZ domains (42). Loss of the most C-terminal amino acid of SR-BI abolishes its interaction with the PDZ1 domain of PDZK1 (36).
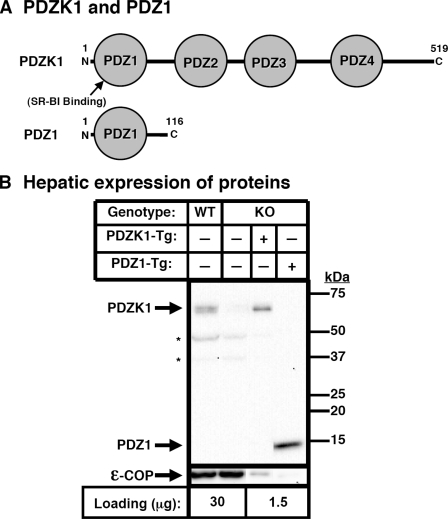
FIGURE 1.
Hepatic expression of PDZK1 and PDZ1 transgenes. A, schematic diagrams of the proteins encoded by the PDZK1 (top) and PDZ1 (bottom) transgenes. B, immunoblot analysis of hepatic expression of PDZK1 and PDZ1 proteins in PDZK1 and PDZ1 transgenic mice. Hepatic protein levels are shown in nontransgenic WT and PDZK1 KO mice (left two lanes) and PDKZI KO[PDZK1-Tg] and PDZK1 KO[PDZ1-Tg] mice (right two lanes). Liver lysates from mice with the indicated genotypes and transgenes (30 or 1.5 μg of protein/lane) were analyzed by immunoblotting, and the bands corresponding to either PDZK1 (∼60 kDa) or PDZ1 (∼14 kDa) were visualized by chemiluminescence. Size markers are indicated on the right. ε-COP (∼34 kDa band, bottom) was used as a loading control. Replicate experiments with multiple exposures and sample loading were used to determine the relative levels of expression of these proteins. Note the faint ε-COP band visible in the last lane. Samples shown were all from a single gel but reordered for presentation purposes. *, background bands seen in all samples.
PDZK1 is most highly expressed in the kidney and is also expressed, as is the case for SR-BI, in the liver and intestines but has very low expression in the adrenal gland, ovary, and testis, in which SR-BI is abundantly expressed (3, 31). Strikingly, PDZK1 knock-out (KO) mice show a dramatic, >95% decrease in SR-BI expression in the liver and a partial decrease in the small intestine but no change in steroidogenic tissues (32). As a consequence of the decrease in hepatic SR-BI protein, PDZK1 KO mice exhibit abnormally high plasma cholesterol levels (∼1.7-fold) transported in abnormally large HDL particles (32). These features are similar to, although not as severe as, those found in SR-BI KO mice (19). We have previously shown that hepatic overexpression of an SR-BI transgene in SR-BI/PDZK1 double knock-out mice leads to SR-BI expression on the cell surface and normal function (43). Thus, when SR-BI is on the cell surfaces of hepatocytes at steady state levels similar to those in wild-type hepatocytes, it can function normally without PDZK1 (43). The mechanism by which PDZK1 regulates the abundance and/or localization of hepatic SR-BI in nontransgenic animals remains unclear.
PDZK1 includes three PDZ domains (PDZ2, PDZ3, and PDZ4) and a 66-residue C-terminal tail (Fig. 1A), which are not involved in direct binding to SR-BI (31, 33). The role(s) of these domains in the regulation of hepatic SR-BI are unknown. Nakamura et al. (35) have shown that the C terminus and the phosphorylation of PDZK1 are required for optimal PDZK1 transgene-mediated induction of increased SR-BI protein levels in cultured cells. It is possible that PDZ2, PDZ3, and/or PDZ4 bind to other, as yet unidentified, cellular components involved in SR-BI regulation. Other hepatic proteins (e.g. OATP1A1) (44) have been shown to interact with the PDZK1 PDZ domains.
Here we have further analyzed the influence of one of the PDZ domains of PDZK1, PDZ1, on hepatic SR-BI abundance, localization and function in vivo. We have compared the effects of transgenes encoding either the full-length PDZK1 or only the SR-BI-binding PDZ1 domain on hepatic SR-BI protein expression and plasma lipoproteins in WT and PDZK1 KO mice. We found that overexpressing full-length PDZK1 in PDZK1 KO mice restored normal hepatic SR-BI protein levels and function, yet in WT mice, it had little, if any, effect on hepatic SR-BI levels and function. However, hepatic overexpression of only the PDZ1 domain was not sufficient to restore normal SR-BI function in PDZK1-deficient mice. In addition, in WT mice, overexpression of the PDZ1 domain overcame the activity of the endogenous hepatic PDZK1, resulting in a 75% reduction in hepatic SR-BI protein levels and intracellular mislocalization of the remaining SR-BI. As a consequence the plasma lipoproteins resembled those in PDZK1 KO mice (hypercholesterolemia, large HDL). Our results suggest that the PDZ1 domain can control both the abundance and intracellular localization of hepatic SR-BI and that normal regulation of SR-BI by PDZK1 is likely to require that its other domains interact with other, as yet unidentified, cellular components.
EXPERIMENTAL PROCEDURES
Generation of PDZK1 and PDZ1 Transgenic Mice—PDZK1 and PDZ1 transgenic mice were generated using the pLIV-LE6 plasmid, kindly provided by Dr. John M. Taylor (Gladstone Institute of Cardiovascular Disease, University of California, San Francisco). The pLiv-LE6 plasmid contains the promoter, first exon, first intron, and part of the second exon of the human apoE gene; the polyadenylation sequence; and a part of the hepatic control region of the apoE/C-I gene locus (45).
