Abstract
Background
Tra1 is an essential 437-kDa component of the Saccharomyces cerevisiae SAGA/SLIK and NuA4 histone acetyltransferase complexes. It is a member of a group of key signaling molecules that share a carboxyl-terminal domain related to phosphatidylinositol-3-kinase but unlike many family members, it lacks kinase activity. To identify genetic interactions for TRA1 and provide insight into its function we have performed a systematic genetic array analysis (SGA) on tra1SRR3413, an allele that is defective in transcriptional regulation.
Results
The SGA analysis revealed 114 synthetic slow growth/lethal (SSL) interactions for tra1SRR3413. The interacting genes are involved in a range of cellular processes including gene expression, mitochondrial function, and membrane sorting/protein trafficking. In addition many of the genes have roles in the cellular response to stress. A hierarchal cluster analysis revealed that the pattern of SSL interactions for tra1SRR3413 most closely resembles deletions of a group of regulatory GTPases required for membrane sorting/protein trafficking. Consistent with a role for Tra1 in cellular stress, the tra1SRR3413 strain was sensitive to rapamycin. In addition, calcofluor white sensitivity of the strain was enhanced by the protein kinase inhibitor staurosporine, a phenotype shared with the Ada components of the SAGA/SLIK complex. Through analysis of a GFP-Tra1 fusion we show that Tra1 is principally localized to the nucleus.
Conclusion
We have demonstrated a genetic association of Tra1 with nuclear, mitochondrial and membrane processes. The identity of the SSL genes also connects Tra1 with cellular stress, a result confirmed by the sensitivity of the tra1SRR3413 strain to a variety of stress conditions. Based upon the nuclear localization of GFP-Tra1 and the finding that deletion of the Ada components of the SAGA complex result in similar phenotypes as tra1SRR3413, we suggest that the effects of tra1SRR3413 are mediated, at least in part, through its role in the SAGA complex.
Background
TRA1 is an essential gene in Saccharomyces cerevisiae that encodes a 437 kDa protein product. It is a member of a family of key signaling and regulatory molecules that contain a C-terminal phosphatidylinositol-3-kinase (PI3K) domain [1] and is a component of two multisubunit transcriptional regulatory complexes, the SAGA/SLIK and NuA4 complexes, which also contain the histone acetyltransferase enzymes, Gcn5 and Esa1, respectively [2-4]. Tra1 interacts directly with transcriptional activator proteins and is thought to be critical in recruitment of SAGA/SLIK and NuA4 to their target promoters [5-8].
Previously we identified mutations in the C-terminal PI3K domain of Tra1 that showed defects in transcriptional activation, sensitivity to ethanol and the cell wall destabilizing agent calcofluor white and resulted in shortened telomeres [9]. The pattern of changes neither fully mimicked those seen upon disruption of other SAGA/SLIK nor NuA4 components. For example, unlike strains with deletions of NuA4 components, the tra1 mutant strains were relatively insensitive to DNA damaging agents. We performed an initial systematic genetic array (SGA) analysis with the most pronounced allele, tra1SRR3413. This analysis did not identify any synthetic lethal interactions but did reveal 23 synthetic slow growth interactions, many in combination with deletions of genes involved in cell membrane/wall processes. As the lack of synthetic lethal interactions may have arisen from an incomplete selection against diploids in the automated screens, we repeated the screen in a strain background that selects more strongly against diploids. This analysis identified 114 genes displaying synthetic sick/lethal (SSL) interactions with tra1SRR3413. Genes involved in transcription, RNA processing, mitochondrial function and membrane sorting/protein trafficking were prevalent. The phenotypes and genetic interactions of these strains also point to a role for Tra1 in the cellular response to stress.
Results
SGA analysis of tra1SRR3413
As a component of both SAGA and NuA4 complexes, Tra1 is positioned to play a major role in nuclear processes. Previously we performed an SGA analysis on Tra1 using an allele that partially impairs function [9]. No synthetic lethal interactions were obtained in this analysis. We thus chose to repeat the study using a strain that more strongly selects against the spurious appearance of diploids. tra1SRR3413 was integrated into yeast strain BY7092 and SGA analysis performed using the collection of nonessential yeast knockout strains. Haploids were analyzed on synthetic complete (SC) media at 26°C, 34°C and 36°C with pinnings performed in quadruplicate. 224 double mutant strains, scored as having potential synthetic interactions in each of the screens, were manually tested for growth on YPD media at 30°C and SC media at 33.5°C, after sporulation of the diploids and germination of spore colonies on YPD. For each strain comparisons were made to the relevant single disruption strain. As shown in Table 1, 114 genetic interactions were confirmed as either synthetic lethal or synthetic slow growth on SC or YPD media (SSL interactions). Identified genes are organized according to similarities in associated gene ontology terms. Many cellular functions are represented but the most prominent group were genes linked to membrane sorting/protein trafficking with an emphasis on vacuolar function. An overlapping group included genes associated with cell wall biogenesis and function. Other groups identified initially were chromosomal functions, RNA processing, gene expression, metabolism and biosynthesis and mitochondrial function. A clear subgroup of a larger chromosomal functions group contained the gene encoding the alternative histone H2AZ (HTZ1) and members of the SWR1 complex, which exchange H2AZ for histone H2A within nucleosomes (for example see [10]).
