Abstract
Etomidate, one of the most potent general anesthetics used clinically, acts at micromolar concentrations as an anesthetic and positive allosteric modulator of γ-aminobutyric acid responses, whereas it inhibits muscle-type nicotinic acetylcholine receptors (nAChRs) at concentrations above 10 μm. We report here that TDBzl-etomidate, a photoreactive etomidate analog, acts as a positive allosteric nAChR modulator rather than an inhibitor, and we identify its binding sites by photoaffinity labeling. TDBzl-etomidate (>10 μm) increased the submaximal response to acetylcholine (10 μm) with a 2.5-fold increase at 60 μm. At higher concentrations, it inhibited the binding of the noncompetitive antagonists [3H]tetracaine and [3H]phencyclidine to Torpedo nAChR-rich membranes (IC50 values of 0. 8 mm). nAChR-rich membranes were photolabeled with [3H]TDBzl-etomidate, and labeled amino acids were identified by Edman degradation. For nAChRs photolabeled in the absence of agonist (resting state), there was tetracaine-inhibitable photolabeling of amino acids in the ion channel at positions M2-9 (δLeu-265) and M2-13 (αVal-255 and δVal-269), whereas labeling of αM2-10 (αSer-252) was not inhibited by tetracaine but was enhanced 10-fold by proadifen or phencyclidine. In addition, there was labeling in γM3 (γMet-299), a residue that contributes to the same pocket in the nAChR structure as αM2-10. The pharmacological specificity of labeling of residues, together with their locations in the nAChR structure, indicate that TDBzl-etomidate binds at two distinct sites: one within the lumen of the ion channel (labeling of M2-9 and -13), an inhibitory site, and another at the interface between the α and γ subunits (labeling of αM2-10 and γMet-299) likely to be a site for positive allosteric modulation.
The excitatory nicotinic acetylcholine receptor (nAChR)3 is a member of a superfamily of neurotransmitter-gated ion channels that also includes the inhibitory GABAARs (1). Information about the three-dimensional structure of these receptors is based upon the crystal structures of homopentameric acetylcholine-binding proteins from molluscs that are homologous to a nAChR extracellular domain (2–4) and models of the structure of a muscle-type nAChR derived from cryoelectron microscope images of the Torpedo nAChR (5, 6). For each receptor, five homologous subunits associate at a central axis that is the ion channel. The amino-terminal half of each subunit contributes to the extracellular domain where neurotransmitter binding sites are located at subunit interfaces (α-γ and α-δ in the α2βγδ Torpedo nAChR). The transmembrane domain (TMD) of each subunit is made up of a loose bundle of four α helices (M1–M4), with amino acids from each M2 helix contributing to the lumen of the ion channel, that is the binding site for many nAChR inhibitors (7). There are also pockets in the TMD within each subunit helix bundle and at subunit interfaces that are potential binding sites for allosteric modulators. Drugs that bind to such sites and act as positive allosteric modulators of agonist binding may represent an important class of therapeutic agents as they will enhance the efficacy of endogenous neurotransmitter signaling while avoiding the prolonged, nonphysiological pattern of receptor activation produced by agonists.
Drugs that act as positive allosteric modulators of GABAARs include benzodiazepines, which bind in the extracellular domain at a site equivalent to the transmitter binding sites but at a different subunit interface (8, 9), and general anesthetics of diverse chemical structure, including volatiles, neurosteroids, and intravenous agents such as etomidate and barbiturates (10, 11). General anesthetic binding sites, distinct from the transmitter and benzodiazepine sites, are in the GABAAR TMD in the pockets within subunits or at subunit interfaces (11–16). In contrast, most general anesthetics act as negative allosteric modulators of nAChRs (14). Positive allosteric nAChR modulators have been identified, including natural products such as physostigmine and galantamine, which are active on muscle and neuronal nAChRs (17–19), and compounds identified recently through high throughput drug screens that have selectivity for one or more neuronal nAChR subtypes (for reviews, see Refs. 20 and 21). Although physostigmine and galantamine probably bind in the nAChR extracellular domain (4, 22), the locations of the binding sites for the other modulators are unknown.
We report here that TDBzl-etomidate, a photoreactive general anesthetic developed to provide an improved definition of etomidate binding sites in GABAARs (23) (Fig. 1), acts as a novel positive allosteric modulator of muscle-type Torpedo nAChR. We used photoaffinity labeling, an experimental approach that directly identifies amino acids contributing to a drug binding site without assumptions about the protein points of contact (24, 25), to identify its binding sites in the nAChR TMD. Azietomidate, another photoreactive etomidate analog that is a general anesthetic and positive modulator of GABAARs, inhibits nAChRs similarly to etomidate (26). Photoaffinity labeling established that [3H]azietomidate binds in the nAChR ion channel (27), whereas in GABAARs it binds in the TMD at the interface between β and α subunits that contains the γ-aminobutyric acid binding sites in the extracellular domain (16). Upon photoactivation, azietomidate, an aliphatic diazirine, forms a carbonium ion that reacts preferentially with acidic side chains and with nucleophilic residues (tyrosine and methionine) but not with aliphatic side chains. In contrast, TDBzl-etomidate, an aryl diazirine, forms a carbene intermediate that reacts efficiently with aliphatic and most other side chains (24). Based upon the pharmacological specificity of residue labeling and residue location in the nAChR structure, TDBzl-etomidate binds at two distinct sites in the transmembrane domain, one located at the interface between the α and γ subunits that is likely to be the site for positive allosteric modulation and the other within the lumen of the ion channel that is an inhibitory site.
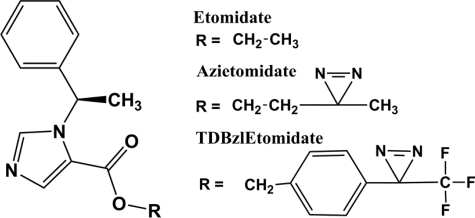
FIGURE 1.
Structures of etomidate and its photoreactive derivatives.
EXPERIMENTAL PROCEDURES
Materials—Membranes rich in nAChRs, containing 1–2 nmol of [3H]acetylcholine (ACh) binding sites/mg of protein, were isolated from Torpedo californica electric organs (Aquatic Research Consultants, San Pedro, CA) as described previously (28). Nonradioactive TDBzl-etomidate and [3H]TDBzl-etomidate (16 Ci/mmol) were synthesized as described previously (23). (R)(+)-etomidate was a gift from Dr. David Gemmell (Organon Laboratories Ltd., Cambridge, UK). [3H]Phencyclidine (PCP; 27 Ci/mmol) was from PerkinElmer Life Sciences, and [3H]tetracaine (30 Ci/mmol) was from Sibtech. [3H]ACh (1.9 Ci/mmol) was synthesized from choline and [3H]acetic anhydride. Staphylococcus aureus glutamyl endopeptidase Glu-C (V8 protease) was from ICN Biomedical, and endoproteinase Lys-C (EndoLys-C) was from Roche Applied Sciences. l-1-Tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin was from Worthington.
Electrophysiology—Standard two-electrode voltage clamp (oocyte clamp OC-725B, Warner Instruments) techniques were used to study the effects of etomidate and TDBzl-etomidate on wild-type Torpedo α2βγδ nAChRs expressed in Xenopus oocytes as described previously (29). TDBzl-etomidate was prepared at 0.1 m in methanol and diluted into low calcium ND96 recording solution (96 mm NaCl, 2 mm KCl, 0.3 mm CaCl2, 1 mm MgCl2, 5 mm HEPES, pH 7.6) containing 1 μm atropine.
Radioligand Binding Assays—The concentration-dependent effects of TDBzl-etomidate on the equilibrium binding of [3H]ACh, [3H]tetracaine, or [3H]PCP to Torpedo nAChR-rich membranes in Torpedo physiological saline (TPS; 250 mm NaCl, 5 mm KCl, 3 mm CaCl2, 2 mm MgCl2, 5 mm NaPO4, pH 7.0) were studied by centrifugation binding assays in a TOMY TX201 centrifuge as described previously (30). TDBzl-etomidate was prepared at 50 mm in methanol and then diluted into TPS. For [3H]tetracaine (19 nm) or [3H]PCP (6 nm; ±1 mm carbamylcholine (Carb)), membrane suspensions (0.2 ml, 750 μg of protein/ml) were equilibrated with the drugs for 2 h prior to centrifugation. For the [3H]ACh binding assay, membrane suspensions were pretreated with diisopropylphosphofluoridate (∼0.5 mm) to inhibit acetylcholinesterase activity, then [3H]ACh (11 nm) and drugs were equilibrated with dilute membrane suspensions (1 ml, 80 μg of protein/ml, 40 nm ACh binding sites) for 45 min before centrifugation. Nonspecific binding was determined in the presence of 1 mm Carb for [3H]ACh, in the presence of 0.2 mm tetracaine for [3H]tetracaine, and in the presence of 1 mm proadifen (+Carb) or 1 mm tetracaine (-Carb) for [3H]PCP. At 1 mm TDBzl-etomidate, the highest concentration tested, the methanol concentration was 2%, which in control experiments altered [3H]ACh and [3H]PCP binding by <5% and reduced [3H]tetracaine binding by 20%.
Data Analysis—The concentration dependence of etomidate inhibition of ACh-induced currents or TDBzl-etomidate inhibition of radioligand binding was fit to the single site binding equation f(x) = f0/[1 + (x/IC50)] + fns where f(x) is the current or total radioligand bound in the presence of inhibitor concentration x, f0 is the current or specific radioligand bound in the absence of inhibitor, fns is the leak current in the absence of ACh or nonspecific radioligand binding, and IC50 is the concentration of inhibitor associated with the inhibition of 50% of ACh-induced currents or radioligand binding.
