Abstract
Palmitoylation of Sonic Hedgehog (Shh) is critical for effective long- and short-range signaling. Genetic screens uncovered a potential palmitoylacyltransferase (PAT) for Shh, Hhat, but the molecular mechanism of Shh palmitoylation remains unclear. Here, we have developed and exploited an in vitro Shh palmitoylation assay to purify Hhat to homogeneity. We provide direct biochemical evidence that Hhat is a PAT with specificity for attaching palmitate via amide linkage to the N-terminal cysteine of Shh. Other palmitoylated proteins (e.g. PSD95 and Wnt) are not substrates for Hhat, and Porcupine, a putative Wnt PAT, does not palmitoylate Shh. Neither autocleavage nor cholesterol modification is required for Shh palmitoylation. Both the Shh precursor and mature protein are N-palmitoylated by Hhat, and the reaction occurs during passage through the secretory pathway. This study establishes Hhat as a bona fide Shh PAT and serves as a model for understanding how secreted morphogens are modified by distinct PATs.
Hedgehog (Hh) and Sonic Hedgehog (Shh) are members of a family of secreted signaling proteins that mediate growth and patterning during embryonic development (1, 2). These proteins act as morphogens to form signaling gradients for long- and short-range interactions (3). In Drosophila, Hh mediates pattern formation in the wing and eye. Vertebrates express three family members, Sonic, Indian, and Desert, of which Shh is the best studied (1). Shh plays a critical role in developmental patterning of the brain in mice and humans, as the absence of Shh induces holoprosencephaly and cyclopia (4, 5). Shh also regulates limb development as well as cellular proliferation and differentiation in both neuronal and non-neuronal cells. In addition, aberrant Shh expression and/or signaling is implicated in the biogenesis of an increasing number of human cancers, including medulloblastoma, melanoma, liver, pancreatic, and urogenital tumors (6, 7).
Hh/Shh proteins undergo a unique set of post-translational processing reactions (8). Hh/Shh is synthesized as a 45-kDa precursor that traffics through the secretory pathway. After the signal sequence is cleaved, Hh/Shh undergoes autocleavage to generate a 19-kDa N-terminal signaling molecule (HhN/ShhN). During this reaction, the C terminus of HhN is modified by covalent attachment of a molecule of cholesterol (9). In addition, the N-terminal cysteine residue of Hh/Shh is modified by palmitoylation (10). Unlike nearly all other known palmitoylated proteins, the palmitate moiety is attached via an amide bond to the N terminus of Hh/Shh.
Palmitoylation of Hh and Shh is critical for signaling. Mutation of the Shh N-terminal Cys to Ser results in an Shh mutant (C24S) with reduced patterning activity in the mouse limb; the equivalent Hh mutant (C84S) has little to no detectable activity in Drosophila (11–15). Fatty acylated forms of Shh are far more active than unacylated Shh as determined by in vitro differentiation assays (10, 15). Studies of chemically modified Shh reveal that the hydrophobic nature of the N-terminal palmitate is an essential feature for regulation of the strength of Shh signaling (16). It is also important to note that, in addition to N-palmitoylation, cholesterol attachment to the C termini of Hh and Shh contributes to signaling capacity (12, 17–19). The interrelationship between these two lipophilic modification reactions has not been established.
A putative Hh palmitoylacyltransferase has been identified in flies, termed Rasp, and in vertebrates, termed Hhat (Hedgehog acyltransferase) (20–22). In mutant Rasp flies, Hh is synthesized but not palmitoylated, and Hh signaling is defective. Knock-out mice lacking Hhat synthesize non-palmitoylated Shh and exhibit defects in neural tube and limb development, indicative of defective Shh signaling (13). Rasp and Hhat are members of a family of multipass transmembrane proteins termed MBOAT (membrane-bound O-acyltransferase) (23). Most of the MBOAT family members transfer fatty acids and other lipids onto hydroxyl groups of membrane-embedded lipids (23–25). Besides Hhat/Rasp, only two other MBOAT proteins, Porcupine (Porc) and GOAT, have been implicated in the transfer of fatty acids to proteins. Porc is a putative palmitoylacyltransferase (PAT)2 for acylation of Wnt/Wg, another family of secreted morphogens, whereas GOAT is the transferase mediating attachment of octanoate to proghrelin, the appetite-stimulating hormone. The presence of multiple transmembrane domains in MBOAT family proteins has limited their purification and molecular characterization.
To date, there is no direct biochemical evidence that Hhat functions as an Shh palmitoylacyltransferase, and it is not known how, when, or where the enzyme recognizes its substrate. To address these issues, we established an in vitro palmitoylation assay system to monitor the mechanism of Shh palmitoylation. Here, we report the purification of Hhat to homogeneity and show that purified Hhat is a bona fide palmitoylacyltransferase with specificity for N-palmitoylation of Shh. Palmitoyl-CoA is the fatty acid donor substrate; neither palmitate as free fatty acid nor palmitate esterified to phospholipid serves as a donor. The reaction is dependent on the presence of a free amino group on the N-terminal cysteine residue, and neither alanine nor serine will substitute for cysteine. Shh palmitoylation is independent of C-terminal cholesterol attachment and prior Shh autocleavage, and the first 11 amino acids of the mature Shh protein are sufficient for Hhat-mediated palmitoylation in vitro. Finally, we show that Hhat and Shh colocalize in the endoplasmic reticulum (ER) and Golgi and that N-palmitoylation of Shh requires passage of Shh through the secretory pathway.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies—Fatty acyl-CoAs, CoA, CoA synthetase, octyl glucoside, anti-FLAG and anti-hemagglutinin (HA) antibodies, FLAG M2-agarose, and 3XFLAG peptide were purchased from Sigma. Enterokinase was purchased from New England Biolabs. [125I]NaI and [3H]palmitate were obtained from PerkinElmer Life Sciences. [3H]Dipalmitoylphosphatidylethanolamine was from American Radioactive Chemicals. The following antibodies were purchased from the indicated manufacturers: Shh and green fluorescent protein (GFP), Santa Cruz Biotechnology; protein-disulfide isomerase, StressGen; and mannosidase II (Abcam). Anti-Fyn serum was generated as described previously (26). Nickel-nitrilotriacetic acid (Ni-NTA) resin was from Qiagen. His-tagged PSD95 and PSD95 Cys → Ser, C-terminally His-tagged myristoylated Gαi, and prenylated glutathione S-transferase-H-Ras constructs were generous gifts from Dr. Maurine Linder (Washington University, St. Louis, MI). Wnt3A and Wnt7A were purchased from R&D Systems.
Mammalian Expression Plasmids—A plasmid encoding HhatΔ91–155 was a generous gift from Dr. Yutaka Kawakami (Keio University, Tokyo, Japan). A single C-terminal HA tag was introduced by PCR, and the PCR product was ligated into pcDNA3.1 (Novagen) using BamHI and EcoRI sites. Full-length Hhat-HA constructs were generated from this parent construct by iterative rounds of PCR primer extension. FLAG-His tags were added to the C terminus by PCR. Hhat H379A constructs were generated by site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene). A plasmid encoding full-length human Shh was a generous gift from Dr. Jessica Treisman (New York University). Shh C24A, C24S, and H270A constructs were generated using the QuikChange mutagenesis kit. Shh-GFP chimeras encoding residues 1–44 or 24–44 of Shh were generated by PCR; the PCR products were ligated into EcoRI- and BamHI-cut pEGFP-N1. Shh-(1–44) C24S was generated using the QuikChange mutagenesis kit. A plasmid encoding Drosophila melanogaster Porc (a gift from Dr. Mary Baylies, Sloan-Kettering Institute) was tagged with HA-FLAG-His using a PCR-based strategy. The Porc-HA-FLAG-His PCR product was ligated into EcoRI- and BamHI-cut pcDNA3.1. See the supplemental “Experimental Procedures” for additional cloning details. All constructs and mutations were confirmed by DNA sequencing.
Cell Culture and Transfection—COS-1 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 50 units/ml penicillin, and 50 μg/ml streptomycin. 293FT cells (Invitrogen) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 50 units/ml penicillin, 50 μg/ml streptomycin, 500 μg/ml Geneticin, 1 mm GlutaMAX (Invitrogen), 1 mm sodium pyruvate, and 0.1 mm nonessential amino acids. Transfections were carried out using Lipofectamine (Invitrogen).
Synthesis of [125I]Iodopalmitate and [3H]Palmitate Analogs—Radioiodination of iodopalmitate with [125I]NaI and synthesis of [125I]iodopalmitoyl-CoA and [3H]palmitoyl-CoA derivatives using CoA synthetase were carried out as described previously (27, 28). The final concentrations of purified [125I]iodopalmitoyl-CoA and [3H]palmitoyl-CoA were determined from the absorbance at 260 nm using the extinction coefficient for palmitoyl-CoA.