To generate the full-length PDZK1 transgenic mouse, a 2.3-kilobase DNA fragment spanning the wild-type murine PDZK1 cDNA was subcloned between the KpnI and XhoI sites of pLIV-LE6 plasmid. The PDZK1-coding region was confirmed by DNA sequencing. The new construct harboring the PDZK1 gene, as well as the above mentioned apoE/apoC-I control locus elements, was linearized by SacII/SpeI digestion, and the resulting 7.3-kilobase fragment was used to generate transgenic mice using standard procedures (46).
To generate the PDZ1 transgenic mouse, a 400-bp DNA fragment containing the first 116 amino acids of PDZK1, including its first PDZ domain, was generated by polymerase chain reaction and subcloned between the KpnI and XhoI sites of pLIV-LE6 plasmid. The sequence of the resulting plasmid was confirmed by DNA sequencing. The new construct harboring PDZ1 as well as the apoE/apoC-I control locus elements was linearized by SacII/SpeI digestion, and the resulting 5.4-kilobase fragment was used to generate transgenic mice using standard procedures (46).
Founder animals for the PDZK1 or PDZ1 transgenes in an FVB/N genetic background were identified by PCR performed on tail DNA using the following oligonucleotide primers: full-length PDZK1 transgene, one at the 3′-end of the PDZK1 cDNA (GCAGATGCCTGTTATAGAAGTGTGC), and one corresponding to the 3′-end of the human apoE gene sequence included in the cloning vector (AGCAGATGCGTGAAACTTGGTGA); PDZ1 transgene, one located within the first PDZ domain of PDZK1 cDNA (CAATGGTGTCTTTGTCGACAAG), and the same 3′-primer used for the full length PDZK1 transgene. Founders expressing the PDZK1 or PDZ1 transgenes were crossed with PDZK1 knock-out mice (129SvEv background). Heterozygous pups expressing the PDZK1 or PDZ1 transgene were crossed with wild-type and PDZK1 knock-out mice to obtain mice with PDZK1 or PDZ1 transgene expression and nontransgenic wild-type and knock-out control mice, thus ensuring that the mixed genetic backgrounds of experimental and control mice in each founder line are matched. Genotyping of PDZK1 knock-out mice was performed as previously described (47).
Transgene expression levels were determined in liver, kidney, adrenal gland, and small intestines by immunoblotting as described below. Expression was only observed in the liver and kidney, with hepatic transgene expression ∼4-fold higher than that seen in the kidney. Expression of the PDZK1 and PDZ1 transgene did not alter the undetectable level of SR-BI in the kidney.
Animals—All mice on a 25:75 FVB/N:129Sv genetic background were maintained on a normal chow diet (47), and ∼6–13-week-old male mice were used for experiments. Four PDZK1-Tg and three PDZ1-Tg founder lines were generated, all showing similar effects on plasma cholesterol levels, respectively. Observations from animals derived from two of the four PDZK1-Tg and two of the three PDZ1-Tg expressing founders were pooled, because transgene-driven protein expression levels for these two sets of founders were similar. All procedures on transgenic and nontransgenic mice were performed in accordance with the guidelines of the Beth Israel Deaconess Medical Center and the Massachusetts Institute of Technology Committee on Animal Care.
Blood and Tissue Sampling and Processing—After a 4-h fast, mice were anesthetized by intraperitoneal injection of 1.25% Avertin, and blood was collected by heart puncture with a heparinized syringe. Plasma was separated from cells by two sequential centrifugations (15 and 10 min, 2500 × g) and stored at 4 °C prior to analysis. Tissue samples, including liver, kidney, adrenal, and small intestine were harvested and immediately frozen on dry ice and stored at -80 °C. Frozen tissue was finely chopped with a blade in SDS buffer (4% SDS, 20% glycerol, 100 mm Tris, pH 6.8) and then sonicated on ice, boiled at 100 °C for 5 min, and subjected to centrifugation at 16,000 × g for 5 min at room temperature. The protein concentrations of these lysate supernatants were then determined using the bicinchoninic acid assay (Pierce). Lysates were then denatured at 100 °C for 10 min in the presence of 2% SDS and 100 mm β-mercaptoethanol immediately before fractionation by gel electrophoresis and analysis by immunoblotting.
Analysis of Plasma Cholesterol and Lipoproteins—Plasma samples from individual mice were collected and analyzed for total cholesterol levels using a commercial kit (Wako Chemical USA, Inc., Richmond, VA) before and after size fractionation by fast performance liquid chromatography (FPLC). FPLC cholesterol profiles shown are from representative individual mice. Total plasma cholesterol data from founders with similar hepatic transgene expression levels were pooled.
Immunoblotting and Immunoperoxidase Analysis of Tissues—For immunoblotting, protein samples (∼30 or 1.5 μg) from total tissue lysates were fractionated by SDS-PAGE, transferred to nitrocellulose or polyvinylidene difluoride membranes, and incubated with either a rabbit polyclonal anti-SR-BI antipeptide antibody (mSR-BI495–112) (1:1000) or a rabbit polyclonal anti-PDZK1 antipeptide antibody (1:5000–1:10,000), followed by an anti-rabbit IgG conjugated to horseradish peroxidase (1:10,000; Invitrogen), and visualized by ECL chemiluminescence (catalog number RPN2132; GE Healthcare). The rabbit polyclonal anti-mSR-BI495–112 antipeptide antibody (prepared by Invitrogen) was generated using similar procedures and against the same peptide as a previously described anti-mSR-BI495 antibody and gives the same results as the anti-mSR-BI495 antibody (3, 43). The rabbit polyclonal antibody was prepared against a 30-mer amino-terminal peptide from the murine PDZK1 protein sequence, coupled to keyhole limpet hemocyanin (Genemed Synthesis, South San Francisco, CA), and affinity-purified. Immunoblotting using polyclonal anti-ε-COP (1:5000) (48) or anti-actin (1:500) (Sigma) antibodies were used as loading controls. The relative amounts of proteins were determined by quantitation using an Eastman Kodak Co. Image Station 440 CF and Kodak 1D software. Multiple independent determinations of band intensities from serially diluted samples were made to ensure that the intensities were both in the linear range of the detector and reproducible.