Photolabeling of nAChR-rich Membranes with [3H]TDBzl-etomidate—Frozen nAChR-rich Torpedo membranes were thawed, diluted 3-fold with TPS, and pelleted by centrifugation. The membrane pellets were then resuspended at 2 mg of protein/ml (∼2.4 μm ACh binding sites) in TPS with oxidized glutathione (1 mm) added to serve as an aqueous scavenger. [3H]TDBzl-etomidate was added to the membrane suspension and mixed by gentle agitation for about 10 min prior to the addition of other ligands. For photolabeling on an analytical scale, the membrane samples in polypropylene microcentrifuge tubes were equilibrated with 0.8 μm [3H]TDBzl-etomidate and additional ligands for 90 min at 4 °C. Following this, aliquots of 130 μl from each sample were placed in a 96-well microtiter plate. For photolabeling on a preparative scale, the membrane suspensions (10 mg of protein/condition) at 2 mg/ml concentration were incubated in 6-cm plastic Petri dishes for 45 min at 4 °C with 1.25 μm [3H]TDBzl-etomidate in the presence or absence of additional ligands. Membrane suspensions were then irradiated for 30 min on ice at 360 nm in a horizontal photochemical chamber reactor (Rayonet RPR-200, Southern New England Ultraviolet Co.) using RPR-3500 bulbs. Following irradiation, the samples were dissolved in sample buffer for SDS-PAGE.
SDS-Polyacrylamide Gel Electrophoresis and Proteolytic Digestions—Photolabeled membranes, solubilized in sample buffer, were separated on 1.5-mm-thick acrylamide gels (31). The resolved polypeptides were visualized by staining with Coomassie Blue R-250 stain (analytical scale experiments) or with GelCode® Blue stain reagent (Pierce) (preparative scale experiments). The stained analytical gels were prepared for fluorography using Amplify (Amersham Biosciences), and the dried gels were exposed to film (Eastman Kodak Co. X-Omat) at -80 °C for various times (4–8 weeks). In parallel experiments, the incorporation of 3H into individual nAChR subunits was quantified by liquid scintillation counting of excised gel slices containing the polypeptide bands of interest (28). For preparative scale photolabeling, the gel bands containing the nAChR β, γ, and δ subunits were excised and passively eluted for 3 days into 12 ml of elution buffer (100 mm NH4HCO3, 0.1% SDS, 2.5 mm dithiothreitol). The α subunit bands were excised and placed in the wells of 15-cm-long, 15% acrylamide “mapping” gels with a 5-cm-long 4.5% acrylamide stacker for limited “in-gel” digestion with S. aureus V8 protease (31). After electrophoresis, the mapping gel was stained with Coomassie Blue R-250, and the proteolytic fragments of 20 kDa (αV8-20), 18 kDa (αV8-18), and 10 kDa (αV8-10) were excised and eluted. The eluates were then filtered and concentrated to <400 μl by centrifugal filtration (Vivaspin 15 Mr 5,000 concentrators, Vivascience Inc., Edgewood, NY). nAChR subunits or subunit fragments were precipitated with 75% acetone (12 h at -20 °C) and then resuspended in 200 μl of resuspension buffer (12 mm Tris, 0.5 mm EDTA, 0.1% SDS, pH 8.6). Based upon the MicroBCA Protein Assay (Pierce), 200–400 μg of nAChR β, γ, and δ subunits and 30–50 μg of αV8-20, αV8-18, and αV8-10 were isolated from 10 mg of membranes.
The αV8-20 fragment (∼40 μg) and δ subunit (∼200 μg) were digested with EndoLys-C (0.5 units/sample) for 14 days. Aliquots of the γ and δ subunits in resuspension buffer were digested for 3 days with V8 protease (100%, w/w). Aliquots of the β subunit or αV8-10 in resuspension buffer supplemented with Genapol C-100 (0.5%) were digested with trypsin (10 units/sample) for 2 days. All digests were carried out at room temperature. The digests of αV8-20 and αV8-10 as well as the V8 protease digests of the γ and δ subunits were fractionated directly by reversed-phase HPLC, whereas the trypsin digests of β subunits and the EndoLys-C digests of δ subunits were further separated on a 1.5-mm-thick, 16.5% T, 6% C Tricine SDS-PAGE gel (32, 33). The Tricine gels were sectioned into 5-mm slices, the polypeptides in each slice were eluted, and the eluates containing the peaks of 3H were concentrated and fractionated by reversed-phase HPLC.
Reversed-phase HPLC—Samples were fractionated by reversed-phase HPLC on an Agilent 1100 binary HPLC system at 40 °C using a Brownlee C4-Aquapore 7-μm (100 × 2.1 mm) column with an upstream C2 guard column. The elution of peptides was monitored by absorbance at 214 nm. Solvent A was 0.08% trifluoroacetic acid; solvent B consisted of 60% acetonitrile, 40% 2-propanol, and 0.05% trifluoroacetic acid. A stepwise linear gradient was used: 0 min, 25% solvent B; 15 min, 28% solvent B; 30 min, 37% solvent B; 45 min, 52% solvent B; 60 min, 73% solvent B; 75 min, 100% solvent B; 80 min, 100% solvent B; 85 min 25% solvent B; and 90 min, 25% solvent B. The flow rate was 200 μl/min with 0.5-ml fractions collected.
Sequence Analysis—Material for sequence analyses was isolated from four independent preparative photolabelings: Ia/b and IIa/b (control versus tetracaine), IIIa/b/c (control, +Carb, +proadifen), and IVa/b/c (+Carb, +PCP, +Carb+PCP). Amino-terminal sequencing was performed on an Applied Biosystems Procise 492 protein sequencer that was modified so that ⅙ of each cycle was analyzed for PTH derivative quantitation and ⅚ was collected for scintillation counting. HPLC fractions were usually drop-loaded at 45 °C onto BiobrenePlus®-treated glass fiber filters (Applied Biosystems catalog number 601111). Fractions containing αM4 or δM1, which are not sequenced efficiently on glass fiber supports, were immobilized on polyvinylidene filters using Prosorb® sample preparation cartridges (Applied Biosystems catalog number 401959). In some instances, the sequencing run was interrupted, and the sample on the filter was treated with o-phthalaldehyde (OPA), which reacts preferentially with primary amines over secondary amines (i.e. proline) and hence can be used to block Edman degradation of any peptide not containing an amino-terminal proline at that cycle (28, 34). The quantity of amino acids released was determined by peak heights, and the amount of each peptide was obtained from a nonlinear least squares fit (Sigma Plot, SPSS) of the equation f(x) = I0 × Rx where f(x) is pmol of the peptide residue in cycle x, I0 is the initial amount of peptide (in pmol), and R is the average repetitive yield. PTH derivatives known to have poor recovery (Ser, Cys, His, and Trp) were omitted from the fit because of known problems with their quantitations. The efficiency of photolabeling of amino acid residues (cpm/pmol) was calculated as (cpmx - cpm(x - 1))/(5 × I0 × Rx).
Molecular Modeling—A homology model of the T. californica nAChR, constructed using the Discovery Studio (Accelerys, Inc.) software package from the structural model of the Torpedo marmorata nAChR (Protein Data Bank code 2BG9), is essentially that of T. marmorata nAChR because the sequences of the four subunits differ between species only by 38 amino acid substitutions, including 16 in the transmembrane helices, without any insertions or deletions. CDOCKER was used to dock 50 energy-minimized structures of TDBzl-etomidate (volume, 284 Å3) into potential binding sites in the nAChR TMD defined by a sphere of 15-Å radius centered within the ion channel or of 12-Å radius centered at each of the five subunit interfaces or within the helical bundle of the δ subunit. The spheres within the ion channel and at the subunit interfaces extended from the level of M2-6 to M2-23, whereas for the δ subunit intrahelical bundle the sphere extended approximately from slightly below M2-13 and above M2-23. Because of the atom limitations of CDOCKER, the nAChR model was “trimmed” by removal of the M4 helices, the cytoplasmic extensions, and extracellular regions >14 Å above the membrane. Each set of 50 solutions was evaluated using CalculateBindingEnergies, and for visualization, each TDBzl-etomidate binding pocket was represented by the Connolly surface of the ensemble of the 10 docking solutions with the most favorable binding energies (Fig. 8 and supplemental Figs. S5 and S6). For the binding site in the channel, the 10 orientations were contained within a pocket of ∼900 Å3, extending from M2-9 to M2-23. For the binding site at the interface between γ and α subunits, the pocket had a volume of ∼580 Å3 and did not extend below M2-9. TDBzl-etomidate binding was also predicted within less constrained pockets at the β-α (∼1380 Å3), α-γ (∼890 Å3), and α-δ (1120 Å3) interfaces that were less well defined and extended into the ion channel and the lipid interface. No stable binding was predicted at the δ-β interface or within the δ subunit helix bundle.
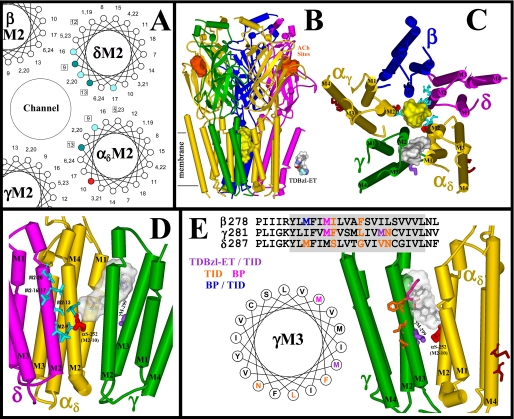
FIGURE 8.