In Vivo Palmitate Labeling—COS-1 cells expressing Shh, Fyn, or Shh-GFP fusions and Hhat were starved for 1 h in Dulbecco's modified Eagle's medium containing 2% dialyzed fetal calf serum, followed by incubation with 10–20 μCi/ml [125I]IC16 (27) for 4 h at 37 °C. Cells were washed twice with 2 ml of ice-cold STE buffer (100 mm NaCl, 10 mm Tris, and 1 mm EDTA (pH 7.4)) and lysed in 500 μl of radioimmune precipitation assay (RIPA) buffer (150 mm NaCl, 50 mm Tris, (pH 7.4), 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and 1 mm EDTA). Lysates were clarified by ultracentrifugation at 100,000 × g for 15 min in a Beckman T100.2 rotor. Protein levels were determined by SDS-PAGE and Western blot analysis. Immunoprecipitations were performed by incubating clarified lysates with 5 μl of the appropriate antibody and 50 μl of protein A/G+-agarose beads (Santa Cruz Biotechnology) at 4 °C for 16 h. The beads were washed twice with 500 μl of RIPA buffer. The final bead pellets were resuspended in 40 μl of 2× SDS-PAGE sample buffer containing 40 mm dithiothreitol (DTT). Immunoprecipitated samples were run on a 12.5% SDS-polyacrylamide gel, dried, and exposed by phosphorimaging for 2–3 days. Screens were analyzed on a FUJIFILM FLA-7000 bioimaging analyzer. Labelings were performed in duplicate and repeated three times. For hydroxylamine treatment, gels were soaked in either 1 m Tris or hydroxylamine (pH 8.0) for 1 h and then dried and analyzed as described above.
Expression and Purification of Recombinant Shh—N-terminally His6-tagged human Shh-(24–197) with an enterokinase cleavage site immediately upstream of residue 24 was amplified using full-length Shh as a template. The purified PCR product was ligated in NcoI- and BamHI-cut pET19b (Novagen). C24S and C24A constructs were generated by site-directed mutagenesis using the QuikChange mutagenesis kit. All mutations were confirmed by sequencing. His-tagged Shh-(24–197) constructs were expressed in Escherichia coli BL21-CodonPlus(DE3) (Novagen), purified on Ni-NTA-agarose resin, and dialyzed (20 mm Tris-HCl (pH 8.0), 350 mm NaCl, and 1 mm β-mercaptoethanol) in the presence of enterokinase. The dialyzed product was further purified by size exclusion chromatography on a Superdex 75 column (GE Healthcare). Pooled fractions after size exclusion chromatography were concentrated to 3.0–3.5 mg/ml in 20 mm HEPES (pH 7.3), 100 mm NaCl, and 1 mm DTT. The protein concentration was measured using the DC protein assay (Bio-Rad). The N termini of both wild-type and mutant proteins were confirmed by Edman degradation.
Hhat-HA-FLAG-His Purification—Plates (20 × 100 mm) of 293FT cells were transfected with Hhat-HA-FLAG-His or pcDNA3.1 empty vector. 48 h post-transfection, the cells were placed on ice, washed twice with 5 ml of ice-cold STE buffer, and then scraped into 5 ml of STE buffer/plate. Cells were pelleted by centrifugation at 1000 × g for 10 min. Cell pellets were resuspended in 8 ml of cold hypotonic lysis buffer (0.2 mm MgCl2 and 10 mm HEPES (pH 7.3)). After a 15-min incubation on ice, cells were lysed by 30 up/down strokes in a Dounce homogenizer with a tight fitting pestle. After lysis, 2 ml of 1.25 m sucrose was added to yield 10 ml of total cell lysate. The lysate was separated into soluble (S100) and membrane (P100) fractions by ultracentrifugation at 100,000 × g for 45 min in a Beckman Ti-70.1 fixed angle rotor. After centrifugation, the supernatant was saved, and the P100 pellets were resuspended in 10 ml of hypotonic Lysis buffer plus 0.25 m sucrose and recentrifuged as described above. The resultant supernatant was combined with the supernatant from the first spin for a total of 20 ml of S100. The P100 membranes were again resuspended in 10 ml of hypotonic lysis buffer plus 0.25 m sucrose and recentrifuged as described above. The supernatant was discarded, and the pellets were resuspended in 10 ml of solubilization/wash buffer (20 mm HEPES (pH 7.3), 350 mm NaCl, 1% octyl glucoside, and 1% glycerol) and incubated on ice for 1 h, followed by centrifugation at 100,000 × g. The resultant pellet was discarded; the supernatant (detergent-soluble fraction) was transferred to a 15-ml tube; and 500 ml of FLAG M2 resin (Sigma) was added. Following a 1-h incubation, the FLAG resin was pelleted by centrifugation at 1000 × g and washed four times with 5 ml of solubilization/wash buffer. Hhat-HA-FLAG-His was eluted with 1.5 ml of solubilization/wash buffer supplemented with 300 ng/ml 3XFLAG peptide. The purified sample was concentrated, and buffer was exchanged to a final volume of 0.5–1.0 ml in 20 mm HEPES (pH 7.3), 100 mm NaCl, 1% octyl glucoside, and 1% glycerol. Protein concentrations were determined using the DC protein assay. The concentration of the final FLAG eluate was determined from the absorbance at 280 nm using an extinction coefficient of 193,045 cm-1 m-1. Samples of the final purified fraction were subjected to SDS-PAGE and silver staining.
In Vitro Palmitoylation Assay—The in vitro assay was performed by incubating 10 μl of Hhat-HA-FLAG-His in 20 mm HEPES (pH 7.3), 100 mm NaCl, 1% octyl glucoside, and 1% glycerol with 10 μl of recombinant Shh (0.2–0.4 mg/ml in 20 mm MES (pH 6.5), 1 mm EDTA, and 1 mm DTT), followed by the addition of 30 μl of reaction buffer (167 mm MES (pH 6.5), 1.7 mm DTT, 0.083% Triton X-100, and 167 μm [125I]iodopalmitoyl-CoA). The reaction was stopped by the addition of 50 μl of 2× sample buffer with 40 mm DTT. Samples were electrophoresed on 12.5% SDS-polyacrylamide gels, which were stained with Coomassie Blue, dried, and exposed to phosphorimaging for 12–18 h. After phosphorimaging, each Shh-containing gel band was excised. [125I]Iodopalmitate incorporation was measured by counting in a PerkinElmer γ-counter. Nonenzymatic incorporation of [125I]iodopalmitate into Shh was corrected for by subtraction of counts from matched pcDNA3.1 mock purification controls.
Immunofluorescence and Confocal Microscopy—48 h post-transfection, COS-1 cells were washed with phosphate-buffered saline (PBS) and fixed with 4% (v/v) paraformaldehyde in PBS for 15 min at room temperature. The cells were washed with PBS and permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature, followed by two washes with PBS. Cells were incubated for 30 min in blocking buffer (3% bovine serum albumin in PBS) and then incubated for 1.5 h with primary antibody diluted in blocking buffer. Cells were washed four times with PBS, followed by a 45-min incubation with secondary antibody. Cells were incubated for 5 min in a 1:5000 dilution of Hoechst dye in PBS and washed with PBS, and coverslips were mounted on slides using ProLong Gold mounting solution (Invitrogen). Images were collected on a Zeiss LSM 510 microscope using a 63× water immersion objective.
Biotinylated Shh Peptide Pulldown Assays—C-terminally biotinylated peptides corresponding to the first 11 amino acids of Shh (CGPGRGFGKRR), N-terminally acetylated Shh (AcCGPGRGFGKRR), and Shh C24A (AGPGRGFGKRR) were synthesized by the Sloan-Kettering Microchemistry Core Facility. Purified peptides were palmitoylated in vitro as outlined above, except that the final Shh peptide concentration was 100 μm. After incubation, 400 μl of RIPA buffer and 50 μl of streptavidin-agarose beads were added, and the mixture was incubated for 1 h at 4 °C with continuous mixing. Biotinylated peptides were pelleted by centrifugation at 1000 × g for 5 min. Pellets were washed twice with 500 ml of RIPA buffer. [125I]Iodopalmitate incorporation was determined by γ-counting. Samples were incubated in either 1 m Tris (pH 8.0) or hydroxylamine (pH 8.0) for 1 h at room temperature, followed by two washes with RIPA buffer.