For immunoperoxidase analysis, livers were harvested, fixed, and frozen. Sections (5 μm) were stained with the anti-mSR-BI495–112 antibody and biotinylated anti-rabbit IgG, visualized by immunoperoxidase staining, and counterstained with Harris modified hematoxylin, as described previously (32).
Enzymatic Digestion of Tissue Lysates—Liver lysates (30 μg of protein) from WT animals with or without the PDZ1 transgene were denatured at 100 °C for 10 min in the presence of 0.5% SDS and 1% β-mercaptoethanol and subsequently treated with 187.5 units of peptide:N-glycosidase F (in the presence of 50 mm sodium phosphate and 1% Nonidet P-40), 1250 units of endoglycosidase H (in the presence of 50 mm sodium citrate), or no enzyme at 37 °C for 3.5 h (New England Biolabs). The treated lysates were then analyzed by immunoblotting using the anti-mSR-BI495–112 polyclonal antibody with chemiluminescence visualization of the bands as described above.
Statistical Analysis—Data are shown as the means ± S.D. Statistically significant differences were determined by either pairwise comparisons of values using the unpaired t test, with (if variances differed significantly) or without Welch's correction, or by one-way analysis of variance followed by the Tukey-Kramer multiple comparison post-test when comparing three or more sets of data with each other. Mean values for experimental groups are considered statistically significantly different for p < 0.05 for both types of tests.
RESULTS
To determine the effects of expressing the full-length or a truncated form (PDZ1 domain) of the PDZK1 protein on hepatic SR-BI protein levels and function, we generated two cDNA constructs that encode either the full-length PDZK1 protein (designated “PDZK1-Tg”) or only the N terminus and PDZ1 domain (“PDZ1-Tg”) (Fig. 1A). These constructs were used to generate transgenic mice with liver- and kidney-specific expression in both wild-type (WT[PDZK1-Tg] or WT[PDZ1-Tg]) and PDZK1 KO (PDZK1 KO[PDZK1-Tg] or PDZK1 KO[PDZ1-Tg]) mice on a mixed FVB/129SvEv background. Transgene expression in the kidney did not alter the undetectable expression of SR-BI in this organ (see “Experimental Procedures”).
Hepatic levels of PDZK1 and PDZ1 proteins in nontransgenic and transgenic mice from two founders each were determined by immunoblotting (typical result in Fig. 1B). Quantitative analysis of multiple immunoblots showed that the steady state levels of transgene-encoded proteins were greater by ∼21-fold in PDZK1-Tg-expressing and 24-fold in PDZ1-Tg-expressing mice than that of endogenous PDZK1 in WT nontransgenic mice (Fig. 1B; note there was lower total protein loading for samples from transgenic animals). Analysis by immunoblotting also showed that the levels of endogenous hepatic PDZK1 in WT[PDZ1-Tg] mice were similar to those in nontransgenic WT mice (83 ± 18% of WT levels of endogenous PDZK1; p = 0.339).
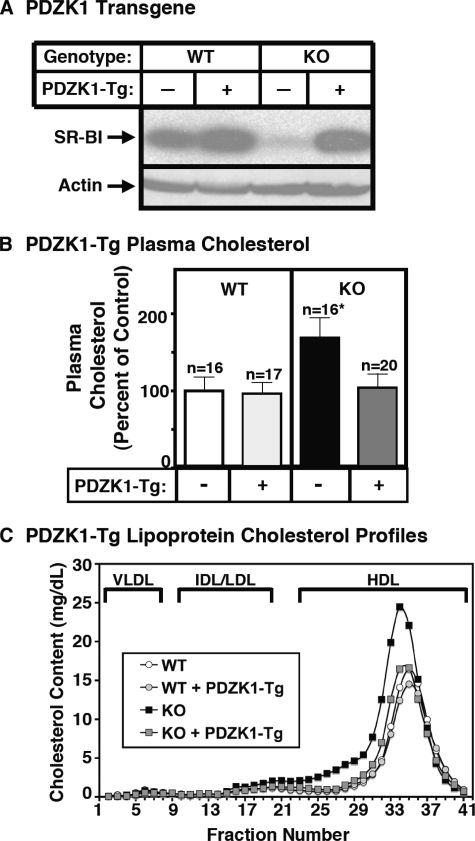
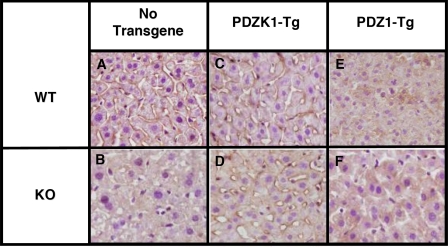
Effects of the PDZK1-Tg on Hepatic SR-BI Expression and Lipoprotein Metabolism—In WT mice, hepatic overexpression of PDZK1 had little effect on the steady state levels (immunoblotting, Fig. 2A, left half of the panel) or cell surface localization (immunohistochemistry; Fig. 3, A and C) of hepatic SR-BI protein. As a consequence, in WT mice, there also was no influence of the PDZK1-Tg on plasma cholesterol levels (Fig. 2B, left) or the size distribution of lipoproteins (FPLC lipoprotein cholesterol profiles), in which the bulk of the cholesterol is found in normal sized HDL particles (Fig. 2C, white and gray circles). In contrast, in PDZK1 KO mice, hepatic overexpression of PDZK1 corrected the previously reported abnormal phenotypes related to SR-BI and lipoprotein metabolism. In PDZK1 KO mice, the PDZK1-Tg restored from <5% of wild-type (32) to normal wild-type levels the steady-state abundance of hepatic SR-BI protein (immunoblotting; Fig. 2A, right half), which was properly localized on the surfaces of the hepatocytes (immunohistochemistry; Fig. 3, B and D). As a consequence, in PDZK1 KO mice, the PDZK1-Tg restored plasma cholesterol levels from the ∼1.7-fold elevation seen in nontransgenic PDKZ1 KO mice to those in wild-type controls (Fig. 2B, right). The PDZK1-Tg also restored the size distribution of lipoproteins from that with abnormally large HDL particles in the nontransgenic PDZK1 KO animals to that with normal sized HDL seen in the nontransgenic WT mice (Fig. 2C, black and gray squares). Note that the peak in the PDZK1 KO sample is shifted to the left relative to that in WT plasma, indicating larger lipoproteins. Similar results were obtained with an adenoviral construct used to overexpress full-length PDZK1 in the livers of mice on a pure 129SvEv genetic background (not shown). Therefore, although hepatic overexpression of PDZK1 has little effect on SR-BI and lipoprotein metabolism in WT mice, it appears to fully complement the PDZK1 deficiency in PDZK1 KO mice with respect to hepatic SR-BI protein levels, cellular localization, and function in lipoprotein metabolism.