The binding sites for TDBzl-etomidate in the nAChR transmembrane domain. A, an α-helical wheel diagram (100°/residue) of the nAChR M2 channel-lining helices (specifically δM2 and αM2) illustrating the positions in the nAChR structure (Protein Data Bank code 2BG9) of the M2 α-carbons relative to the channel lumen. Residues labeled by [3H]TDBzl-etomidate in the resting state and inhibited by tetracaine are dark cyan, whereas all cyan residues were labeled in the desensitized state (+Carb). Residue αM2-10, labeling of which was not inhibited by tetracaine but was potentiated by PCP or proadifen, is colored red. Residues photolabeled by [3H]tetracaine (43) are boxed. B and C, views of the T. californica nAChR homology model (α, gold; β, blue; γ, green; δ, magenta) from perspectives parallel to the membrane surface (B) or looking down the channel from the extracellular side (C). In C, residues labeled by [3H]TDBzl-etomidate are shown in stick format color-coded forαM2-10 (red), the M2 channel lumen (cyan), γM3 (purple), andαM4 (brown). The volumes defined by the ensemble of the 10 lowest energy orientations of TDBzl-etomidate docked within the ion channel (yellow; 900 Å3) or in the pocket between the γ and α (gray; 580 Å3) subunits are shown in Connolly surface representations. In B, the locations of the agonist sites are denoted in orange, the approximate limits of the membrane are noted, and a Connolly surface representation of TDBzl-etomidate (284 Å3) is included for scale. D and E, views of the transmembrane domains of the δ, α, and γ subunits from the channel lumen (D) or from the lipid interface (E) with the amino acids identified that are labeled by TDBzl-etomidate in the ion channel (cyan), in the pocket at the γ-α interface (αSer-252 (αM2-10, red) and γMet-299 (purple)), and in αM4 (brown). D, the TDBzl-etomidate binding pocket between γ and α is shown in gray with a TDBzl-etomidate (stick representation, carbon, black; oxygen, red; nitrogen, blue; fluorine, light blue; hydrogen, white) docked in its lowest energy orientation. Also included in E are the γM3 amino acids photolabeled by [125I]TID (orange; γPhe-292, γLeu-296, and γAsn-300 (33)) or by [3H]benzophenone (magenta; γMet-291 (39)). A helical wheel diagram of γM3 and an alignment of the βM3, γM3, and δM3 sequences, with the same color coding of the photolabeled amino acids, are included to illustrate the difference in location between the amino acids labeled by TID at the lipid interface and those labeled by TDBzl-etomidate (ET) or benzophenone (BP) at the γ-α subunit interface.
RESULTS
TDBzl-etomidate, a Positive Modulator of ACh Responses—We used two-electrode voltage clamp to compare the effects of TDBzl-etomidate and etomidate on the ACh current responses for Torpedo nAChRs expressed in Xenopus oocytes (Fig. 2, A and B). Neither etomidate nor TDBzl-etomidate, when applied in the absence of agonist, produced any detectable current response (i.e. <0.1% of the maximal response for ACh). As reported previously (26), when coapplied in the presence of 10 μm ACh, a concentration producing ∼20% of the maximal response, etomidate produced a dose-dependent inhibition characterized by an IC50 of 20 μm. In contrast, TDBzl-etomidate at concentrations above 10 μm increased the ACh response with a 2.5-fold increase seen at the highest concentration studied (60 μm). At that concentration the methanol concentration was 0.05%, but in control experiments we established that methanol at 0.5% altered the ACh response by less than 5%. Consistent enhancement of the ACh response was seen when TDBzl-etomidate was applied in either increasing or decreasing concentrations, and the enhancement was fully reversible when the ACh response was measured minutes after removal of TDBzl-etomidate.
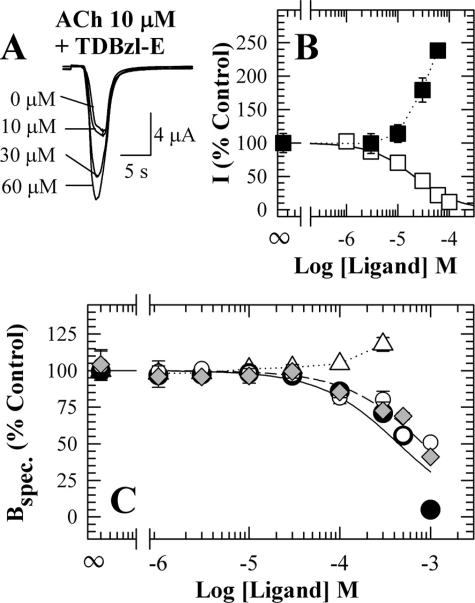
FIGURE 2.
TDBzl-etomidate potentiation of ACh responses of the Torpedo nAChR. A, representative current responses measured by two-electrode voltage clamp from a single oocyte superfused with 10 μm ACh alone, which produces ∼20% of the maximal response, or coapplied with TDBzl-etomidate. B, the currents (I) elicited by 10 μm ACh in the presence of varying concentrations of TDBzl-etomidate (▪) or etomidate (□) normalized to the current in the absence of either ligand. For TDBzl-etomidate the responses are the means ± S.E. (vertical bars) for data from three oocytes. For etomidate, the concentration dependence of the inhibition of ACh responses was fit to a single site model with IC50 = 20 ± 1 μm. C, the effects of TDBzl-etomidate on the specific binding to Torpedo nAChR-rich membranes of [3H]tetracaine (⋄; 19 nm), [3H]PCP (6 nm) in the absence (○) or presence of 1 mm Carb (•), or [3H]ACh (▵; 11 nm). In the absence of Carb, TDBzl-etomidate inhibition of either [3H]tetracaine or [3H]PCP binding was characterized by an IC50 of 0.9 ± 0.1 mm. The inhibition of [3H]PCP binding (+Carb) was characterized by an IC50 of 0.5 ± 0.2 mm. For each radioligand, the data were normalized to the specific binding in the absence of TDBzl-etomidate, which for [3H]tetracaine was 34,600 ± 700 cpm (specific) and 4250 ± 40 cpm (nonspecific). For [3H]PCP, the specific binding was 5100 ± 240 cpm (+Carb) and 840 ± 50 cpm (-Carb) with nonspecific binding of 300 ± 20 cpm (+Carb) and 500 ± 30 cpm (-Carb). For [3H]ACh, the total and nonspecific binding were 8500 ± 250 and 80 ± 15 cpm. In a parallel experiment, 100 μm proadifen potentiated [3H]ACh binding by 180% of control. spec., specific.
We also measured the effects of TDBzl-etomidate on the equilibrium binding of [3H]ACh and drugs that bind in the ion channel preferentially in the resting state ([3H]tetracaine (35)) or desensitized state ([3H]PCP (36)) (Fig. 2C). When [3H]ACh equilibrium binding was measured at a concentration sufficient to occupy ∼20% of sites, TDBzl-etomidate increased binding by ∼20% at the highest concentration studied (300 μm). In the absence of agonist it inhibited [3H]tetracaine or [3H]PCP binding with an IC50 of 0.9 mm, and in the presence of agonist it inhibited [3H]PCP binding with an IC50 of 0.5 mm.
Photoincorporation of [3H]TDBzl-etomidate into nAChR-rich Membranes—We used SDS-PAGE followed by fluorography (Fig. 3A) or liquid scintillation counting of excised gel bands (Fig. 3, B and C) to provide an initial characterization of the pattern and pharmacological specificity of nAChR subunit photolabeling. Membranes were photolabeled after equilibration with [3H]TDBzl-etomidate (0.8 μm) in the absence of other drugs or in the presence of the agonist Carb and/or drugs that bind in the ion channel preferentially in the desensitized state (proadifen (37) or PCP) or in the resting state (tetracaine).
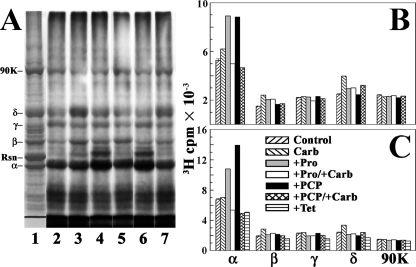
FIGURE 3.
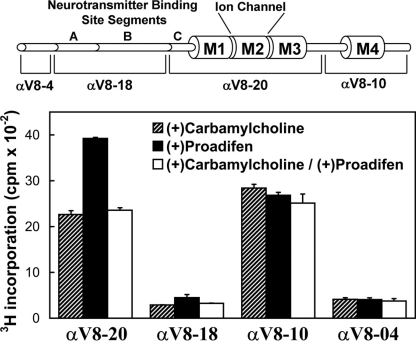
[3H]TDBzl-etomidate photoincorporation into Torpedo nAChR-rich membranes. A, membranes (260 μg of protein, 400 pmol of ACh binding sites in 120 μl of TPS supplemented with 1 mm oxidized glutathione) were equilibrated with 0.8 μm [3H]TDBzl-etomidate alone (control; lane 2) or with 1 mm Carb (lane 3), 0.1 mm proadifen (lane 4), 1 mm Carb and 0.1 mm proadifen (lane 5), 0.1 mm PCP (lane 6), or 1 mm Carb and 0.1 mm PCP (lane 7). After irradiation at 365 nm for 30 min, the polypeptides were resolved by SDS-PAGE and visualized by Coomassie Blue stain (lane 1), and the gel was processed for fluorography (lanes 2–8; 30-day exposure at -80 °C). The stained polypeptide bands corresponding to the nAChR α, β, γ and δ subunits, rapsyn (Rsn), and the α subunit of the Na+/K+-ATPase (90K) are indicated. B, in a parallel experiment, the polypeptide gel bands were excised from the stained gel, and the 3H incorporation was determined by liquid scintillation counting. C, in a separate experiment, the effect of tetracaine (Tet; 0.1 mm) on photoincorporation of [3H]TDBzl-etomidate was compared with Carb, proadifen (Pro), and PCP.