RESULTS
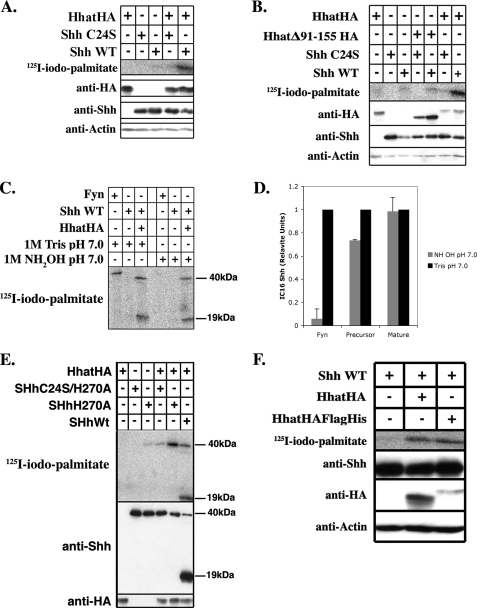
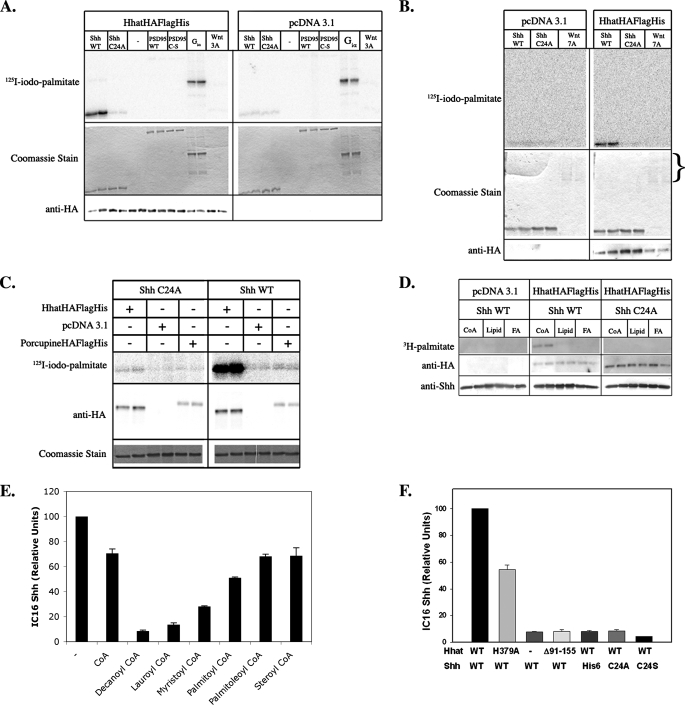
Reconstitution of Shh Palmitoylation in Vivo—Given the inherent difficulty involved in purifying polytopic membrane proteins in an active conformation, we first sought to reconstitute the Shh palmitoylation reaction in tissue culture cells. COS-1 cells were cotransfected with plasmids encoding full-length Shh and either empty pcDNA3.1 vector or HA-tagged Hhat in pcDNA3.1. Cells were labeled for 3–4 h with [125I]IC16 palmitate (herein referred to as [125I]iodopalmitate), a radioiodinated palmitate analog that accurately monitors protein palmitoylation (26, 27). Shh was immunoprecipitated from cell lysates, and the amount of radiolabel incorporated was determined by phosphorimaging analysis after SDS-PAGE. 10-Fold more [125I]iodopalmitate was incorporated into Shh in cells coexpressing Shh and Hhat compared with cells expressing Shh and empty vector (Fig. 1A), indicating that expression of Hhat stimulates Shh palmitoylation. Similar results were obtained when Shh and Hhat were coexpressed in 293FT or 3T3 cells (data not shown). By contrast, Shh C24S, an Shh mutant in which the cysteine modification site was changed to serine, did not incorporate [125I]iodopalmitate when Hhat was overexpressed (Fig. 1A). These findings suggest that Hhat-induced palmitoylation of Shh occurs at the biologically relevant site and that Shh palmitoylation can be faithfully reconstituted in COS-1 cells.
FIGURE 1.
Reconstitution of Shh palmitoylation in vivo. COS-1 cells were transfected with the indicated constructs and labeled with [125I]iodopalmitate for 4 h. Cell lysates were analyzed directly by Western blotting or after Shh immunoprecipitation. A, B, E, and F: upper panels, [125I]iodopalmitate incorporation into immunoprecipitated Shh as detected by phosphorimaging; lower panels, Western blots of the same extracts probed with anti-HA, anti-Shh, and anti-actin antibodies. C: immunoprecipitated samples containing [125I]iodopalmitate-labeled Fyn or Shh loaded in duplicate on a 12.5% SDS-polyacrylamide gel. After electrophoresis, the gel was split in half. Each half was incubated in either 1 m Tris or NH2OH for 1 h at 20°C and then analyzed by phosphorimaging. D: quantification of the experiment in C, performed three times. Phosphorimaging signals were normalized for levels of protein expression; NH2OH sensitivity is expressed as a percentage of Tris-treated controls. WT, wild-type.
Hhat has at least two alternatively spliced variants. When the shorter splice variant of Hhat lacking amino acids 91–155 (HhatΔ91–155) was coexpressed with Shh, no stimulation of Shh palmitoylation above the pcDNA3.1 vector control was detected (Fig. 1B). The expression levels of HhatΔ91–155 were similar to or greater than those of full-length Hhat. This indicates that residues 91–155 of Hhat are required for Hhat function and provides further evidence that full-length Hhat is a PAT for Shh.
Palmitoylation of Shh in Hhat-transfected Cells Occurs via Amide Linkage—Palmitate is attached to Shh via amide linkage to the N-terminal cysteine (N-palmitoylation). To confirm that Shh was N-palmitoylated when coexpressed with Hhat, we treated the palmitoylated samples with hydroxylamine (NH2OH). Thioester linkages are sensitive, whereas amide linkages are resistant to NH2OH treatment. Lysates from [125I]iodopalmitate-labeled cells expressing Fyn, an S-palmitoylated protein, or Shh and Hhat were immunoprecipitated with anti-Shh or anti-Fyn antibody and subjected to SDS-PAGE. The gels were treated with either 1 m Tris or NH2OH and then dried and analyzed by phosphorimaging. Nearly all the radiolabel incorporated into Fyn was lost after treatment with NH2OH, as expected for thioester-linked palmitate. By contrast, Shh retained essentially all of the radiolabel after NH2OH treatment, consistent with palmitate being attached via amide linkage (Fig. 1, C and D).
Shh Palmitoylation Does Not Require Autoprocessing or Cholesterol Incorporation—We next determined whether N-palmitoylation of Shh is dependent on Shh autoprocessing and cholesterol modification at the C terminus. Incorporation of [125I]iodopalmitate radiolabel was evident in the mature 19-kDa form of Shh as well as a band of 40 kDa (Fig. 1C), which likely represents the Shh precursor prior to autocleavage and cholesterol addition. To unambiguously determine whether the uncleaved Shh precursor could be palmitoylated, [125I]iodopalmitate labeling was performed in cells coexpressing Hhat and Shh H270A, an Shh mutant that is defective in autoprocessing and cholesterol incorporation (29). Shh H270A efficiently incorporated [125I]iodopalmitate, and the reaction was dependent on the presence of Hhat and Cys24 (Fig. 1E). Thus, Hhat-mediated palmitoylation of Shh does not require prior autoprocessing, and cholesterol modification is not a prerequisite for N-palmitoylation. Taken together, these data predict that reconstitution of Shh palmitoylation in vitro should be feasible using mature recombinant Shh.
Hhat Mediates Shh Palmitoylation in Vitro—To determine whether Hhat is sufficient for Shh palmitoylation, we set out to purify Hhat and reconstitute Shh palmitoylation in vitro. Because successful purification of multipass membrane-bound enzymes is often achieved using a tandem affinity approach, FLAG and His6 tags were added after the C-terminal HA tag of Hhat to generate Hhat-HA-FLAG-His. [125I]Iodopalmitate labeling of Shh in cells expressing Hhat-HA-FLAG-His revealed that the additional affinity tags did not interfere with Hhat activity (Fig. 1F).
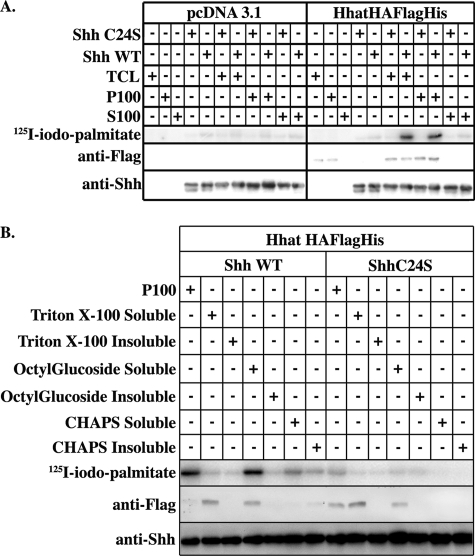
We next established an in vitro Shh palmitoylation assay system using purified recombinant Shh as a source of mature Shh protein substrate and [125I]iodopalmitoyl-CoA as the fatty acyl-CoA donor. Cell lysates derived from COS-1 cells transfected with either Hhat-HA-FLAG-His or empty vector were used initially as a crude source of enzyme. After a 1-h incubation, the reaction was separated by SDS-PAGE, and incorporation of [125I]iodopalmitate into Shh was determined. Little to no detectable radiolabel was incorporated into Shh when incubated with lysates from cells transfected with empty vector (Fig. 2A). By contrast, the Hhat-HA-FLAG-His-containing lysate strongly stimulated in vitro palmitoylation of wild-type Shh, but not Shh C24S, consistent with the results obtained with transfected COS-1 cells in vivo.
FIGURE 2.