FIGURE 2.
Effect of expression of the PDZK1 transgene on hepatic SR-BI protein levels (A) and plasma lipoprotein cholesterol (B and C) in WT and PDZK1 KO mice. A, liver lysates (∼30 μg of protein) from mice with the indicated genotypes, with (+) or without (-) the PDZK1 transgene, were analyzed by immunoblotting, and SR-BI (∼82 kDa band) was visualized by chemiluminescence. Actin (∼40 kDa) was used as a loading control. B and C, plasma was harvested from mice with the indicated genotypes and PDZK1 transgene. B, total plasma cholesterol levels were determined in individual samples by an enzymatic assay; results from the indicated numbers of animals (n) were pooled by genotype and normalized to the mean value for WT plasma cholesterol (100% = 116.9 mg/dl). Independent WT and KO control animals for each founder line were generated to ensure that the mixed genetic backgrounds for experimental and control mice were matched. *, the PDZK1 KO plasma cholesterol levels were statistically significantly different (p ≤ 0.001) from those plasma cholesterol levels of WT, WT[PDZK1-Tg], and PDZK1 KO[PDZK1-Tg] mice. C, plasma samples (harvested as in B) from individual animals were size-fractionated by FPLC, and the total cholesterol content of each fraction was determined by an enzymatic assay. The chromatograms are representative of multiple individually determined profiles. Approximate elution positions of native very low density lipoprotein (VLDL), intermediate density lipoprotein/low density lipoprotein (IDL/LDL), and HDL particles are indicated by brackets and were determined as previously described (19).
FIGURE 3.
Immunohistochemical analysis of hepatic SR-BI in WT and PDZK1 KO nontransgenic (A and B), PDZK1 transgenic (C and D), and PDZ1 transgenic (E and F) mice. Livers from mice of the indicated genotypes and transgenes were fixed, frozen, and sectioned. The sections were then stained with a polyclonal anti-SR-BI antibody and a biotinylated anti-rabbit IgG secondary antibody and visualized by a immunoperoxidase staining (magnification, ×600).
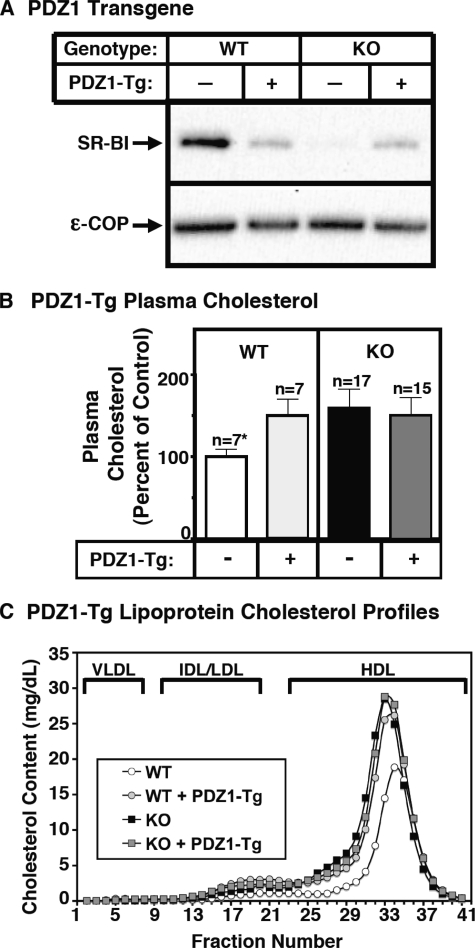
Effects of the PDZ1-Tg on Lipoprotein Metabolism and Hepatic SR-BI Expression—In PDZK1 KO mice, 24-fold hepatic overexpression of the truncated PDZ1 protein had virtually no effect on the abnormally high plasma cholesterol levels (Fig. 4B, right) or the size distribution of lipoproteins (FPLC lipoprotein cholesterol profiles), in which the bulk of the cholesterol is found in abnormally large HDL particles (Fig. 4C, black and gray squares). Thus, the N-terminal PDZ1 domain-containing region of PDZK1 is not sufficient to restore normal SR-BI function in PDZK1 KO mice, suggesting that other portions of the PDZK1 protein must play critical roles in regulating hepatic SR-BI. Strikingly, in WT mice, we found that the PDZ1-Tg elevated plasma cholesterol levels (Fig. 4B, left) and increased the size of the HDL particles (Fig. 4C, white and gray circles) to those observed in nontransgenic PDZK1 KO mice. We therefore conclude that PDZ1-Tg acts as in a dominant negative fashion with respect to SR-BI function in WT mice.
FIGURE 4.