In the absence of other drugs, [3H]TDBzl-etomidate was photoincorporated into each nAChR subunit. The most prominent effect of the drugs was an ∼80% increase in α subunit labeling in the presence of proadifen or PCP that was not seen for nAChRs stabilized in the desensitized state by Carb or in the presence of tetracaine. Within the β and δ subunits, Carb increased labeling by ∼50% compared with control, and that increased labeling was reduced by proadifen or PCP. The 4000 cpm PCP/proadifen enhanced labeling within the α subunit indicated labeling of ∼0.1% of subunits at 0.8 μm [3H]TDBzl-etomidate.
To provide an initial localization of the proadifen-sensitive labeling within the nAChR α subunit, nAChR-rich membranes were photolabeled in three conditions (+Carb, +proadifen, and +Carb+proadifen). The nAChR α subunits were then isolated by SDS-PAGE and subjected to in-gel proteolytic digestion to generate large, non-overlapping subunit fragments that contain either the extracellular domain or TMD (Fig. 4). Digestion with V8 protease generates fragments of 20 kDa (αV8-20) beginning at αSer-173 and containing ACh binding site segment C and the M1, M2, and M3 membrane-spanning helices, an 18-kDa fragment (αV8-18) beginning at αThr-52 and containing ACh binding site segments A and B, a 10-kDa fragment (αV8-10) beginning at αAsn-338 and containing αM4, and a 4-kDa fragment (αV8-4) beginning at αSer-1 (31). For membranes labeled in the presence of Carb, ∼90% of 3H was recovered equally distributed between αV8-20 and αV8-10, whereas the proadifen enhancement of labeling was restricted to αV8-20. The 3H incorporation within αV8-18, αV8-10, and αV8-4 was the same in the three labeling conditions. The pharmacological specificity of the labeling of these large subunit fragments differed from that seen for [3H]azietomidate for which there was proadifen-inhibitable labeling in αV8-20 in the presence of Carb due to labeling of αM2-20 (αGlu-262) at the extracellular end of the ion channel (27), and in αV8-18 there was Carb-inhibitable labeling resulting from labeling of αTyr-93 in the ACh binding site.
FIGURE 4.
Distribution of [3H]TDBzl-etomidate photoincorporation within large fragments of the nAChR α subunit. nAChR-rich membranes (0.4 mg of protein at 2 mg/ml) were photolabeled with 1.3 μm [3H]TDBzl-etomidate in the presence of 1 mm Carb, 0.1 mm proadifen, or 1 mm Carb and 0.1 mm proadifen. After photolabeling, the samples were subjected to SDS-PAGE, and the polypeptide bands were visualized by Coomassie Blue stain. For each condition, the band corresponding to the nAChR α subunit was excised, transferred to the well of a 15% mapping gel, and digested in gel by S. aureus V8 protease as described under “Experimental Procedures.” Following electrophoresis, the mapping gels were stained with Coomassie Blue to visualize the distinct α subunit polypeptide fragments generated by V8 protease digest (Ref. 31; shown diagrammatically in the upper panel). Each gel was then cut into 5-mm strips, and the 3H incorporation was quantified by scintillation counting. The peaks of 3H were associated with the αV8-20 and αV8-10 fragments, which contain the M1, M2, and M3 segments (αV8-20) or M4 (αV8-10). The data shown are the average and range (vertical bars) of two experiments.
To identify the amino acids photolabeled by [3H]TDBzl-etomidate in the nAChR in the resting state, membranes were photolabeled on a preparative scale on two occasions in the absence and presence of tetracaine, a closed state channel blocker. To compare photolabeling patterns between the resting and desensitized states, in a third photolabeling experiment, membranes were photolabeled in the absence of other drugs, in the presence of Carb, or in the presence of proadifen. To further characterize labeling in the desensitized state, in a fourth experiment, membranes were photolabeled on a preparative scale in the presence of Carb, in the presence of PCP, or in the presence of Carb and PCP. The experimental identification of the amino acids labeled in the nAChR transmembrane domain is presented by representative sequence data in the figures, and the data from multiple labelings and sequencing experiments are combined in Table 1 to compare the efficiencies of labeling of the different positions in the different receptor states.
TABLE 1.
Pharmacological specificity of [3H]TDBzl-etomidate labeling within αM2 and δM2 (cpm/pmol of PTH derivative) The 3H incorporation in each residue was calculated from the observed 3H release as described under “Experimental Procedures,” and the mass was calculated from the initial and repetitive yields. The results are from multiple photolabeling experiments, including data from Figs. 5, 6, 7. When multiple samples were sequenced, the data are presented as the mean and range. ND, not determined; NQ, not quantifiable.
|
Amino acid
|
Resting state
|
Desensitized state
|
+PCP
|
+Proadifen
|
||
|---|---|---|---|---|---|---|
| Control | +Tetracaine | +Carb | +Carb/+PCP | |||
| αM2-9 Leu-251 | 1 | <0.4 | 11 ± 3 | 1 | <2 | <2 |
| αM2-10 Ser-252 | 16 ± 3 | 14 ± 2 | 11 ± 3 | 19 | 255 | 225 |
| αM2-13 Val-255 | 3 ± 2 | 0.6 ± 0.3 | 7 ± 1 | 5 | NQ | NQ |
| δM2-9 Leu-265 | 0.5 ± 0.1 | 0.2 ± 0.1 | 5 ± 1 | 0.8 | 0.6 | 10 |
| δM2-13 Val-269 | 7 ± 2 | 0.4 ± 2 | 3 ± 1 | 0.9 | 1.2 | 0.5 |
| δM2-16 Leu-272 | <0.2 | <0.2 | 2 | 1.5 | <0.2 | <0.3 |
| δM2-17 Leu-273 | <0.2 | <0.2 | 5 ± 1 | 3 | <0.3 | <0.3 |
| δM2-20 Gln-276 | <0.3 | <0.3 | 1.5 ± 0.5 | 1 | <0.1 | <0.3 |
| γM3 Met-299 | 4 ± 1 | ND | 3 ± 1 | 4 | 3 | 4 ± 1 |
| αM4 Cys-412 | NQ | NQ | ND | 7 | ND | ND |
| αM4 Met-415 | NQ | NQ | ND | 5 | ND | ND |
State-dependent Photoincorporation in αM2—To identify the labeled amino acids within the α subunit, the labeled αV8-20 fragments were isolated by in-gel digestion with V8 protease and then digested with EndoLys-C, which generates fragments that can be separated by reversed-phase HPLC that begin at αHis-186 and contain ACh binding site segment C and αM1, and that begin at αMet-242 and contain αM2 and αM3 (27). HPLC fractionation of the digests revealed that ∼60% of 3H was recovered in the hydrophobic fractions expected to contain the αM2 fragment (supplemental Fig. S1), and ∼20% was recovered in the column flow-through fractions.
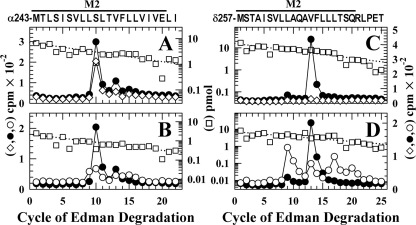
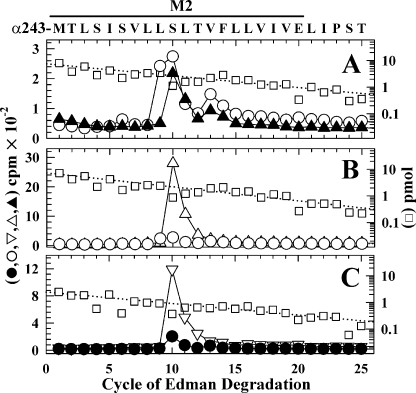
For nAChRs labeled in the absence of other drugs (Fig. 5A), amino-terminal sequencing of the hydrophobic 3H peak from the HPLC established that the primary sequence began at αMet-243 and that there were peaks of 3H release in cycles 10 (260 cpm) and 13 (50 cpm) consistent with labeling of αSer-252 (αM2-10) and αVal-255 (αM2-13). Based upon the results from two separate labeling experiments (Table 1), tetracaine reduced the labeling at αM2-10 by <15%, whereas the labeling at αM2-13 was reduced by ∼80%. For nAChRs labeled in the desensitized state (+Carb) (Fig. 5B), there was labeling of αM2-9 (αLeu-252), which was not labeled in the resting state, as well as labeling of αM2-10 and αM2-13. Based upon sequence analyses of samples from two independent labelings of nAChRs in the resting and desensitized states (Table 1), αM2-9 and αM2-10 were labeled at similar efficiencies (11 cpm/pmol)4 in the desensitized state, and the labeling of αM2-10 was reduced by ∼30% in the desensitized state compared with the resting state.
FIGURE 5.