Partially purified Hhat-HA-FLAG-His stimulates Shh palmitoylation in vitro. A, COS-1 cells transfected with Hhat-HA-FLAG-His or empty pcDNA3.1 vector were lysed and separated into S100 and P100 fraction as described under “Experimental Procedures.” 10 μl of each fraction was reacted with 2 μg of recombinant wild-type (WT) Shh or Shh C24A and 100 μm [125I]iodopalmitoyl-CoA in reaction buffer (20 mm HEPES (pH 7.3), 200 mm NaCl, 1 mm ATP, 1 mm MgCl2, 1 mm DTT, and 0.05% Triton X-100) for 1 h at room temperature and then separated on 12.5% SDS-polyacrylamide gels. TCL, total cell lysate. B, P100 membranes derived from Hhat-HA-FLAG-His- or pcDNA3.1-expressing cells, as described above, were solubilized in buffer containing 1% octyl glucoside, Triton X-100, or CHAPS. Following ultracentrifugation, detergent-soluble and -insoluble fractions were tested for PAT activity as described above. In A and B, the upper panels are phosphorimages showing [125I]iodopalmitate incorporation into Shh. The lower panels are Western blots of the same samples probed with anti-FLAG and anti-Shh antibodies.
We next separated the lysate into cytosolic (S100) and membrane (P100) fractions by ultracentrifugation at 100,000 × g. The P100 fraction contained all of the Hhat-HA-FLAG-His and all of the Shh palmitoylation activity (Fig. 2A). To determine whether additional cofactors were present in the soluble fraction, increasing amounts of the S100 fraction were added to the P100 fraction. No further enhancement of Shh palmitoylation was observed, indicating that the P100 membrane fraction is sufficient to promote Shh palmitoylation in vitro.
Several different detergents were tested for their ability to solubilize Hhat from the P100 membrane fraction. Only octyl glucoside was capable of solubilizing Hhat-HA-FLAG-His in active form (Fig. 2B). Octyl glucoside does not absorb in the UV range and has a high critical micelle concentration, features that are desirable for membrane protein purification. We therefore selected this nonionic detergent for Hhat solubilization and subsequent purification steps.
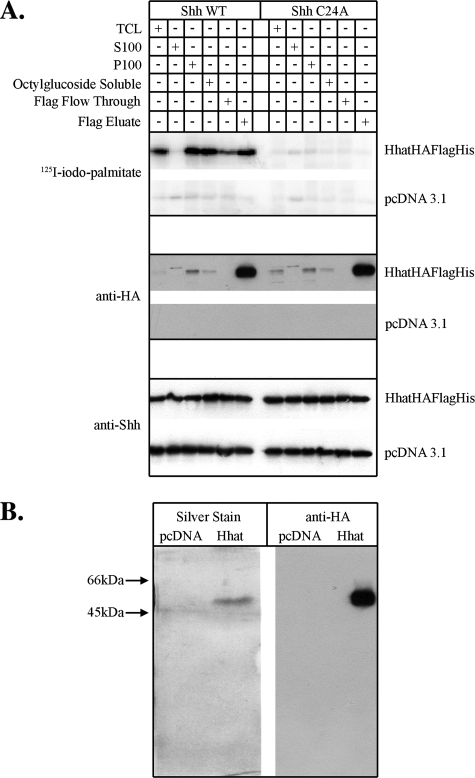
Purified Hhat Is a PAT That Mediates Shh Palmitoylation—Several cell lines were tested in an effort to maximize Hhat expression. 293FT cells transfected with Hhat-HA-FLAG-His cDNA expressed ∼3–5-fold more Hhat-HA-FLAG-His protein/plate compared with transfected COS-1 cells, and Hhat-HA-FLAG-His was efficiently solubilized from 293FT membranes by octyl glucoside in active form. Initial attempts to bind Hhat-HA-FLAG-His to Ni-NTA resin were unsuccessful. We therefore turned to anti-FLAG immunoaffinity chromatography, which allowed us to successfully purify Hhat (Table 1). Octyl glucoside-solubilized Hhat-HA-FLAG-His was bound to an anti-FLAG M2-agarose column and eluted with solubilization buffer containing 3XFLAG peptide (Fig. 3A). Eluted samples were concentrated and analyzed by SDS-PAGE and silver staining. A single prominent silver-stained band, migrating with a molecular mass of ∼50 kDa, was present after FLAG affinity purification (Fig. 3B). Western blot analysis with anti-HA and anti-FLAG antibodies indicated that this band is the Hhat-HA-FLAG-His construct (Fig. 3B and data not shown). As a negative control, a mock sample was prepared from cells transfected with empty vector and carried through the entire purification protocol; no bands were detected in either the silver stain or Western blot (Fig. 3B). These results indicate that Hhat has been purified to essential homogeneity.
TABLE 1.
Hhat purification from 293FT cells
| Fraction | Protein | Total activity | Specific activity | Purification |
|---|---|---|---|---|
| mg | pmol/min | pmol/min/mg | -fold | |
| Total cell lysate | 20.2 | 3543.8 | 175.4 | 1 |
| Total membranes | 10.6 | 3556.23 | 335.5 | 1.9 |
| Octyl glucoside-soluble | 6.0 | 7013.5 | 1168.9 | 6.7 |
| FLAG eluate | .05 | 2547.6 | 50,952 | 290.5 |
FIGURE 3.
Purification of Hhat to homogeneity. A, 293FT cells transfected with Hhat-HA-FLAG-His or pcDNA3.1 vector were lysed, and Hhat-HA-FLAG-His was purified as described under “Experimental Procedures.” 10 μl of the indicated fraction was assayed for PAT activity as described in the legend to Fig. 2. Upper panels, phosphorimages showing [125I]iodopalmitate incorporation into Shh; middle and lower panels, Western blots of the same samples probed with anti-HA and anti-Shh antibodies. WT, wild-type; TCL, total cell lysate. B, shown are silver stain and Western blot analysis of purified Hhat-HA-FLAG-His. The FLAG eluate fraction was concentrated 5-fold and then electrophoresed on a 12.5% SDS-polyacrylamide gel. Gels were either fixed and silver-stained or Western-blotted with Anti-HA antibodies. The entire length of each gel is shown.
Purified Hhat-HA-FLAG-His was tested for PAT activity in the in vitro Shh palmitoylation assay. Specific incorporation of [125I]iodopalmitate was detected into wild-type Shh, but not Shh C24A (Fig. 3A). Little to no radiolabel incorporation occurred when empty vector-transfected samples were used. Together with the data depicted in Fig. 3B, this result supports the hypothesis that Hhat is a bona fide Shh PAT.
Characterization of Hhat-mediated Shh Palmitoylation—The FLAG column eluate fraction had lower activity than expected based on the increase in Hhat protein levels (Fig. 3A). To determine whether a cofactor had been lost during purification, increasing amounts of the FLAG column flow-through fraction were added to the FLAG eluate. No further stimulation of Shh palmitoylation was observed, suggesting that the loss of specific activity was not due to the loss of a cofactor in the FLAG affinity purification.
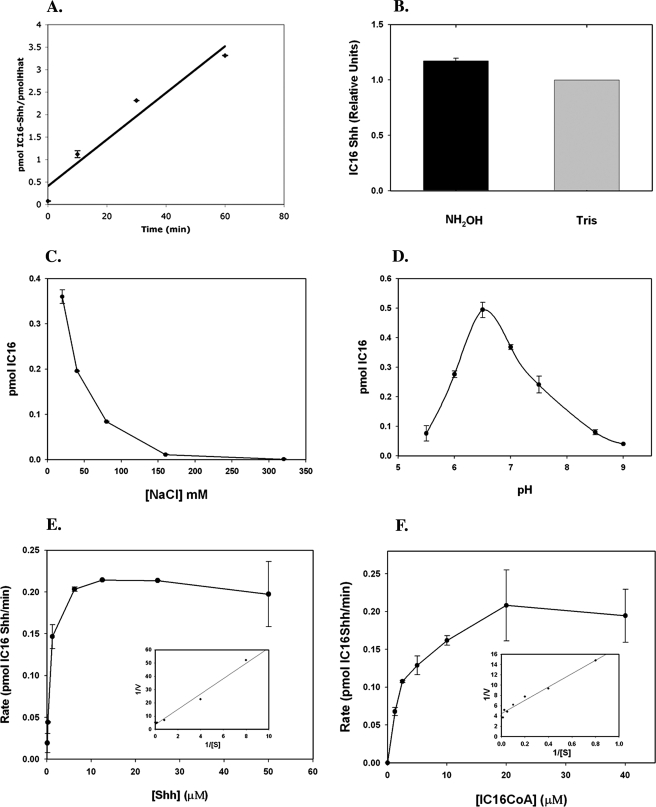
We hypothesized that purified Hhat might be acutely sensitive to components in the assay mixture and therefore set out to optimize the reaction conditions. Incorporation of [125I]iodopalmitate into Shh was linear for at least 1 h at room temperature (Fig. 4A). At this time point, a ratio of 3.5 mol of palmitoylated Shh/mol of Hhat was reached, indicating that Hhat-mediated palmitoylation of Shh is catalytic. Nearly all of the radiolabel incorporated into Shh by purified Hhat was resistant to NH2OH treatment, consistent with palmitate attachment occurring via amide linkage (Fig. 4B). Hhat activity was strongly inhibited by increasing NaCl concentrations (Fig. 4C), and this likely accounts for the reduced activity in the original FLAG eluate fraction (which contained 187 mm NaCl). Hhat exhibited optimal activity at pH 6.5 (Fig. 4D). This matches the pKa for histidine and is consistent with the presence of a highly conserved histidine residue in MBOAT family members thought to be involved in catalysis (23). Additional optimization was performed (Table 2), leading to final optimized in vitro reaction conditions as follows: 20 mm NaCl, 100 mm MES (pH 6.5), 1 mm DTT, 0.2% octyl glucoside, 0.2% glycerol, 0.05% Triton X-100, and 2 mm EDTA. Under these conditions, a maximal stoichiometry of ∼0.7 mol of iodopalmitate/mol of Shh was achieved.