Effect of expression of the PDZ1 transgene on hepatic SR-BI protein levels (A) and plasma lipoprotein cholesterol (B and C) in WT and PDZK1 KO mice. A, liver lysates (∼30 μg of protein) from mice with the indicated genotypes, with (+) or without (-) the PDZ1 transgene, were analyzed by immunoblotting, and SR-BI (∼82-kDa band) was visualized by chemiluminescence. ε-COP (∼34 kDa) was used as a loading control. Note the faint SR-BI band in the nontransgenic PDZK1 KO lane. B and C, plasma was harvested from mice with the indicated genotypes and PDZ1 transgene. B, total plasma cholesterol levels were determined in individual samples by enzymatic assay, and results from the indicated numbers of animals (n) were pooled by genotype and normalized to the mean value for WT plasma cholesterol (100% = 131.7 mg/dl). Independent WT and KO control animals for each founder line were generated to ensure that the mixed genetic backgrounds for experimental and control mice are matched. *, the nontransgenic WT plasma cholesterol levels were statistically significantly different (p ≤ 0.001) from those plasma cholesterol levels of WT[PDZ1-Tg], PDZK1 KO, and PDZK1 KO[PDZ1-Tg] mice. C, plasma samples (harvested as in B) from individual animals were size-fractionated by FPLC, and the total cholesterol content of each fraction was determined by an enzymatic assay. The chromatograms are representative of multiple individually determined profiles. Approximate elution positions of native very low density lipoprotein (VLDL), intermediate density lipoprotein/low density lipoprotein (IDL/LDL), and HDL particles are indicated by brackets and were determined as previously described (19).
We then examined the effects of the PDZ1-Tg on hepatic SR-BI protein abundance and localization. Immunoblotting analysis (Fig. 4A, left portion) shows that in WT mice, the PDZ1-Tg lowered the steady-state hepatic SR-BI protein levels to 24 ± 5% of that in nontransgenic WT mice, yet the levels remained >5-fold higher than that in nontransgenic PDZK1 KO mice. Thus, unlike the full-length PDZK1-Tg, in WT mice the PDZ1-Tg partially suppressed hepatic SR-BI protein expression. Initially it was somewhat surprising that this partial suppression of hepatic SR-BI protein levels was accompanied by changes in plasma lipoproteins equivalent to those seen in nontransgenic PDZK1 KO mice that have <5% of WT hepatic SR-BI. However, immunohistochemical localization of the residual SR-BI in WT[PDZ1-Tg] mice (Fig. 3E) showed that most of the residual SR-BI was intracellular, with no detectable receptor on the surfaces of hepatocytes. This result was confirmed by immunofluorescence microscopy (data not shown). Thus, the absence of substantial amounts of SR-BI on the sinusoidal surfaces of hepatocytes in WT[PDZ1-Tg] mice appears to explain the similarity of the lipoprotein levels in these mice and nontransgenic PDZK1 KO mice. In both cases, there was virtually no surface SR-BI to interact with circulating lipoproteins.
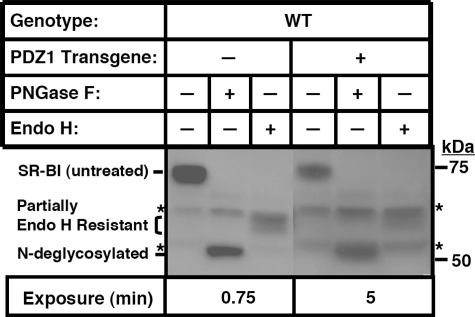
To determine if the intracellular mislocalization of the residual SR-BI in WT[PDZ1-Tg] mice were due to a block in SR-BI trafficking in the early stages of the secretory pathway (endoplasmic reticulum to medial Golgi), we compared the glycosylation state of the SR-BI protein in nontransgenic WT and WT[PDZ1-Tg] mice. We have previously shown that SR-BI is heavily glycosylated (11 N-linked glycosylation sites) (49, 50). N-Linked oligosaccharide chains on newly synthesized glycoproteins in the endoplasmic reticulum are initially fully sensitive to cleavage by the enzyme endoglycosidase H, but enzymatic modifications that occur as the glycoprotein passes through the medial Golgi convert many, but not all, of these chains to an endoglycosidase H-resistant form (51). Virtually all N-linked oligosaccharide chains on glycoproteins, both endoglycosidase H-sensitive and endoglycosidase H-resistant, can be removed by the glycosidase peptide:N-glycosidase F (51). The fully matured hepatic SR-BI protein from nontransgenic WT mice (immunoblotting of untreated and glycosidase-treated liver lysates (Fig. 5, left) exhibited a pattern of glycosidase sensitivity similar to that previously reported for SR-BI in cultured cells and the adrenal gland (49). This SR-BI protein was fully sensitive to peptide:N-glycosidase F (reduction in apparent molecular mass from ∼82 kDa (untreated) to ∼55 kDa), yet only partially resistant to hydrolysis by endoglycosidase H (electrophoretic mobility intermediate between the untreated and fully N-deglycosylated forms). (Unidentified background bands are indicated with asterisks.) The results for glycosidase analysis of hepatic SR-BI from WT[PDZ1-Tg] mice were virtually identical to those from nontransgenic WT mice (although the lower SR-BI expression required longer exposure times and correspondingly stronger background bands). Essentially all of the SR-BI protein was partially endoglycosidase H-resistant and thus had passed through the medial Golgi. Therefore, we conclude that the intracellular mislocalization of the residual hepatic SR-BI in WT[PDZ1-Tg] mice was not a consequence of a block in endoplasmic reticulum to medial Golgi intracellular trafficking.
FIGURE 5.
Immunoblot analysis of the effects of the PDZ1 transgene on the N-glycosylation of hepatic SR-BI in WT mice. Liver lysates (30 μg of protein) from WT animals with (+) or without (-) the PDZ1 transgene were treated as indicated with peptide:N-glycosidase F (PNGase F) (187.5 units), endoglycosidase H (Endo H) (1250 units), or no enzyme at 37 °C for 3.5 h and then analyzed by immunoblotting using an anti-SR-BI polyclonal antibody with chemiluminescence visualization of the bands. *, background bands seen in all samples, including in samples of nontransgenic PDZK1 KO lysates (not shown). The lanes for samples from the PDZ1 transgenic mice (right) contained lower levels of SR-BI protein and were therefore subjected to longer exposure times (5 versus 0.75 min). As a consequence, the intensities of the background bands were higher than for the nontransgenic samples on the left.