State-dependent [3H]TDBzl-etomidate photoincorporation within αM2 and δM2. 3H (•, ○, and ⋄) and PTH-amino acids (□) released during sequence analysis of α and δ subunit fragments beginning at the amino terminus of αM2 (A and B) or δM2 (C and D) are shown. The primary amino acid sequences are shown above the top panels. A and C, to identify amino acids labeled in the resting state, nAChR-rich membranes were photolabeled with 1 μm [3H]TDBzl-etomidate (75 μCi) on a preparative scale (10 mg of protein/condition) in the absence (•) or presence of 0.1 mm tetracaine (⋄). B and D, to compare labeling in the resting and desensitized states, membranes were photolabeled in the absence (•) or presence of 1 mm Carb (○) (or with 0.1 mm proadifen (see Fig. 6B)). As described under “Experimental Procedures,” the fragments containing αM2 were isolated by reversed-phase HPLC from EndoLys-C digests of αV8-20 (supplemental Fig. S1A). To isolate the fragments beginning at the amino terminus of δM2, EndoLys-C subunit digests were fractionated by Tricine SDS-PAGE, and the major peak of 3Hat 12–14 kDa was further purified by reversed-phase HPLC (supplemental Fig. S2). A, the primary sequence began at αMet-243 (I0 = 7 pmol, - and +tetracaine). The peaks of 3H release in cycles 10 and 13 indicated labeling of αSer-252 and αVal-255 at 19 and 5 cpm/pmol, respectively, with the labeling of those amino acids reduced by 30 and 80% in the presence of tetracaine. B, the primary sequence began at αMet-243 (□; -Carb, I0 = 3.7 pmol; +Carb, I0 = 2.2 pmol). For nAChRs labeled in the absence of agonist, the 3H releases in cycles 10 and 13 indicated labeling of αSer-252 and αVal-255 at 25 and 6 cpm/pmol, whereas in +Carb the 3H releases in cycles 9, 10, and 13 indicated labeling of αLeu-251, αSer-252, and αVal-255 at 8, 8, and 6 cpm/pmol. C, the primary sequence began at δMet-257 (control, I0 = 23 pmol; □, +tetracaine, I0 = 16 pmol). The peak of 3H release in cycle 13 indicated labeling of δVal-269 at 9 cpm/pmol that was reduced by 95% in the presence of tetracaine. The small peak of 3H release in cycle 9 indicated tetracaine-inhibitable labeling of δLeu-265 at ∼0.6 cpm/pmol. D, the primary sequence began at δMet-257 (□; -Carb, I0 = 6.7 pmol; +Carb, I0 = 7.1 pmol). For nAChRs labeled in the absence of agonist, the 3H releases in cycles 9 and 13 indicated labeling of δLeu-265 and δVal-269 at 1 and 13 cpm/pmol. For nAChRs labeled in +Carb, the 3H releases in cycles 9 and 13 indicated labeling of δLeu-265 and δVal-269 at 5 and 2.5 cpm/pmol, and the 3H releases in cycles 16, 17, and 20 indicated labeling in the desensitized state of δLeu-272, δLeu-273, and δGln-276 at 2, 4, and 1 cpm/pmol.
State-dependent Labeling within δM2—To identify the labeled amino acids within the δ subunit, the photolabeled subunits were digested with EndoLys-C, which produces a fragment of ∼21 kDa (δEKC-21) beginning at δHis-20/δGlu-48 and containing ACh binding site segments D, E, and F, and fragments of 10–14 kDa (δEKC-10/14), one beginning at δMet-257 at the amino terminus of δM2 and extending through δM3 and another beginning at δPhe-206 and containing δM1 (38). When the digests were fractionated by Tricine SDS-PAGE (supplemental Fig. S2A), there was a major peak of 3Hat ∼10–14 kDa that was reduced for nAChRs labeled in the presence of tetracaine, and less than 5% of the 3H was recovered in the gel band containing δEKC-21. When the material eluted from the 10–14-kDa gel bands was fractionated by reversed-phase HPLC (supplemental Fig. S2B), ∼60% of 3H was recovered in the hydrophobic fractions expected to contain the fragment beginning at δMet-257, and ∼10% of 3H eluted in the fractions containing the δM1 fragment.
For nAChRs labeled in the resting state, sequence analysis of the HPLC fractions containing the fragment beginning at δMet-257 (Fig. 5C) revealed a major peak of 3H release in cycle 13 (400 cpm) with lower level release in cycle 9 (30 cpm) consistent with labeling of δVal-269 (δM2-13) and δLeu-265 (δM2-9), respectively. For nAChRs labeled in the presence of tetracaine, the labeling of δM2-13 was reduced by 95%. For nAChRs labeled in the desensitized state (+Carb), the labeling of δM2-9 was increased by 400% compared with the resting state (Fig. 5D), whereas the labeling of δM2-13 was reduced by 80%. In addition, the increases of 3H release in cycles 16, 17, and 20 indicated that δLeu-272 (δM2-16), δLeu-273 (δM2-17), and δGln-276 (δM2-20) are also labeled in the desensitized state at efficiencies similar to those of δM2-9 and δM2-13 (2–5 cpm/pmol; Table 1).
Photolabeling in αM2 in the Presence of PCP or Proadifen—For nAChRs labeled in the desensitized state (+Carb) (Fig. 6A), PCP inhibited the labeling of αLeu-251 (αM2-9) and αVal-255 (αM2-13) by >90% and ∼50%, respectively. Surprisingly the labeling of αSer-252 (αM2-10) was essentially the same in the absence and presence of PCP.4 Additional evidence that the pharmacological specificity of labeling of αM2-10 was quite distinct from that of αM2-9 and αM2-13 was seen in nAChRs photolabeled in the presence of PCP or proadifen but in the absence of Carb. PCP selectively increased the labeling of αM2-10 by 10-fold compared with that seen in +Carb (Fig. 6B), and proadifen also increased the labeling of αM2-10 by 10-fold compared with the labeling in the resting state with no detectable increase in the labeling of αM2-13 (Fig. 6C).5
FIGURE 6.
[3H]TDBzl-etomidate labeling within αM2 in the presence of PCP or proadifen. 3H (○, ▴, ▵, ▿, and •) and PTH-amino acids (□) released during sequencing of α subunit fragments beginning at the amino terminus of αM2 are shown. The primary amino acid sequence is shown above the top panel. A and B, nAChR-rich membranes were photolabeled with 1 μm [3H]TDBzl-etomidate (75 μCi) on a preparative scale (10 mg of protein/condition) in +Carb (○), +Carb+PCP (▴), or PCP alone (▵). The fragments beginning at αMet-243 were purified by reversed-phase HPLC from EndoLys-C digests of αV8-20 (+Carb, I0 = 7.5 pmol (A, □); +Carb+PCP, I0 = 5.8 pmol; +PCP, I0 = 7.3 pmol (B, □)). A, for nAChRs labeled in +Carb (○), the 3H releases in cycles 9, 10, and 13 indicated labeling of αLeu-251, αSer-252, and αVal-255 at 14, 13, and 8 cpm/pmol. Addition of PCP (▴) reduced the labeling of αLeu-251 to 1 cpm/pmol, whereas labeling of αSer-252 (18 cpm/pmol) was not inhibited. B, for nAChRs labeled in +PCP (▵), the 3H release in cycle 10 indicated labeling of αSer-252 at 250 cpm/pmol, i.e. at 20-fold higher efficiency than the labeling in +Carb (○). C, for nAChRs labeled in +proadifen (▿), the fragment beginning at αMet-243 was present at 2.3 pmol (□), and the 3H release in cycle 10 indicated labeling of αSer-252 at 225 cpm/pmol, which was 10-fold higher than the labeling in the resting state (•; from Fig. 5B).
Photolabeling in δM2 in the Presence of PCP or Proadifen—Photolabeling was quantified for nAChRs equilibrated with PCP in the presence of Carb and in its absence as well as for proadifen in the absence of Carb (Table 1 and supplemental Fig. S3). For nAChRs labeled in the desensitized state (+Carb), PCP reduced the labeling efficiency of δLeu-265 (δM2-9) and δVal-269 (δM2-13) by 85% and labeling of δLeu-273 (δM2-17) by ∼40%. For nAChRs labeled in the absence of Carb, the presence of PCP resulted in a labeling pattern quantitatively different from that seen in the presence of proadifen. In the presence of PCP, the labeling at δM2-9 was only 10% of the efficiency seen in the +Carb labeling, whereas in the presence of proadifen it was 200%. In the presence of PCP or proadifen, the labeling at δM2-13 was 50 or 20% of that seen in +Carb. In contrast to the photolabeling pattern seen in +Carb, in the presence of PCP or proadifen alone there was little if any [3H]TDBzl-etomidate photoincorporation at δM2-17 or δM2-20 at the extracellular end of the ion channel.
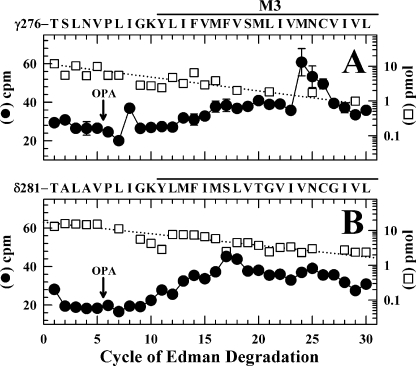
Labeling in γM3 but Not δM3—Potential 3H incorporation within the M2-M3 loops or M3 helices of the γ and δ subunits was determined as described previously (39) by sequence analysis of fractions from HPLC purifications of V8 protease digests of the labeled subunits. V8 protease cleaves the subunits at γGlu-275/δGlu-280 near the carboxyl termini of the M2 helices, generating fragments beginning at γThr-276 and δThr-281 that extend through M3. Although HPLC fractionation does not separate those fragments from other hydrophobic fragments, the 3H incorporation within those fragments can be determined by Edman degradation by taking advantage of the presence of a proline (γPro-281/δPro-286) in the sixth cycle of Edman degradation and of a sequencing protocol in which o-phthalaldehyde, which reacts with primary amines but not proline, is applied prior to a sequencing cycle containing a proline to prevent Edman degradation of all peptides not containing a proline (28, 34). For nAChRs labeled in the absence of agonist, sequence analysis established that the fragment beginning at γThr-276 was present at 13 pmol, and the peak of 3H release in cycle 24 (25 cpm) indicated labeling of γMet-299 at 4 cpm/pmol (Fig. 7A) with labeling at an efficiency similar to that seen in +Carb (data not shown). In contrast, there was no evidence of labeling in the corresponding region of the δ subunit for nAChRs labeled in the absence (Fig. 7B) or presence of Carb (not shown). Based upon the mass levels of the sequenced fragments and the observed 3H release, labeling of any amino acid in δM3, if it occurred, would be at less than 0.3 cpm/pmol.