FIGURE 4.
Characterization of Hhat PAT activity. A, purified Shh (70 pmol) was incubated with purified Hhat-HA-FLAG-His (2. 5 pmol) or FLAG eluate from mock-transfected samples in the presence of 100 μm [125I]iodopalmitoyl-CoA for the indicated time points.B, the 1-h time point reaction was analyzed by SDS-PAGE, and gels were incubated in either 1 m Tris or NH2OH for 1 h and dried. Phosphorimaging signals were normalized for protein expression levels; NH2OH sensitivity is expressed as a percentage of Tris-treated controls. C and D, the reaction was performed for 1 h at the indicated NaCl concentrations or pH, respectively. E, Shh at the indicated concentrations was incubated with purified Hhat-HA-FLAG-His (2.5 pmol) in the presence of 100 μm [125I]iodopalmitoyl-CoA for 1 h. F, [125I]iodopalmitoyl-CoA at the indicated concentrations was incubated with purified Hhat-HA-FLAG-His (2.5 pmol) in the presence of 50 μm Shh. The insets for E and F represent Lineweaver-Burk plots. For each panel, Shh protein bands were excised from dried gels, and the amount of [125I]iodopalmitate incorporation was determined by γ-counting. Graphs represent the average of three experiments corrected for nonspecific incorporation of [125I]iodopalmitate as described under “Experimental Procedures.”
TABLE 2.
Hhat palmitoylation: reaction conditions and kinetics
|
Optimization of in vitro palmitoylation reaction conditions
| ||||||
|---|---|---|---|---|---|---|
| pH | [NaCl] | [DTT] | [Triton X-100] | [ATP] | [Divalent cation] | |
| Initial | 100 mm HEPES (pH 7.3) | 200 mm | 1 mm | 0.05% | 1 mm | 1 mm MgCl2 |
| Final | 100 mm MES (pH 6.5) | 20 mm | 1 mm | 0.05% | 0 | 0 |
| Required | Yes | Yes | No | No | ||
|
Kinetic measurements
| |||
|---|---|---|---|
|
Shh titration at 100 μm
[125I]iodopalmitoyl-CoA
|
[125I]Iodopalmitoyl-CoA titration at 50 μm Shh
|
||
| Vmax | Km | Vmax | Km |
| pmol/min | μm | pmol/min | μm |
| 0.25 ± 0.03 | 1.25 ± 0.26 | 0.21 ± 0.03 | 3.0 ± 0.28 |
Kinetic analyses were performed by titrating the levels of both Shh and [125I]iodopalmitoyl-CoA while keeping the Hhat concentration constant. The rate of Hhat-mediated Shh palmitoylation increased with increasing concentrations of each individual substrate and then reached saturation (Fig. 4, E and F), a feature consistent with an enzymatic reaction. Although Shh palmitoylation involves two substrates and therefore will not strictly obey Michaelis-Menten kinetics, we derived apparent Vmax and Km values by titrating Shh at a maximal iodopalmitoyl-CoA concentration and vice versa. These values are reported in Table 2.
Hhat PAT Exhibits Apparent Specificity for Shh—We tested the ability of Hhat to stimulate palmitate incorporation into other proteins known to be S-palmitoylated. No increase in palmitoylation of PSD95 (Fig. 5A) or prenylated H-Ras (data not shown) was detected in the presence of purified Hhat compared with the FLAG eluate prepared from mock-transfected cells. Moreover, Hhat had no effect on the amount of [125I]iodopalmitate incorporated into myristoylated Gαi (Fig. 5A), a protein that undergoes nonenzymatic palmitoylation (30).
FIGURE 5.
Hhat PAT activity is specific for Shh. A and B, purified Hhat-HA-FLAG-His (2. 5 pmol) was incubated either alone (-) or with 2 μg of wild-type (WT) Shh, Shh C24A, wild-type PSD95, PSD95 Cys → Ser, Gαi, or Wnt7A or 0.2 μg or Wnt3A and 100 μm [125I]iodopalmitoyl-CoA for 1 h at room temperature. [125I]Iodopalmitate incorporation was detected by phosphorimaging (upper panels). Substrate proteins were detected by Coomassie Blue staining (middle panels). Hhat-HA-FLAG-His levels were detected by Western blotting with anti-HA antibody (lower panels). Wnt proteins are glycosylated at multiple sites, which makes analysis of Wnt7A levels by SDS-PAGE and Coomassie Blue staining problematic (note multiple diffuse bands). However, protein assays confirmed that Wnt7A was present at levels similar to Shh. C, P100 membranes derived from 293FT cells expressing Hhat-HA-FLAG-His, Porc-HA-FLAG-His, or empty pcDNA3.1 vector were incubated with 2 μg of wild-type Shh or Shh C24A along with 100 μm [125I]iodopalmitoyl-CoA for 1 h at room temperature. Detection was performed as described above. D, purified Hhat-HA-FLAG-His (2.5 pmol) was incubated with 2 μg of wild-type Shh or Shh C24A along with 20 μm either [3H]palmitoyl-CoA (CoA), [3H]dipalmitoylphosphatidylethanolamine (Lipid), or [3H]palmitic acid (FA) for 1 h at room temperature. [3H]Palmitate incorporation was detected by fluorography (upper panel). Hhat-HA-FLAG-His and Shh were detected by Western blotting with anti-HA antibody (middle panel) or anti-Shh antibody (lower panel). E, purified Hhat-HA-FLAG-His (2.5 pmol) was incubated with 2 μg of purified wild-type Shh and 50 μm [125I]iodopalmitoyl-CoA alone (-) or with the indicated CoA (100 μm) for 1 h at room temperature. [125I]Iodopalmitate incorporation was detected by phosphorimaging (IC16-Shh). Graphs represent the average of three experiments. F, 10 μl (2.5 pmol) of purified Hhat-FLAG-His (WT), Hhat-FLAG-His-H379A (H379A), Hhat-HA-His-FLAGΔ91–155 (Δ91–155), or FLAG eluate from mock-transfected samples (-) was incubated with 2 μg of wild-type Shh, Shh C24A, Shh C24S, or N-terminally His6-tagged wild-type Shh (His6) for 1 h at room temperature. [125I]Iodopalmitate incorporation was detected by phosphorimaging (IC16-Shh). Graphs represent the average of three experiments.
Wnt family proteins, such as Shh, are secreted morphogens that are also palmitoylated. Wnt palmitoylation is likely mediated by the MBOAT family protein Porc. Given these similarities, we evaluated cross-reactivity between the two systems. No increase in palmitoylation of purified Wnt3A was detected in the presence of purified Hhat (Fig. 5A). However, commercially purified Wnt3A was available only in short supply, and the levels of Wnt3A used in the assay were 10-fold lower than those of Shh. The in vitro assay was then repeated using Wnt7A, which is available in larger quantities. Even at protein levels equal to those of Shh, no increase in Wnt7A palmitoylation was observed in the presence of Hhat (Fig. 5B).
We next determined whether Porc could palmitoylate Shh. P100 membranes were prepared from cells expressing either Hhat-HA-FLAG-His or Porc-HA-FLAG-His. Only Hhat-HA-FLAG-His-containing membranes were able to induce Shh palmitoylation in vitro (Fig. 5C). These findings suggest that there is little to no cross-reactivity between the Wnt/Porc and Shh/Hhat systems and imply that Hhat is specific for Shh.
Free Fatty Acid and Phospholipids Do Not Act as Palmitate Donors in Vitro—The identity of the palmitate donor was determined by performing in vitro palmitoylation assays in the presence of molar equivalents of [3H]palmitoyl-CoA, [3H]dipalmitoylphosphatidylethanolamine, or [3H]palmitic acid. Radiolabel incorporation from [3H]palmitoyl-CoA, but not [3H]dipalmitoylphosphatidylethanolamine or [3H]palmitic acid, was detected in Shh (Fig. 5D), indicating that fatty acyl-CoA is the likely palmitate donor.