Although in PDKZ1 KO mice the PDZ1-Tg did not influence plasma cholesterol levels or the lipoprotein profile (Fig. 4, B (right) and C (squares)), it did reproducibly increase the steady state levels of hepatic SR-BI protein from less than 5% to 21 ± 6% of nontransgenic WT levels (Fig. 4A, right). Fig. 3F shows that this residual SR-BI was located intracellularly rather than on the cell surface and thus cannot function efficiently as a receptor for circulating HDL. The mechanism by which hepatic overexpression of PDZ1 in PDZK1 KO mice partially restores the steady state level of SR-BI protein yet results in its mislocalization remains uncertain (see “Discussion”). It is noteworthy that the level of SR-BI protein was virtually the same in WT[PDZ1-Tg] (24 ± 5% of WT) and PDZK1 KO[PDZ1-Tg] (21 ± 6% of WT) mice. This similarity raises the possibility that the overexpressed PDZ1 domain completely displaced the endogenous PDKZ1 protein's binding to SR-BI in WT[PDZ1-Tg] livers.
DISCUSSION
The first PDZ domain, PDZ1, of the cytoplasmic adaptor protein PDZK1 binds to the C terminus of SR-BI, and as a consequence, PDZK1 controls hepatic SR-BI protein expression and lipoprotein metabolism (31, 32, 36). Expression of full-length PDZK1 in the liver of PDZK1 KO mice corrects their dramatic decrease in hepatic SR-BI protein levels and hypercholesterolemia, without apparently altering SR-BI expression in other tissues. This suggests that the effect of PDZK1 on lipoprotein metabolism is mediated principally by its control of SR-BI in liver.
When a transgene encoding a truncated form of PDZK1 primarily comprising only the SR-BI-binding PDZ1 domain (PDZ1-Tg; Fig. 1A) was overexpressed in WT mice, it exhibited dominant negative activity, and the mice, in several respects, resembled PDZK1 KO mice (dramatically reduced cell surface expression of hepatic SR-BI protein, hypercholesterolemia). The residual hepatic SR-BI protein in these transgenic animals (∼25% of WT controls), which was 5-fold greater than that in nontransgenic PDZK1 KO mice (<5% of control), was redistributed from the cell surface to the intracellular region. We infer based on PDZ1 binding to SR-BI (31, 36), but have not demonstrated directly, that these effects were consequences of the recombinant, truncated PDZ1 domain competing with endogenous PDZK1 for binding to SR-BI. These results suggest that those portions of the PDZK1 molecule C-terminal to the PDZ1 domain that are not known to bind directly to SR-BI play a critical role in mediating the influence of PDZK1 on SR-BI. It seems likely that these other regions of PDZK1 interact with additional cellular components to properly control SR-BI activity. It is possible that the phosphorylated C terminus of PDZK1 (35) or the other three PDZ domains (PDZ2–PDZ4) and their binding partners may play a critical role in regulating SR-BI. Additional putative PDZK1-interacting components apparently are either not present in limiting amounts or cannot interact with PDZK1 efficiently unless it is bound to SR-BI, because overexpression of PDZK1 protein in the liver of WT mice had virtually no effect on normal hepatic SR-BI levels and lipoprotein metabolism (no dominant negative phenotypes observed; the excess PDZK1 did not titrate out other critical components).
There are several potential mechanisms by which full-length PDZK1 or truncated PDZ1 might influence both SR-BI protein abundance and intracellular localization. For example, PDZK1 might inhibit degradation of hepatic SR-BI by binding to its C terminus, directly preventing access to proteolytic pathways (e.g. ubiquitination and/or proteasomes), or indirectly by controlling the intracellular trafficking of SR-BI (e.g. directing post-medial Golgi sorting to the cell surface, inhibiting endocytosis from the cell surface, or promoting normal endocytic recycling back to the cell surface) and preventing delivery to sites of degradation or both. The effects of expression of the PDZ1-Tg in PDZK1 KO mice are compatible with both of these mechanisms. The PDZ1-Tg increased by 5-fold the steady state levels of SR-BI compared with those in the nontransgenic PDZK1 KO, raising the possibility that binding to the SR-BI C terminus may have interfered with degradation. In addition, the residual SR-BI in the livers of both WT[PDZ1-Tg] and PDZK1 KO[PDZ1-Tg] mice was mislocalized intracellularly, possibly due to the inability of the truncated protein·SR-BI complex to interact with cellular components that only bind to the complex when additional domains of PDZK1 are present.
In summary, PDZK1 appears to affect SR-BI protein levels both through binding to the C terminus and through interactions with other cellular components. Future studies will be required to identify these putative cellular components, the domains in PDZK1 responsible for these interactions, and the mechanism by which they cooperate with PDZK1 to control SR-BI function.
Acknowledgments
We thank Joel Lawitts from the BIDMC transgenic facility, S. Xu for generating and characterizing the anti-mSR-BI495-112 antibody, and H. Lodish and D. Sabatini for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants HL077780 (to O. K.) and HL64737, HL52212, and HL66105 (to M. K.). This work was also supported by FONDECYT Grant 1070634 (to A. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HDL, high density lipoprotein; SR-BI, scavenger receptor, class B, type I; PDZ, postsynaptic density protein (PSD-95)/Drosophila disc large tumor suppressor (dlg)/tight junction protein (ZO1); Tg, transgenic; KO, knockout; WT, wild-type; FPLC, fast performance liquid chromatography; apo, apolipoprotein.