FIGURE 7.
[3H]TDBzl-etomidate photolabels γM3 but not δM3. 3H(•) and PTH-amino acids (□) released during sequencing of fragments from V8 protease digests of γ (A) and δ (B) subunits isolated from nAChRs from the photolabeling experiment used in Fig. 5, B and D (labeling in the absence of Carb), are shown. The V8 protease digests were fractionated by reversed-phase HPLC, fractions containing the peak of 3H(∼70% solvent B) were pooled and separated into two aliquots that were sequenced individually, and the samples were treated with OPA at cycle 6. The primary amino acid sequences are shown above each panel. A, after OPA, sequencing continued for the fragment beginning at γThr-276 (I0 = 12 pmol) and for the equivalent, contaminating fragment from the δ subunit (δThr-281; I0 = 4 pmol). In the cycles before OPA, sequences were also present beginning at γIle-209 and γAsn-439 (I0 values of ∼5 pmol), but treatment with OPA reduced those levels by >90%. The plotted 3H release is the average from two sequencing experiments with vertical lines indicating the ranges when greater than the symbol sizes. The 3H release in cycle 24 corresponds to labeling of γMet-297 at 4 cpm/pmol because for the δ subunit sample (B) there was no release above background in cycle 24. B, after treatment with OPA in cycle 6, sequencing continued for the fragment beginning atδThr-281 (I0 = 20 pmol). In the cycles before OPA, sequences were also present beginning at δIle-192 and δVal-443 (I0 values of ∼15 pmol), but treatment with OPA reduced those levels by >90%. Although beginning in cycle 10 there was a general increase in background 3H release from 20 to 40 cpm, there was no release above background in cycle 24, and any labeling within δM3 would be at <0.3 cpm/pmol.
No Labeling within δM1 or the δM2-M3 Loop—Two small hydrophobic photoreactive nAChR noncompetitive antagonists, [125I]3-(trifluoromethyl)-3-(m-iodophenyl)diazirine ([125I]TID) and [3H]benzophenone, photolabel in the presence of agonist, but not in the resting state, amino acids in δM1 (δPhe-232/δCys-236) and in the δM2-δM3 loop (δPro-286 and/or δIle-288) (38–40) that contribute to a state-dependent binding pocket at the extracellular end of the δ subunit transmembrane domain four-helix bundle. We found no evidence of [3H]TDBzl-etomidate photoincorporation in these regions in either the absence or presence of Carb. We looked for 3H incorporation within δM1 by sequencing for 35 cycles the fragment beginning at δPhe-206 (7 pmol) that elutes with a minor peak of 3H in the HPLC fractionation of the EndoLys-C digest of labeled δ subunits (Ref. 38 and supplemental Fig. S2B). No 3H release exceeded by more than 10 cpm the background release (7–10 cpm) in each cycle of Edman degradation (data not shown), indicating that labeling of any amino acid, if it occurred, would be at less than 0.5 cpm/pmol. Sequence analysis of the fragment beginning at δThr-281 revealed no detectable labeling of δPro-286 or other amino acids in the δM2-M3 loop in the absence of Carb (Fig. 7B) or in +Carb (not shown).
Photolabeling in αM4—Amino acids in the M4 α-helices in contact with lipid have been identified by photolabeling with the hydrophobic probes [125I]TID, [3H]diazofluorene, and [3H]benzophenone (33, 39, 41). We characterized [3H]TDBzl-etomidate labeling in αM4 by sequencing the α subunit fragment beginning at αTyr-401 that can be isolated from trypsin digests of αV8-10 by reversed-phase HPLC (33) (supplemental Figs. S1B and S4). For nAChRs labeled in the presence of Carb and PCP, the peaks of 3H release in cycles 12 (160 cpm) and 15 (50 cpm) indicated labeling of αCys-412 and αMet-415, amino acids also labeled by [125I]TID and [3H]diazofluorene as well as [3H]benzophenone (αMet-415). In a further experiment, we found that the presence of tetracaine did not alter the efficiency of labeling of αCys-412 or αMet-415 (not shown).
DISCUSSION
In this work, we demonstrate that [3H]TDBzl-etomidate acts as a positive allosteric nAChR modulator, and we identify its binding sites in the nAChR. At concentrations from 10 to 60 μm, TDBzl-etomidate potentiated the response to 10 μm ACh, a concentration producing 20% of the maximal response. TDBzl-etomidate at higher concentrations should inhibit ACh responses because it inhibits the binding of the channel blockers [3H]tetracaine (resting state) and [3H]PCP (desensitized state) with IC50 values of 0.9 and 0.4 mm, and it photolabels amino acids in the channel lumen. Etomidate and azietomidate, which only inhibit ACh responses, are both more potent as inhibitors of [3H]PCP binding (IC50 values of 30 and 70 μm) (27). Because neither etomidate nor azietomidate potentiates responses of the Torpedo nAChR, we did not begin this work expecting to find that TDBzl-etomidate acts as a potentiator. Interestingly the structure of TDBzl-etomidate, a substituted (1-phenylethyl-5-carboxylate)imidazole, has similarities to the substituted (2-amino-5-keto)thiazoles recently introduced as potentiators of several neuronal, but not muscle, nAChRs (42), but it is quite distinct from previously described allosteric modulators of muscle-type nAChRs such as physostigmine or galantamine, which activate via a site distinct from the agonist site (17, 19, 22). Although studies using higher resolution electrophysiological techniques will be necessary to determine the gating parameters altered by TDBzl-etomidate, the observed potentiation could result from a decrease of ACh Kapp from 25 to 10 μm because the ACh response was characterized by a Kapp of 25 μm and a Hill coefficient of ∼1.6.
Photolabeling in αM2 and δM2—Comparison of [3H]TDBzl-etomidate photoincorporation within the M2 helices in the absence and presence of positively charged channel blockers revealed that the pharmacological specificity of αM2-10 photolabeling differed from that of M2-9 or -13 in the α or δ subunits (summarized in Table 1). Photolabeling of αM2-10 in the resting state was not inhibited by tetracaine, a resting state selective channel blocker that itself photolabels M2-9 and/or -13 in each subunit (43); and in the desensitized state, photolabeling of αM2-10 was not inhibited by PCP, which also binds in the ion channel although at an undetermined site. In contrast, tetracaine reduced photolabeling of αM2-13 and δM2-13 by >80%, and in the desensitized state PCP reduced labeling of αM2-9 and δM2-9 by >80%. Photolabeling of αM2-10 was, in fact, increased 10-fold in the presence of proadifen, which also binds in the ion channel preferentially in the desensitized state.
[3H]TDBzl-etomidate photoincorporation within αM2 and δM2 was also state-dependent. Within δM2, labeling of δM2-13 (δVal-269) was reduced by 50%, and labeling of δM2-9 was increased by 5-fold in the desensitized compared with the resting state. In addition, δLeu-272, δLeu-273, and δGln-276 (δM2-16, -17, and -20) were labeled in the desensitized state but not in the resting state. Within αM2, the labeling of αM2-10 was reduced by 50%, and αM2-9 (αLeu-251) labeling was increased by ∼5-fold in the desensitized state compared with the resting state.
PCP and proadifen each bind with ∼10-fold higher affinity in the nAChR ion channel in the presence of agonist than in the absence, and in the absence of agonist they stabilize the nAChR in a desensitized state with high affinity for agonist although structurally distinct from that stabilized by agonist (36, 37, 44). The differential TDBzl-etomidate labeling patterns seen in the presence of Carb compared with PCP or proadifen may be evidence that those drugs stabilize a desensitized state with a channel structure distinct from that in the presence of Carb. However, most of the differences in labeling are for residues that contribute to the lumen of the ion channel and are expected to be in close proximity to bound PCP or proadifen. The 10-fold increase in labeling of αM2-10 seen in the presence of PCP or proadifen, but not in the presence of Carb, could be accounted for by a subtle reorientation of αM2-10 with those drugs bound in the channel compared with the unoccupied channel in the desensitized state.
Two Binding Sites for TDBzl-etomidate in the nAChR Transmembrane Domain—The pharmacological specificity of labeling of αM2-10 requires that TDBzl-etomidate binds in the nAChR TMD in proximity to αM2-10 even when tetracaine or PCP is bound within the ion channel. With the amino acid positions for the M2 helices displayed as a helical wheel (Fig. 8A), position M2-10 is oriented ∼90° from positions 5, 9, and 13 that are labeled by [3H]tetracaine in αM2. Within δM2, the amino acids photolabeled by [3H]TDBzl-etomidate in the desensitized state (δM2-9, -13, -16, -17, and -20) lie on a common, broad surface (δ arc 120°) with PCP inhibiting labeling of δM2-9 by the greatest extent (∼85%). Early mutational analyses had predicted that position M2-10 contributed to the lumen of the open ion channel because substitutions at M2-6 and -10 affected the potency of QX-222, a quaternary ammonium channel blocker (45). However, recent results based upon lysine- and arginine-scanning substitutions in M2 provide strong evidence that the positions 2, 6, 9, and 13 are oriented toward the lumen of the open channel (46).