We next examined the ability of different chain length fatty acyl-CoAs to compete with [125I]iodopalmitoyl-CoA. Nonradioactive fatty acyl-CoAs with acyl chain lengths ranging from 10 to 18 carbons were added to the in vitro reaction at a 2-fold molar excess to [125I]iodopalmitoyl-CoA. The addition of palmitoyl-CoA inhibited Hhat-mediated [125I]iodopalmitate incorporation into Shh by 55%, whereas monounsaturated palmitoleoyl-CoA (16:1) or the longer chain stearoyl-CoA (18:0) was less effective (Fig. 5E). Surprisingly, shorter acyl chain length CoAs (myristoyl-CoA (14:0), lauroyl-CoA (12:0), and decanoyl-CoA (10:0)) were substantially better competitors than palmitoyl-CoA (Fig. 5E). These data suggest that Hhat might be able to bind to a wide variety of acyl-CoAs. However, because palmitoyl-CoA is the predominant fatty acyl-CoA in vivo and the levels of free short chain acyl-CoAs are extremely low (31, 32), chain length specificity is probably dictated by control of acyl-CoA availability.
Mechanistic Basis for Hhat-mediated Palmitoylation of Shh—The molecular mechanism of Shh palmitoylation was explored in further detail. MBOAT family members contain a highly conserved histidine that is believed to be important for catalysis. Mutation of the corresponding residue in Hhat, His379, to alanine resulted in a nearly 50% loss of activity (Fig. 5F), consistent with this histidine playing a role in Hhat catalysis. The Hhat splice variant HhatΔ91–155-HA was devoid of activity in the in vitro assay, indicating that these 65 residues are required for Hhat activity. We then examined the requirements within the Shh sequence for N-palmitoylation. Neither Ala nor Ser could substitute for Cys24 in the Shh palmitoylation reaction (Fig. 5F). Moreover, a free N terminus was required, as N-terminally His-blocked Shh was not a substrate for Hhat, despite the presence of a free thiol group on Cys24 (Fig. 5F). These data imply that an N-terminal cysteine with a free N terminus is a requirement for Hhat-mediated Shh palmitoylation.
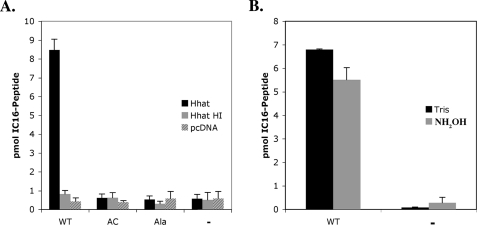
The minimal sequence for N-palmitoylation of Shh is not known. Because all Hedgehog proteins contain a conserved sequence, CGPGR, at their N termini and positions 10 and 11 are arginine residues in Shh, Indian, and Desert Hh, we reasoned that a peptide containing the first 11 amino acids of mature Shh might be sufficient to serve as a substrate in the in vitro PAT assay. A peptide with the wild-type Shh sequence incorporated [125I]iodopalmitate via an amide bond in an Hhat-dependent manner, whereas a peptide with an N-terminal Ala did not (Fig. 6, A and B). An N-terminal cysteine with a free N terminus was required because a wild-type peptide containing a blocked, acetylated N terminus did not incorporate [125I]iodopalmitate (Fig. 6A). These data further support the hypothesis that Hhat-mediated palmitoylation of Shh requires a Cys residue with a free amino terminus and indicate that the first 11 amino acids of Shh are sufficient for recognition by Hhat.
FIGURE 6.
A peptide containing the first 11 amino acids of Shh is palmitoylated by Hhat. A, 10 μl (5 pmol) of purified Hhat-HA-FLAG-His that was untreated (Hhat) or heat-inactivated (95 °C, 5 min; Hhat HI) or FLAG eluate from mock-transfected samples (pcDNA) was incubated either alone (-) or with 100 μm wild-type biotinylated Shh peptide (WT), N-terminally acetylated biotinylated Shh peptide (AC), or biotinylated Shh peptide (Ala) for 1 h at room temperature. Biotinylated peptides were precipitated using streptavidin-agarose. [125I]Iodopalmitate incorporation was determined byγ-counting. B, hydroxylamine sensitivity was determined by soaking wild-type and no-substrate (-) samples in 0.1 m Tris or NH2OH for 18 h at room temperature. Graphs represent the average of three experiments.
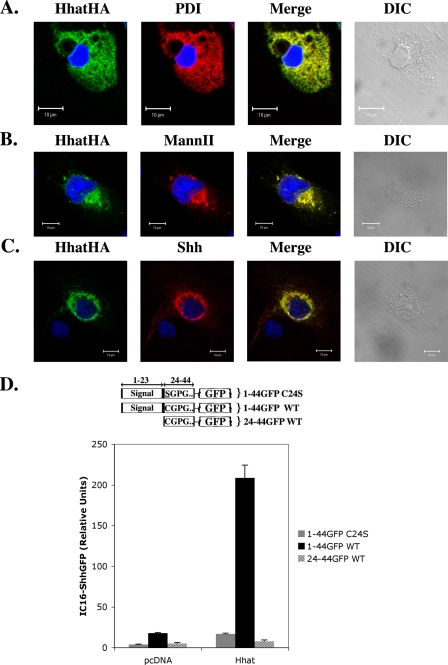
Hhat-mediated Palmitoylation of Shh Occurs within the Secretory Pathway—Palmitoylation of Shh has been proposed to occur in a luminal compartment in the secretory pathway (22). To test this hypothesis, the subcellular localization of both Hhat and Shh was determined. COS-1 cells expressing Hhat-HA were probed with antibodies directed against HA and either protein-disulfide isomerase, an ER marker, or mannosidase II, a Golgi marker, and then analyzed by indirect immunofluorescence. In ∼70% of the cells, a compact perinuclear staining was observed that colocalized with mannosidase II (Fig. 7B). In the remaining ∼30% of the cells, Hhat-HA formed a diffuse reticulate pattern that colocalized with protein-disulfide isomerase (Fig. 7A). When COS-1 cells coexpressing Hhat-HA and Shh were analyzed, colocalization of Hhat-HA and Shh was observed. The highest degree of colocalization occurred in a compact perinuclear region, most likely the Golgi (Fig. 7C). These observations are consistent with Hhat-mediated palmitoylation of Shh occurring during transit through the secretory pathway.
FIGURE 7.
Hhat-mediated Shh palmitoylation occurs within the secretory pathway. A–C, COS-1 cells transfected with Hhat-HA alone or with full-length Shh were fixed and processed for indirect immunofluorescence. Hhat-HA colocalization with protein-disulfide isomerase (PDI) (A), mannosidase II (MannII) (B), and Shh (C) is depicted. D, COS-1 cells cotransfected with the indicated Shh-GFP fusion protein (schematized above graph) and either Hhat-HA or empty pcDNA3.1 vector were labeled with [125I]iodopalmitate. Fusion proteins were immunoprecipitated with anti-GFP antibody, and [125I]iodopalmitate incorporation was detected by phosphorimaging. Graphs represent the average of three experiments. WT, wild-type.
To directly test whether luminal access is required for Shh palmitoylation, we generated fusion constructs containing the first 21 amino acids (positions 24–44) of mature Shh, with or without the Shh signal peptide, fused to GFP (Fig. 7D). When coexpressed with Hhat, a Shh-GFP fusion construct containing the Shh signal peptide incorporated [125I]iodopalmitate in a Cys24- and Hhat-dependent manner (Fig. 7D). By contrast, a construct lacking the signal peptide was unable to incorporate palmitate (Fig. 7D). This result supports the hypothesis that Hhat-mediated palmitoylation requires entry into the secretory pathway and implies that palmitoylation occurs within the lumen of either the ER and/or Golgi.
DISCUSSION
In this work, we combined an in vitro assay for Shh palmitoylation with affinity chromatography of epitope-tagged Hhat to purify enzymatically active Hhat to apparent homogeneity. This is the first demonstration of Shh palmitoylation by Hhat in a purified system. Several lines of evidence strongly support the hypothesis that Hhat is both necessary and sufficient for Shh palmitoylation. First, genetic studies revealed that Hh/Shh is not palmitoylated when produced by Hhat/Rasp null cells (20–22). Second, N-palmitoylation of purified Shh can be recapitulated in vitro, and the reaction is stimulated 10–20-fold by the addition of purified Hhat. Third, Hhat deletion (Δ91–155) and substitution (H379A) mutants exhibit no or reduced Shh palmitoylation activity in vivo and in vitro, respectively, and Shh palmitoylation activity is destroyed when purified Hhat is first subjected to heat denaturation. Fourth, purified Hhat exhibits specificity for palmitoylation of the N-terminal cysteine residue of Shh. The Hhat reaction is catalytic and achieves near-stoichiometric incorporation of palmitate into Shh. These findings directly implicate Hhat as a PAT with specificity for Shh.
Fatty Acylation of Secreted Proteins by MBOAT Family Members—The MBOAT family was originally defined as a family of O-acyltransferases that catalyze attachment of hydrophobic moieties to lipid substrates (23). To date, three MBOAT family members have been proposed to mediate fatty acid attachment to protein targets: Hhat, Porc, and GOAT (22, 33–35). The data presented herein provide the first biochemical demonstration that a purified MBOAT family member is capable of attaching a fatty acid to a protein substrate rather than to another lipid.