References
- 1.Rigotti, A., Miettinen, H. E., and Krieger, M. (2003) Endocr. Rev. 24 357-387 [DOI] [PubMed] [Google Scholar]
- 2.Acton, S. L., Scherer, P. E., Lodish, H. F., and Krieger, M. (1994) J. Biol. Chem. 269 21003-21009 [PubMed] [Google Scholar]
- 3.Acton, S., Rigotti, A., Landshulz, K. T., Xu, S., Hobbs, H. H., and Krieger, M. (1996) Science 271 518-520 [DOI] [PubMed] [Google Scholar]
- 4.Out, R., Kruijt, J. K., Rensen, P. C., Hildebrand, R. B., de Vos, P., Van Eck, M., and Van Berkel, T. J. (2004) 279 18401-18406 [DOI] [PubMed]
- 5.Out, R., Hoekstra, M., de Jager, S. C., de Vos, P., van der Westhuyzen, D. R., Webb, N. R., Van Eck, M., Biessen, E. A., and Van Berkel, T. J. (2005) J. Lipid Res. 46 1172-1181 [DOI] [PubMed] [Google Scholar]
- 6.Fu, T., Kozarsky, K. F., and Borensztajn, J. (2003) J. Biol. Chem. 278 52559-52563 [DOI] [PubMed] [Google Scholar]
- 7.Van Eck, M., Hoekstra, M., Out, R., Bos, I. S., Kruijt, J. K., Hildebrand, R. B., and Van Berkel, T. J. (2008) J. Lipid Res. 49 136-146 [DOI] [PubMed] [Google Scholar]
- 8.Glass, C., Pittman, R. C., Weinstein, D. W., and Steinberg, D. (1983) Proc. Natl. Acad. Sci. U. S. A. 80 5435-5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein, Y., Dabach, Y., Hollander, G., Halperin, G., and Stein, O. (1983) Biochim. Biophys. Acta 752 98-105 [DOI] [PubMed] [Google Scholar]
- 10.Reaven, E., Chen, Y. I., Spicher, M., and Azhar, S. (1984) J. Clin. Invest. 74 1384-1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji, Y., Jian, B., Wang, N., Sun, Y., de la Llera Moya, M., Phillips, M. C., Rothblat, G. H., Swaney, J. B., and Tall, A. R. (1997) J. Biol. Chem. 272 20982-20985 [DOI] [PubMed] [Google Scholar]
- 12.Jian, B., de la Llera-Moya, M., Ji, Y., Wang, N., Phillips, M. C., Swaney, J. B., Tall, A. R., and Rothblat, G. H. (1998) J. Biol. Chem. 273 5599-5606 [DOI] [PubMed] [Google Scholar]
- 13.Zimetti, F., Weibel, G. K., Duong, M., and Rothblat, G. H. (2006) J. Lipid Res. 47 605-613 [DOI] [PubMed] [Google Scholar]
- 14.Treguier, M., Moreau, M., Sposito, A., Chapman, M. J., and Huby, T. (2007) Biochim. Biophys. Acta 1771 129-138 [DOI] [PubMed] [Google Scholar]
- 15.Kozarsky, K. F., Donahee, M. H., Rigotti, A., Iqbal, S. N., Edelman, E. R., and Krieger, M. (1997) Nature 387 414-417 [DOI] [PubMed] [Google Scholar]
- 16.Arai, T., Wang, N., Bezouevski, M., Welcch, C., and Tall, A. R. (1999) J. Biol. Chem. 274 12610-12615 [DOI] [PubMed] [Google Scholar]
- 17.Ueda, Y., Wang, N., Bezouevski, M., Welch, C., and Tall, A. R. (2000) J. Biol. Chem. 274 2366-2371 [DOI] [PubMed] [Google Scholar]
- 18.Kozarsky, K. F., Donahee, M. H., Glick, J. M, Krieger, M., and Rader, D. J. (2000) Artioscler. Thromb. Vasc. Biol. 20 721-727 [DOI] [PubMed] [Google Scholar]
- 19.Rigotti, A., Trigatti, B. L., Penman, M., Rayburn, H., Herz, J., and Krieger, M. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 12610-12615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mardones, P., Quinones, V., Amigo, L., Moreno, M., Miquel, J. F., Schwarz, M., Miettinen, H. E., Trigatti, B., Krieger, M., VanPatten, S., Cohen, D. E., and Rigotti, A. (2001) J. Lipid Res. 42 170-180 [PubMed] [Google Scholar]
- 21.Trigatti, B., Rayburn, H., Vinals, M., Braun, A., Miettinen, H., Penman, M., Hertz, M., Schrenzel, M., Amigo, L., Rigotti, A., and Krieger, M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 9322-9327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huszar, D., Varban, M. L., Rinninger, F., Feeley, R., Arai, T., Fairchild-Huntress, V., Donovan, M. J., and Tall, A. R. (2000) Arterioscler. Thromb. Vasc. Biol. 20 1068-1073 [DOI] [PubMed] [Google Scholar]
- 23.Van Eck, M., Twisk, J., Hoekstra, M., Van Rij, B. T., Van der Lans, C. A., Bos, I. S., Kruijt, J. K., Kuipers, F., and Van Berkel, T. J. (2003) J. Biol. Chem. 278 23699-23705 [DOI] [PubMed] [Google Scholar]
- 24.Braun, A., Zhang, S., Miettinen, H. E., Ebrahim, S., Holm, T. M., Vasile, E., Post, M. J., Yoerger, D. M., Picard, M. H., Krieger, J. L., Andrews, N. C., Simons, M., and Krieger, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7283-7288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji, Y., Wang, N., Ramakrishnan, R., Sehayek, E., Huszar, D., Breslow, J. L., and Tall, A. R. (1999) J. Biol. Chem. 274 33398-33402 [DOI] [PubMed] [Google Scholar]