The amino acids photolabeled by [3H]TDBzl-etomidate are identified in Fig. 8, B–E, in the model of the Torpedo nAChR structure in the absence of agonist (Protein Data Bank code 2BG9). A side view of the nAChR transmembrane and extracellular domains is shown in Fig. 8B with the ACh and ion channel binding sites indicated as Connolly surfaces. A space-filling model of TDBzl-etomidate (volume, 284 Å3) is also included for comparison. The labeled amino acids in αM2, δM2, γM3, and αM4 are included in a view of the nAChR transmembrane domain from the base of the extracellular domain (Fig. 8C), in a view from the lumen of ion channel toward δ-α-γ (Fig. 8D), and in a view from the lipid interface toward γ-α (Fig. 8E). Also included in the images are Connolly surface representations of two of the predicted drug binding pockets for TDBzl-etomidate (see “Experimental Procedures”), one (yellow; 900 Å3) within the lumen of the ion channel in proximity to positions M2-9 and -13 and the second (gray; 580 Å3) at the interface of the γ and α subunits (in proximity to αM2-10). Expanded views are presented in stereo of TDBzl-etomidate docked in the pocket at the interface between the γ and α subunits (supplemental Fig. S5) and with the amino acids that project into the pocket from αM1, αM2, γM2, and γM3 highlighted (supplemental Fig. S6).
A TDBzl-etomidate Binding Site at the γ-α Interface—M2-10 is potentially in proximity to the lumen of the ion channel and to the interface between the γ-α (or β-α) subunits. However, the fact that neither tetracaine nor PCP inhibits photolabeling of αM2-10 when they inhibit labeling of M2-9 and M2-13 establishes that TDBzl-etomidate must be binding to a site outside the channel when it photolabels αM2-10. Thus, TDBzl-etomidate must be binding at the γ-α and/or β-α subunit interface where within the model of nAChR structure there are pockets that can readily accommodate a molecule the size of TDBzl-etomidate. The predicted binding site at the γ-α interface is also in proximity to γMet-299, the residue in γM3 photolabeled by [3H]TDBzl-etomidate at an efficiency similar to that in the absence or presence of Carb or proadifen.
γMet-299 is one of the four amino acids in γM3 labeled by [125I]TID although at only 30% of the efficiency of γAsn-300 (33). Comparison of the location of γMet-299 in the nAChR structure with the other TID-labeled amino acids (Fig. 8E) reveals that γMet-299 is partially accessible from the lipid interface, but more importantly, it is the only one of the TID-labeled amino acids that is also accessible from the γ-α interface. The other TID-labeled amino acids define a surface more fully exposed to the lipid. Interestingly γMet-291, which is labeled by [3H]benzophenone in an agonist-sensitive manner (39), is also oriented toward the γ-α interface, two helical turns above γMet-299.
Because of its high hydrophobicity, it is not possible to quantify directly the reversible, specific binding of [3H]TDBzl-etomidate to the nAChR. However, because proadifen or PCP increased photolabeling of αM2-10 by 1 μm [3H]TDBzl-etomidate by a factor of 10, its interactions with the novel binding site at the subunit interface are likely to be of low affinity. TDBzl-etomidate binding within the ion channel (M2-9 and M2-13) is also of low affinity based upon the millimolar concentrations required to inhibit [3H]tetracaine or [3H]PCP binding. Therefore, at the micromolar concentrations of [3H]TDBzl-etomidate used for photolabeling, the two sites are unlikely to be occupied simultaneously within a single nAChR. Further photolabeling studies at higher [3H]TDBzl-etomidate concentrations will be required to determine the relative affinities for the two sites.
Site of Potentiation by TDBzl-etomidate—We predict that the site at the subunit interface is the site that is occupied when TDBzl-etomidate potentiates the response to ACh and that binding to the site in the lumen of the ion channel results in inhibition. For the GABAAR, recent photolabeling studies (16) have demonstrated that [3H]azietomidate and etomidate, which potentiate GABAAR responses, bind in the TMD at the interface of subunits that contains the transmitter binding sites in the extracellular domain (equivalent to the α-γ and α-δ interfaces in the nAChR).
In addition to the proposed binding site for TDBzl-etomidate at the γ-α subunit interface, the nAChR structure also predicts the existence of potential binding sites at three of the four other subunit interfaces (but not the δ-β interface (“Experimental Procedures”)). As discussed above, photolabeling of γMet-299 provides evidence that the labeling of αM2-10 results from TDBzl-etomidate binding at the γ-α interface, but αM2-10 labeling may also result from TDBzl-etomidate binding at the β-α interface. Because the enhanced labeling of the α subunit in the presence of proadifen or PCP was accounted for by the 10-fold increase of αM2-10 labeling and neither proadifen nor PCP enhanced labeling of the other subunits, there is no evidence for an equivalent TDBzl-etomidate binding at other subunit interfaces. However, because M2-10 is a serine in the α subunit and an alanine in other subunits, the lack of M2-10 labeling in the other subunits may be a consequence of the reduced reactivity of alanine compared with serine (47). Our results provide strong evidence that within the nAChR in the resting and desensitized states αM2-10 is in proximity to TDBzl-etomidate when it binds at a site at the interface between subunits and that this site is distinct from its binding site within the ion channel lumen.
TDBzl-etomidate Binding in the Ion Channel—[3H]TDBzl-etomidate photolabeling of α and δ M2-9 and/or -13 for nAChRs in the resting state was fully inhibited by tetracaine, consistent with the location of the binding site of tetracaine in the lumen of the ion channel as defined by photolabeling (43). The PCP binding site within the ion channel in the desensitized state has not been determined, but based upon photolabeling studies with [3H]chlorpromazine (48), it is likely to bind at the level of M2-2, -6, and -9. This would explain why PCP fully inhibited [3H]TDBzl-etomidate labeling at M2-9 but only partially inhibited labeling at M2-13, -16, -17, or -20. Presumably for nAChRs in the desensitized state, TDBzl-etomidate can bind either near the level of M2-9 and -13 or further toward the extracellular end of the channel, and hence, in the presence of PCP it can still bind near the extracellular end of the ion channel.
TDBzl-etomidate Does Not Bind in the δ Subunit Helix Bundle—The absence of labeling of δM2-18/22 or of amino acids in δM1 or in the δM2-M3 loop indicates that a molecule the size of [3H]TDBzl-etomidate (volume, 284 Å3) does not bind within the δ subunit four-helix bundle in contrast to the agonist-dependent binding there for two smaller hydrophobic drugs, [125I]TID (volume, 150 Å3) (38, 40) or [3H]benzophenone (volume, 125 Å3) (39). Because photoactivated TDBzl-etomidate forms the same reactive carbene intermediate as does TID, the lack of labeling in this site by TDBzl-etomidate must result from its lack of binding rather than an inability of the reactive intermediate to label aliphatic side chains. Interestingly with the limited sizes of the pockets in each subunit helix bundle in the nAChR structure, TDBzl-etomidate binding was not predicted by the CDOCKER algorithm (“Experimental Procedures”).
nAChR Positive Allosteric Modulators—The paucity of nAChR positive allosteric modulators is in contrast to the structural diversity of GABAAR modulators. Galantamine and physostigmine activate muscle-type nAChRs in the absence of agonist probably by binding to a site in the nAChR extracellular domain (4, 19, 22). nAChR equivalents of the benzodiazepines have not been identified, and most general anesthetics act as nAChR inhibitors. Although it is not known whether the general anesthetics that are nAChR inhibitors can bind to sites in the nAChR TMD other than the ion channel, it is clear that their interactions within the ion channel are of higher affinity than with any other site. TDBzl-etomidate also binds within the ion channel, but in contrast to etomidate and azietomidate, its affinity for the site within the ion channel is reduced sufficiently to allow positive modulation by binding to another TMD site. In the future, it will be important to identify other molecules that bind competitively with [3H]TDBzl-etomidate at its novel site in the nAChR TMD and to determine whether those drugs also act as positive allosteric nAChR modulators.
Supplementary Material
Acknowledgments
We thank Dr. Ayman Hamouda for performing the [3H]tetracaine binding studies and for helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-58448 from the NIGMS. This work was also supported by an award to Harvard Medical School from the Howard Hughes Biomedical Research Support Program for Medical Schools. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
Footnotes
The abbreviations used are: nAChR, nicotinic acetylcholine receptor; ACh, acetylcholine; azietomidate, 2-(3-methyl-3H-diaziren-3-yl)ethyl 1-(phenylethyl)-1H-imidazole-5-carboxylate; TDBzl-etomidate, 4-[3-(trifluoromethyl)-3H-diazirin-3-yl]benzyl-1-(1-phenylethyl)-1H-imidazole-5-carboxylate; TID, 3-(trifluoromethyl)-3-(m-iodophenyl)diazirine; GABAAR, γ-aminobutyric acid type A receptor; Carb, carbamylcholine chloride; V8 protease, S. aureus endopeptidase Glu-C; EndoLys-C, endoproteinase Lys-C; OPA, o-phthalaldehyde; PCP, phencyclidine; TMD, transmembrane domain; HPLC, high pressure liquid chromatography; TPS, Torpedo physiological saline; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; PTH, phenylthiohydantoin.
When the amino acid in cycle x - 1 is unlabeled, the net 3H release in cycle x is quantified as cpmx - cpmx - 1 (see “Experimental Procedures”). However, the increase in 3H release in cycle 9 followed by a further increase in cycle 10 indicates labeling of those two successive amino acids. The experimentally observed release in cycle 10 includes the 3H release from cycle 10 and also 3H originating from cycle 9 due to the lag inherent in Edman degradation and the potentially inefficient transfer of the cleaved, labeled amino acid from the filter through the flask to the fraction collector. In this case, the 3H recovered in cycle 10 originating from cycle 9 is estimated by fitting the releases in cycles 10–12 to an exponential decay and using that fit to calculate the contribution from cycle 9 to the observed release in cycle 10.