Nearly all palmitoylated proteins contain palmitate linked via a thioester bond (36, 37). S-Palmitoylation of proteins is carried out primarily by members of the DHHC family of palmitoylacyltransferases (36, 38). Analysis of the palmitoyl proteome of yeast revealed that there is overlap in the targets for DHHC PATs and that individual S-palmitoylated proteins can be recognized by multiple DHHC PATs (39). We therefore analyzed whether purified Hhat can palmitoylate other proteins that are substrates for either DHHC or MBOAT PATs. The data presented in this study and in a recent study of GOAT indicate that Hhat does not palmitoylate DHHC substrates (PSD95, H-Ras, and Gαs) (Fig. 5) or other MBOAT substrates (such as Wnt proteins (Fig. 5) and proghrelin (34)). Conversely, Shh is not palmitoylated by another MBOAT family protein, Porc (Fig. 5). We conclude that Hhat exhibits specificity for palmitoylating Shh and suggest that this reaction cannot be recapitulated by other PATs.
Mechanism of N-Palmitoylation by Hhat—There are two unique aspects of palmitoylated Shh that distinguish it from other palmitoylated proteins: the modified residue is the N-terminal cysteine (rather than an internal cysteine), and palmitate is linked exclusively via an amide bond (rather than a thioester). Both of these features are recapitulated in the in vitro Shh palmitoylation reaction. Purified Hhat specifically recognizes an N-terminal cysteine residue; neither alanine nor serine can substitute. The inability of serine to serve as a palmitate acceptor implies that the fatty acid cannot be attached via oxyester linkage to the serine hydroxyl group. By contrast, two other proteins that are MBOAT family substrates accept fatty acids onto serine residues. Wnt3A is modified by attachment of palmitoleic acid to Ser209 (in addition to palmitate at Cys77), and proghrelin is modified by octanoate linked to Ser-3 (34, 35).
Why does Shh contain amide-linked rather than thioester-linked palmitate? One possibility, as suggested by Pepinsky et al. (10), is that palmitate is initially attached via thioester linkage to the cysteine sulfhydryl group. Next, an intramolecular S-to-N shift occurs, transferring the fatty acid to the N-terminal amino group (10). This two-step mechanism is operative for nonenzymatic palmitoylation of Shh proteins or peptides (10, 16, 40). However, a complication of this model is that it predicts that Hhat should be capable of attaching two molecules of palmitate to the N-terminal cysteine of Shh: one via an amide bond and one via a thioester bond. A mechanism for removal of the second, thioester-linked palmitate (e.g. a thioesterase) would have to be invoked.
Alternatively, Hhat might catalyze direct attachment of palmitate to the N-terminal cysteine NH2 without a thioester intermediate. Our data favor this mechanism. At all time points tested in the in vitro palmitoylation reaction (2–60 min), the bond between palmitate and Shh was an amide linkage. Neither N-terminally His-tagged Shh nor an N-acetylated Shh peptide was palmitoylated by Hhat, despite the presence of a free sulfhydryl group on the N-terminal cysteine (Figs. 5 and 6). We suggest that Hhat catalyzes Shh palmitoylation via direct amide linkage of palmitate to the free N terminus of Shh. This mechanism is analogous to N-myristoylation, whereby N-myristoyltransferase attaches myristate to the N-terminal glycine of acceptor substrates via amide linkage (41). Additional studies will be needed to determine whether a thioester intermediate transiently forms during Hhat catalysis.
To date, only three proteins have been shown to contain N-linked palmitate: Shh/Hh and Spitz, which are substrates for Hhat, and Gαs (10, 37, 42–44). The mode of attachment of the fatty acid has important implications for signaling function. Thioester-linked palmitate is removed by palmitoyl-protein thioesterases, allowing S-palmitoylated proteins to undergo reversible cycles of palmitoylation/depalmitoylation (45). By contrast, amide-linked palmitate does not undergo turnover, resulting in stable attachment of the fatty acid to Shh and other N-palmitoylated proteins.
Palmitoylation in the Lumen of the Secretory Pathway—At least two lines of evidence support the hypothesis that Hhat-mediated palmitoylation occurs during passage of Shh through the secretory pathway. First, both Shh and Hhat colocalize in the ER and Golgi. Second, the Shh signal sequence is required for Hhat-mediated palmitoylation in vivo of Shh-GFP fusion constructs (Fig. 7). Two other MBOAT proteins, Porc and GOAT, have been proposed to access their substrates in the lumen of the ER. It is interesting to note that the internal pH of the ER is 7.0 and that of the medial/trans-Golgi is 6.58 (46, 47); both values are at or near the pH optimum of 6.5 measured for Hhat. These findings are consistent with the notion that Hhat-mediated palmitoylation of Shh occurs in the lumen of the ER and/or Golgi.
The fatty acid donor in the Hhat palmitoylation reaction is palmitoyl-CoA (Fig. 5), yet long chain acyl-CoAs are not permeable across the ER membrane (48). There are, however, several mechanisms that would allow palmitoyl-CoA to gain access to the lumen of the ER and/or Golgi. Free fatty acids can be specifically transported into the ER lumen, where they are converted by luminal acyltransferases into palmitoyl-CoA (49). Moreover, the ER contains a carnitine palmitoyltransferase that generates palmitoylcarnitine, which is then transported across the ER membrane into the lumen and converted to palmitoyl-CoA (48, 50). Palmitoyl-CoA hydrolase activities have also been detected in the ER lumen, consistent with the presence of luminal palmitoyl-CoA (51, 52). Thus, the ER lumen is a likely source of palmitoyl-CoA for palmitoylation mediated by ER-localized MBOAT PATs such as Hhat and Porc.
It is not known if N-palmitoylation of Shh by Hhat occurs co- or post-translationally and how the reaction is coordinated with signal sequence cleavage. Clearly, the signal sequence must be removed to obtain amide-linked palmitate on mature Shh. Insights into a potential mechanism can be obtained by comparing Shh modification with that of secreted bacterial lipoproteins. The lipoprotein precursor contains a cysteine residue immediately following the signal sequence. This cysteine is first modified by attachment of thioether-linked diacylglyceride. After signal sequence cleavage, the N-terminal lipid-modified cysteine is then further modified by the addition of palmitate to the cysteine amino group (53). Although no additional modifications have been detected on the N-terminal cysteine of Shh (10, 22), it is reasonable to postulate that cleavage of the Shh signal sequence occurs first, followed by Hhat-mediated acylation of the newly exposed Cys24 amino group. Our finding that a free N terminus is required for Shh palmitoylation by Hhat is consistent with this mechanism.
In eukaryotes, two secreted palmitoylated proteins containing a cysteine after the signal sequence have been identified, Shh/Hh and Spitz; both proteins have been shown to be substrates for Hhat/Rasp (42, 43). Flies express at least two other potential Rasp substrates, Keren and Gurken (42), and other Hhat substrates, including the Hedgehog family members Indian and Desert Hedgehog, are likely to be present in mammalian cells. The identification of lipid modifications on Wnt and proghrelin adds to the growing list of secreted proteins that are fatty acylated by MBOAT proteins. Current and future studies of Hhat should therefore shed light on the mechanism of luminal protein fatty acylation and its consequences for the signaling function of lipid-modified secreted proteins.
Supplementary Material
Acknowledgments
We thank Drs. Yutaka Kawakami, Jessica Treisman, Mary Baylies, and Maurine Linder for generous gifts of reagents; Drs. San San Yi and Hediye Erjument-Bromage for peptide synthesis; Drs. Kathryn Anderson and Jessica Treisman for critical reading of the manuscript; and Raisa Louft-Nisenbaum for expert technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants GM57966 and GM008539. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures.”