- 26.Yesilaltay, A., Morales, M. G., Amigo, L., Zanlungo, S., Rigotti, A., Karackattu, S. L., Donahee, M. H., Kozarsky, K. F., and Krieger, M. (2006) Endocrinology 147 1577-1588 [DOI] [PubMed] [Google Scholar]
- 27.Ueda, Y., Royer, L., Gong, E., Zhang, J., Cooper, P. N., Francone, O., and Rubin, E. M. (1999) J. Biol. Chem. 274 7165-7171 [DOI] [PubMed] [Google Scholar]
- 28.Holm, T. M., Braun, A., Trigatti, B. L., Brugnara, C., Sakamoto, M., Krieger, M., and Andrews, N. C. (2002) Blood 99 1817-1824 [DOI] [PubMed] [Google Scholar]
- 29.Meurs, I., Hoekstra, M., van Wanrooij, E. J., Hildebrand, R. B., Kuiper, J., Kuipers, F., Hardeman, M. R., Van Berkel, T. J., and Van Eck, M. (2005) Exp. Hematol. 33 1309-1319 [DOI] [PubMed] [Google Scholar]
- 30.Miettinen, H. E., Rayburn, H., and Krieger, M. (2001) J. Clin. Invest. 108 1717-1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ikemoto, M., Arai, H., Feng, D., Tanaka, K., Aoki, J., Dohmae, N., Takio, K., Adachi, H., Tsujimoto, M., and Inoue, K. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6538-6543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kocher, O., Yesilaltay, A., Cirovic, C., Pal, R., Rigotti, A., and Krieger, M. (2003) J. Biol. Chem. 278 52820-52825 [DOI] [PubMed] [Google Scholar]
- 33.Kocher, O., Comella, N., Tognazzi, K., and Brown, L. (1998) Lab. Invest. 78 117-125 [PubMed] [Google Scholar]
- 34.Yesilaltay, A., Kocher, O., Rigotti, A., and Krieger, M. (2005) Curr. Opin. Lipidol. 16 147-152 [DOI] [PubMed] [Google Scholar]
- 35.Nakamura, T., Shibata, N., Nishimoto-Shibata, T., Feng, D., Ikemoto, M., Motjima, K., Iso-O, N., Tsukamoto, K., Tsujimoto, M., and Arai, H. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 13404-13409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silver, D. L. (2002) J. Biol. Chem. 277 34042-34047 [DOI] [PubMed] [Google Scholar]
- 37.Webb, N. R., de Villiers, W. J., Connell, P. M., de Beer, F. C., and van der Westhuyzen, D. R. (1997) J. Lipid Res. 38 1490-1495 [PubMed] [Google Scholar]
- 38.Webb, N. R., Connell, P. M., Graf, G. A., Smart, E. J., de Villiers, W. J. S., de Beer, F. C., and van der Westhuyzen, D. R. (1998) J. Biol. Chem. 273 15241-15248 [DOI] [PubMed] [Google Scholar]
- 39.Eckhardt, E. R., Cai, L., Shetty, S., Zhao, Z., Szanto, A., Webb, N. R., and Van der Westhuyzen, D. R. (2006) J. Biol. Chem. 281 4348-4353 [DOI] [PubMed] [Google Scholar]
- 40.Eckhardt, E. R., Cai, L., Sun, B., Webb, N. R., and van der Westhuyzen, D. R. (2004) J. Biol. Chem. 279 14372-14381 [DOI] [PubMed] [Google Scholar]
- 41.Zhang, X., Moor, A. N., Merkler, K. A., Liu, Q., and McLean, M. P. (2007) Endocrinology 148 5295-5304 [DOI] [PubMed] [Google Scholar]
- 42.Hung, A. Y., and Sheng, M. (2002) J. Biol. Chem. 277 5699-5702 [DOI] [PubMed] [Google Scholar]
- 43.Yesilaltay, A., Kocher, O., Pal, R., Leiva, A., Quinones, V., Rigotti, A., and Krieger, M. (2006) J. Biol. Chem. 281 28975-28980 [DOI] [PubMed] [Google Scholar]
- 44.Wang, P., Wang, J. J., Xiao, Y., Murray, J. W., Novikoff, P. M., Angeletti, R. H., Orr, G. A., Lan, D., Silver, D. L., and Wolkoff, A. W. (2005) J. Biol. Chem. 280 30143-30149 [DOI] [PubMed] [Google Scholar]
- 45.Simonet, W. S., Bucay, N., Lauer, S. J., and Taylor, J. M. (1993) J. Biol. Chem. 268 8221-8229 [PubMed] [Google Scholar]
- 46.Palmiter, R. D., and Brinster, R. L. (1986) Annu. Rev. Genet. 20 465-499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kocher, O., Pal, R., Roberts, M., Cirovic, C., and Gilchrist, A. (2003) Mol. Cell Biol. 23 1175-1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo, Q., Penman, M., Trigatti, B. L., and Krieger, M. (1996) J. Biol. Chem. 271 11191-11196 [DOI] [PubMed] [Google Scholar]
- 49.Babitt, J., Trigatti, B., Rigotti, A., Smart, E. J., Anderson, R. G., Xu, S., and Krieger, M. (1997) J. Biol. Chem. 272 13242-13249 [DOI] [PubMed] [Google Scholar]
- 50.Vinals, M., Xu, S., Vasile, E., and Krieger, M. (2003) J. Biol. Chem. 278 5325-5332 [DOI] [PubMed] [Google Scholar]
- 51.Maley, F., Trimble, R. B., Tarentino, A. L., and Plummer, T. H., Jr. (1989) Anal. Biochem. 180 195-204 [DOI] [PubMed] [Google Scholar]
- 52.Braun, A., Zhang, S., Miettinen, H. E., Ebrahim, S., Holm, T. M., Vasile, E., Post, M. J., Yoerger, D. M., Picard, M. H., Krieger, J. L., Andrews, N. C., Simons, M., and Krieger, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7283-7288 [DOI] [PMC free article] [PubMed] [Google Scholar]