The progressively declining 3H release in cycles 11–13 following the 1200 cpm released in cycle 10 in Fig. 6C, which results from the sequencing lag from cycle 10, obscures detection of subtle changes in labeling of αM2-13 in the presence of proadifen compared with the resting state.
References
- 1.Changeux, J.-P., and Edelstein, S. J. (2005) Nicotinic Acetylcholine Receptors: from Molecular Biology to Cognition, Odile Jacob Publishing, New York
- 2.Brejc, K., van Dijk, W. J., Klaassen, R., Schuurmans, M., van der Oost, J., Smit, A. B., and Sixma, T. K. (2001) Nature 411 269-276 [DOI] [PubMed] [Google Scholar]
- 3.Hansen, S. B., Sulzenbacher, G., Huxford, T., Marchot, P., Taylor, P., and Bourne, Y. (2005) EMBO J. 24 3635-3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen, S. B., and Taylor, P. (2007) J. Mol. Biol. 369 895-901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyazawa, A., Fujiyoshi, Y., and Unwin, N. (2003) Nature 423 949-958 [DOI] [PubMed] [Google Scholar]
- 6.Unwin, N. (2005) J. Mol. Biol. 346 967-989 [DOI] [PubMed] [Google Scholar]
- 7.Arias, H. R., Bhumireddy, P., and Bouzat, C. (2006) Int. J. Biochem. Cell Biol. 38 1254-1276 [DOI] [PubMed] [Google Scholar]
- 8.Cromer, B., Morton, C. J., and Parker, M. W. (2002) Trends Biochem. Sci. 27 280-287 [DOI] [PubMed] [Google Scholar]
- 9.Ernst, M., Bruckner, S., Boresch, S., and Sieghart, W. (2005) Mol. Pharmacol. 68 1291-1300 [DOI] [PubMed] [Google Scholar]
- 10.Franks, N. P., and Lieb, W. R. (1994) Nature 367 607-614 [DOI] [PubMed] [Google Scholar]
- 11.Hemmings, H. C., Akabas, M. H., Goldstein, P. A., Trudell, J. R., Orser, B. A., and Harrison, N. L. (2005) Trends Pharmacol. Sci. 26 503-510 [DOI] [PubMed] [Google Scholar]
- 12.Mihic, S. J., Ye, Q., Wick, M. J., Koltchines, V. V., Krasowski, M. D., Finn, S. E., Mascia, M. P., Valenzuela, C. F., Hanson, K. K., Greenblatt, E. P., Harris, R. A., and Harrison, N. L. (1997) Nature 389 385-389 [DOI] [PubMed] [Google Scholar]
- 13.Belelli, D., Lambert, J. J., Peters, J. A., Wafford, K., and Whiting, P. J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 11031-11036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamakura, T., Bertaccini, E., Trudell, J. R., and Harris, R. A. (2001) Annu. Rev. Pharmacol. Toxicol. 41 23-51 [DOI] [PubMed] [Google Scholar]
- 15.Hosie, A. M., Wilkins, M. E., Da Silva, H. M. A., and Smart, T. G. (2006) Nature 444 486-489 [DOI] [PubMed] [Google Scholar]
- 16.Li, G.-D., Chiara, D. C., Sawyer, G. W., Husain, S. S., Olsen, R. W., and Cohen, J. B. (2006) J. Neurosci. 26 11599-11605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okonjo, K. O., Kuhlmann, J., and Maelicke, A. (1991) Eur. J. Biochem. 200 671-677 [DOI] [PubMed] [Google Scholar]
- 18.Samochocki, M., Hoffle, A., Fehrenbacher, A., Jostock, R., Ludwig, J., Christner, C., Radina, M., Zerlin, M., Ullmer, C., Pereira, E. F. R., Lubbert, H., Albuquerque, E. X., and Maelicke, A. (2003) J. Pharmacol. Exp. Ther. 305 1024-1036 [DOI] [PubMed] [Google Scholar]
- 19.Akk, G., and Steinbach, J. H. (2005) J. Neurosci. 25 1992-2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertrand, D., and Gopalakrishnan, M. (2007) Biochem. Pharmacol. 74 1155-1163 [DOI] [PubMed] [Google Scholar]
- 21.Romanelli, M. N., Gratteri, P., Guandalini, L., Martini, E., Bonaccini, C., and Gualtieri, F. (2007) ChemMedChem 2 746-767 [DOI] [PubMed] [Google Scholar]
- 22.Schrattenholz, A., Godovac-Zimmermann, J., Schafer, H. J., Albuquerque, E. X., and Maelicke, A. (1993) Eur. J. Biochem. 216 671-677 [DOI] [PubMed] [Google Scholar]
- 23.Husain, S. S., Nirthanan, S., Ruesch, D., Solt, K., Cheng, Q., Li, G. D., Arevalo, E., Olsen, R. W., Raines, D. E., Forman, S. A., Cohen, J. B., and Miller, K. W. (2006) J. Med. Chem. 49 4818-4825 [DOI] [PubMed] [Google Scholar]
- 24.Kotzyba-Hibert, F., Kapfer, I., and Goeldner, M. (1995) Angew. Chem. Int. Ed. Engl. 34 1296-1312 [Google Scholar]
- 25.Vodovozova, E. L. (2007) Biochemistry (Mosc.) 72 1-20 [DOI] [PubMed] [Google Scholar]
- 26.Husain, S. S., Ziebell, M. R., Ruesch, D., Hong, F., Arevalo, E., Kosterlitz, J. A., Olsen, R. W., Forman, S. A., Cohen, J. B., and Miller, K. W. (2003) J. Med. Chem. 46 1257-1265 [DOI] [PubMed] [Google Scholar]
- 27.Ziebell, M. R., Nirthanan, S., Husain, S. S., Miller, K. W., and Cohen, J. B. (2004) J. Biol. Chem. 279 17640-17649 [DOI] [PubMed] [Google Scholar]
- 28.Middleton, R. E., and Cohen, J. B. (1991) Biochemistry 30 6987-6997 [DOI] [PubMed] [Google Scholar]
- 29.Blanton, M. P., Xie, Y., Dangott, L. J., and Cohen, J. B. (1999) Mol. Pharmacol. 55 269-278 [DOI] [PubMed] [Google Scholar]
- 30.White, B. H., Howard, S., Cohen, S. G., and Cohen, J. B. (1991) J. Biol. Chem. 266 21595-21607 [PubMed] [Google Scholar]
- 31.White, B. H., and Cohen, J. B. (1988) Biochemistry 27 8741-8751 [DOI] [PubMed] [Google Scholar]
- 32.Schagger, H., and von Jagow, G. (1987) Anal. Biochem. 166 368-379 [DOI] [PubMed] [Google Scholar]
- 33.Blanton, M. P., and Cohen, J. B. (1994) Biochemistry 33 2859-2872 [DOI] [PubMed] [Google Scholar]
- 34.Brauer, A. W., Oman, C. L., and Margolies, M. N. (1984) Anal. Biochem. 137 134-142 [DOI] [PubMed] [Google Scholar]
- 35.Middleton, R. E., Strnad, N. P., and Cohen, J. B. (1999) Mol. Pharmacol. 56 290-299 [DOI] [PubMed] [Google Scholar]
- 36.Oswald, R. E., Heidmann, T., and Changeux, J.-P. (1983) Biochemistry 22 3128-3136 [DOI] [PubMed] [Google Scholar]
- 37.Boyd, N. D., and Cohen, J. B. (1984) Biochemistry 23 4023-4033 [DOI] [PubMed] [Google Scholar]
- 38.Arevalo, E., Chiara, D. C., Forman, S. A., Cohen, J. B., and Miller, K. W. (2005) J. Biol. Chem. 280 13631-13640 [DOI] [PubMed] [Google Scholar]
- 39.Garcia, G., III, Chiara, D. C., Nirthanan, S., Hamouda, A. K., Stewart, D. S., and Cohen, J. B. (2007) Biochemistry 46 10296-10307 [DOI] [PubMed] [Google Scholar]
- 40.White, B. H., and Cohen, J. B. (1992) J. Biol. Chem. 267 15770-15783 [PubMed] [Google Scholar]
- 41.Blanton, M. P., Dangott, L. J., Raja, S. K., Lala, A. K., and Cohen, J. B. (1998) J. Biol. Chem. 273 8659-8668 [DOI] [PubMed] [Google Scholar]
- 42.Broad, L. M., Zwart, R., Pearson, K. H., Lee, M., Wallace, L., McPhie, G. I., Emkey, R., Hollinshead, S. P., Dell, C. P., Baker, S. R., and Sher, E. (2006) J. Pharmacol. Exp. Ther. 318 1108-1117 [DOI] [PubMed] [Google Scholar]
- 43.Gallagher, M. J., and Cohen, J. B. (1999) Mol. Pharmacol. 56 300-307 [DOI] [PubMed] [Google Scholar]
- 44.Ryan, S. E., Blanton, M. P., and Baenziger, J. E. (2001) J. Biol. Chem. 276 4796-4803 [DOI] [PubMed] [Google Scholar]
- 45.Charnet, P., Labarca, C., Leonard, R. J., Vogelaar, N. J., Czyzyk, L., Gavin, A., Davidsen, N., and Lester, H. A. (1990) Neuron 2 87-95 [DOI] [PubMed] [Google Scholar]
- 46.Cymes, G. D., Ni, Y., and Grosman, C. (2005) Nature 438 975-980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sigrist, H., Mühlemann, M., and Dolder, M. (1990) J. Photochem. Photobiol. B Biol. 7 277-287 [Google Scholar]
- 48.Revah, F., Galzi, J. L., Giraudat, J., Haumont, P.-Y., Lederer, F., and Changeux, J.-P. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 4675-4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.