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: PAT, palmitoylacyltransferase; ER, endoplasmic reticulum; HA, hemagglutinin; GFP, green fluorescent protein; Ni-NTA, nickel-nitrilotriacetic acid; RIPA, radioimmune precipitation assay; DTT, dithiothreitol; MES, 4-morpholineethanesulfonic acid; PBS, phosphate-buffered saline; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
References
- 1.McMahon, A. P., Ingham, P. W., and Tabin, C. J. (2003) Curr. Top. Dev. Biol. 53 1-114 [DOI] [PubMed] [Google Scholar]
- 2.Fuccillo, M., Joyner, A. L., and Fishell, G. (2006) Nat. Rev. Neurosci. 7 772-783 [DOI] [PubMed] [Google Scholar]
- 3.Ho, K. S., and Scott, M. P. (2002) Curr. Opin. Neurobiol. 12 57-63 [DOI] [PubMed] [Google Scholar]
- 4.Chiang, C., Litingtung, Y., Lee, E., Young, K. E., Corden, J. L., Westphal, H., and Beachy, P. A. (1996) Nature 383 407-413 [DOI] [PubMed] [Google Scholar]
- 5.Roessler, E., Belloni, E., Gaudenz, K., Vargas, F., Scherer, S. W., Tsui, L. C., and Muenke, M. (1997) Hum. Mol. Genet 6 1847-1853 [DOI] [PubMed] [Google Scholar]
- 6.di Magliano, M. P., and Hebrok, M. (2003) Nat. Rev. Cancer 3 903-911 [DOI] [PubMed] [Google Scholar]
- 7.Thayer, S. P., di Magliano, M. P., Heiser, P. W., Nielsen, C. M., Roberts, D. J., Lauwers, G. Y., Qi, Y. P., Gysin, S., Fernandez-del Castillo, C., Yajnik, V., Antoniu, B., McMahon, M., Warshaw, A. L., and Hebrok, M. (2003) Nature 425 851-856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mann, R. K., and Beachy, P. A. (2004) Annu. Rev. Biochem. 73 891-923 [DOI] [PubMed] [Google Scholar]
- 9.Porter, J. A., Young, K. E., and Beachy, P. A. (1996) Science 274 255-259 [DOI] [PubMed] [Google Scholar]
- 10.Pepinsky, R. B., Zeng, C., Wen, D., Rayhorn, P., Baker, D. P., Williams, K. P., Bixler, S. A., Ambrose, C. M., Garber, E. A., Miatkowski, K., Taylor, F. R., Wang, E. A., and Galdes, A. (1998) J. Biol. Chem. 273 14037-14045 [DOI] [PubMed] [Google Scholar]
- 11.Lee, J. D., Kraus, P., Gaiano, N., Nery, S., Kohtz, J., Fishell, G., Loomis, C. A., and Treisman, J. E. (2001) Dev. Biol. 233 122-136 [DOI] [PubMed] [Google Scholar]
- 12.Dawber, R. J., Hebbes, S., Herpers, B., Docquier, F., and van den Heuvel, M. (2005) BMC Dev. Biol. 5 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, M. H., Li, Y. J., Kawakami, T., Xu, S. M., and Chuang, P. T. (2004) Genes Dev. 18 641-659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goetz, J. A., Singh, S., Suber, L. M., Kull, F. J., and Robbins, D. J. (2006) J. Biol. Chem. 281 4087-4093 [DOI] [PubMed] [Google Scholar]
- 15.Kohtz, J. D., Lee, H. Y., Gaiano, N., Segal, J., Ng, E., Larson, T., Baker, D. P., Garber, E. A., Williams, K. P., and Fishell, G. (2001) Development (Camb.) 128 2351-2363 [DOI] [PubMed] [Google Scholar]
- 16.Taylor, F. R., Wen, D., Garber, E. A., Carmillo, A. N., Baker, D. P., Arduini, R. M., Williams, K. P., Weinreb, P. H., Rayhorn, P., Hronowski, X., Whitty, A., Day, E. S., Boriack-Sjodin, A., Shapiro, R. I., Galdes, A., and Pepinsky, R. B. (2001) Biochemistry 40 4359-4371 [DOI] [PubMed] [Google Scholar]
- 17.Callejo, A., Torroja, C., Quijada, L., and Guerrero, I. (2006) Development (Camb.) 133 471-483 [DOI] [PubMed] [Google Scholar]
- 18.Gallet, A., Rodriguez, R., Ruel, L., and Therond, P. P. (2003) Dev. Cell 4 191-204 [DOI] [PubMed] [Google Scholar]
- 19.Gallet, A., Ruel, L., Staccini-Lavenant, L., and Therond, P. P. (2006) Development (Camb.) 133 407-418 [DOI] [PubMed] [Google Scholar]
- 20.Lee, J. D., and Treisman, J. E. (2001) Curr. Biol. 11 1147-1152 [DOI] [PubMed] [Google Scholar]
- 21.Micchelli, C. A., The, I., Selva, E., Mogila, V., and Perrimon, N. (2002) Development (Camb.) 129 843-851 [DOI] [PubMed] [Google Scholar]
- 22.Chamoun, Z., Mann, R. K., Nellen, D., von Kessler, D. P., Bellotto, M., Beachy, P. A., and Basler, K. (2001) Science 293 2080-2084 [DOI] [PubMed] [Google Scholar]
- 23.Hofmann, K. (2000) Trends Biochem. Sci. 25 111-112 [DOI] [PubMed] [Google Scholar]
- 24.Chang, C. C., Lee, C. Y., Chang, E. T., Cruz, J. C., Levesque, M. C., and Chang, T. Y. (1998) J. Biol. Chem. 273 35132-35141 [DOI] [PubMed] [Google Scholar]
- 25.Hishikawa, D., Shindou, H., Kobayashi, S., Nakanishi, H., Taguchi, R., and Shimizu, T. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 2830-2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang, X., Nazarian, A., Erdjument-Bromage, H., Bornmann, W., Tempst, P., and Resh, M. D. (2001) J. Biol. Chem. 276 30987-30994 [DOI] [PubMed] [Google Scholar]
- 27.Peseckis, S. M., Deichaite, I., and Resh, M. D. (1993) J. Biol. Chem. 268 5107-5114 [PubMed] [Google Scholar]
- 28.Berthiaume, L., Peseckis, S. M., and Resh, M. D. (1995) Methods Enzymol. 250 454-466 [DOI] [PubMed] [Google Scholar]
- 29.Hall, T. M., Porter, J. A., Beachy, P. A., and Leahy, D. J. (1995) Nature 378 212-216 [DOI] [PubMed] [Google Scholar]
- 30.Duncan, J. A., and Gilman, A. G. (1996) J. Biol. Chem. 271 23594-23600 [DOI] [PubMed] [Google Scholar]
- 31.Bhatnagar, R. S., Schall, O. F., Jackson-Machelski, E., Sikorski, J. A., Devadas, B., Gokel, G. W., and Gordon, J. I. (1997) Biochemistry 36 6700-6708 [DOI] [PubMed] [Google Scholar]
- 32.Schjerling, C. K., Hummel, R., Hansen, J. K., Borsting, C., Mikkelsen, J. M., Kristiansen, K., and Knudsen, J. (1996) J. Biol. Chem. 271 22514-22521 [DOI] [PubMed] [Google Scholar]
- 33.Kadowaki, T., Wilder, E., Klingensmith, J., Zachary, K., and Perrimon, N. (1996) Genes Dev. 10 3116-3128 [DOI] [PubMed] [Google Scholar]
- 34.Yang, J., Brown, M. S., Liang, G., Grishin, N. V., and Goldstein, J. L. (2008) Cell 132 387-396 [DOI] [PubMed] [Google Scholar]
- 35.Takada, R., Satomi, Y., Kurata, T., Ueno, N., Norioka, S., Kondoh, H., Takao, T., and Takada, S. (2006) Dev. Cell 11 791-801 [DOI] [PubMed] [Google Scholar]
- 36.Mitchell, D. A., Vasudevan, A., Linder, M. E., and Deschenes, R. J. (2006) J. Lipid Res. 47 1118-1127 [DOI] [PubMed] [Google Scholar]
- 37.Resh, M. D. (2006) Sci. STKE 2006 RE14. [DOI] [PubMed] [Google Scholar]
- 38.Linder, M. E., and Deschenes, R. (2004) J. Cell Sci. 117 521-526 [DOI] [PubMed] [Google Scholar]
- 39.Roth, A. F., Wan, J., Bailey, A. O., Sun, B., Kuchar, J. A., Green, W. N., Phinney, B. S., Yates, J. R., III, and Davis, N. G. (2006) Cell 125 1003-1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sang, S. L., and Silvius, J. R. (2005) J. Pept. Res. 66 169-180 [DOI] [PubMed] [Google Scholar]
- 41.Farazi, T. A., Waksman, G., and Gordon, J. I. (2001) J. Biol. Chem. 276 39501-39504 [DOI] [PubMed] [Google Scholar]
- 42.Miura, G. I., Buglino, J., Alvarado, D., Lemmon, M. A., Resh, M. D., and Treisman, J. E. (2006) Dev. Cell 10 167-176 [DOI] [PubMed] [Google Scholar]
- 43.Miura, G. I., and Treisman, J. E. (2006) Cell Cycle 5 1184-1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleuss, C., and Krause, E. (2003) EMBO J. 22 826-832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Resh, M. D. (2006) Nat. Chem. Biol. 2 584-590 [DOI] [PubMed] [Google Scholar]
- 46.Kim, J. H., Johannes, L., Goud, B., Antony, C., Lingwood, C. A., Daneman, R., and Grinstein, S. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2997-3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Llopis, J., McCaffery, J. M., Miyawaki, A., Farquhar, M. G., and Tsien, R. Y. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6803-6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gooding, J. M., Shayeghi, M., and Saggerson, E. D. (2004) Eur. J. Biochem. 271 954-961 [DOI] [PubMed] [Google Scholar]
- 49.Rys-Sikora, K. E., and Gill, D. L. (1998) J. Biol. Chem. 273 32627-32635 [DOI] [PubMed] [Google Scholar]
- 50.Sierra, A. Y., Gratacos, E., Carrasco, P., Clotet, J., Urena, J., Serra, D., Asins, G., Hegardt, F. G., and Casals, N. (2008) J. Biol. Chem. 283 6878-6885 [DOI] [PubMed] [Google Scholar]
- 51.Berge, R. K., and Dossland, B. (1979) Biochem. J. 181 119-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mentlein, R., Rix-Matzen, H., and Heymann, E. (1988) Biochim. Biophys. Acta 964 319-328 [DOI] [PubMed] [Google Scholar]
- 53.Pugsley, A. P. (1993) Microbiol. Rev. 57 50-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.