Table 1.
Genes with synthetic slow growth or synthetic lethal interactions with tra1SRR3413
| GENE1 | Orf | SSL2 | GO3 | Location, component4 | phenotype5 | stress6 | #7 |
| MCK1 | YNL307C | SL | 1, 3 | cytoplasm, nucleus | X | 28 | |
| CDC73 | YLR418C | s | 1, 3, 10 | Cdc73/Paf complex, nucleus | T, R | X | 159 |
| CSE2 | YNR010W | S | 1, 3, 10 | mediator complex | R | X | 19 |
| HTZ1 | YOL012C | S | 1, 3, 10 | nucleus | 186 | ||
| LEO1 | YOR123C | s | 1, 3, 10 | Cdc73/Paf complex, nucleus | T | 39 | |
| RAD52 | YML032C | s | 1, 3, 10 | nucleus | X | 196 | |
| SPT8 | YLR055C | s | 1, 3, 10 | SAGA complex, nucleus | 62 | ||
| SRB2 | YHR041C | s | 1, 3, 10 | Srb-mediator complex | T, E | 34 | |
| SWR1 | YDR334W | s | 1, 3, 10 | SWR1 complex, nucleus | R | 112 | |
| VPS71 | YML041C | s | 1, 3, 10, 9 | SWR1 complex, nucleus | E | 97 | |
| VPS72 | YDR485C | s | 1, 3, 10, 9 | SWR1 complex, nucleus | E | 87 | |
| KAR3 | YPR141C | s | 1, 3, 7 | microtubule, spindle pole body | E | 131 | |
| UBI4 | YLL039C | s | 1, 3, 8, 9 | cytoplasm | X | 7 | |
| UBP15 | YMR304W | s | 1, 3, 8, 9 | cytoplasm | 0 | ||
| SWC3 | YAL011W | s | 1, 3, 9, 10 | SWR1 complex, nucleus, mitochondrion | 85 | ||
| DOA1 | YKL213C | s | 1, 9, 8 | cytoplasm, nucleus | X | 26 | |
| TOR1 | YJR066W | S | 1,2,3,4,5,6,7,8,9,10 | cellular membranes | R | X | 27 |
| CKB2 | YOR039W | s | 1,2,3,4,5,7,8,9,10 | protein kinase CK2, CURI complex | X | 14 | |
| SNF1 | YDR477W | s | 1,3,4,5,6,7,8,9,10 | cytoplasm, nucleus, nuclear evelope, mitochondrion, vacuole | R | X | 18 |
| SNF4 | YGL115W | S | 1,3,4,5,6,7,8,9,10 | cytoplasm, nucleus, nuclear envelope, PM | 7 | ||
| CNB1 | YKL190W | s | 1,3,4,5,7,9,10 | calcineurin complex | X | 48 | |
| EOS1 | YNL080C | S | 1,3,4,7 | ER membrane | X | 4 | |
| SIS2 | YKR072C | s | 1,3,5,10 | cytoplasm, nucleus | X | 10 | |
| NUM1 | YDR150W | s | 1,3,7,9 | cell cortex, cellular bud tip, mitochondrion | 57 | ||
| SWI4 | YER111C | s | 1, 3, 7, 10 | nucleus | CW | 69 | |
| UBP6 | YFR010W | S | 1,3,8,9,10 | proteosome | X | 28 | |
| MSC1 | YML128C | s | 1,3,9,6 | ER, mitochondrion | R | 14 | |
| ARP6 | YLR085C | s | 10, 1, 2 | SWR1 complex, cytoplasm | R | 149 | |
| CAF40 | YNL288W | s | 2, 10 | CCR4-NOT complex | R | 8 | |
| NST1 | YNL091W | s | 2, 10 | cytoplasm | X | 2 | |
| PUB1 | YNL016W | s | 2, 10 | cytoplasm, nucleus, hnRNP complex | X | 5 | |
| PIH1 | YHR034C | s | 2, 3, 8, 10 | cytoplasm, nucleus, snRNP complex | 2 | ||
| HMT1 | YBR034C | s | 2, 4 10 | nucleus | 4 | ||
| PML39 | YML107C | SL | 2, 4, 10 | nuclear pore, ribosome | 4 | ||
| HCR1 | YLR192C | s | 2, 8 | translation IF3 complex | T | X | 5 |
| PPQ1 | YPL179W | S | 2, 8 | cytoplasm | 0 | ||
| TMA23 | YMR269W | s | 2, 8 | nucleolus, ribosome | 5 | ||
| RRP6 | YOR001W | SL | 2, 8, 10 | nuclear exosome (RNase complex) | 16 | ||
| SKY1 | YMR216C | s | 2,3,4,8,10 | cytoplasm | 5 | ||
| STP1 | YDR463W | s | 2,4,7,8,10 | nucleus | 2 | ||
| KNS1 | YLL019C | SL | 3 | X | 0 | ||
| BIM1 | YER016W | s | 3, 1, 7 | cytoplasm, microtubule, kinetochore, spindle pole body | 282 | ||
| NBP2 | YDR162C | s | 3, 7 | cytoplasm, nucleus | CW, R | X | 49 |
| PRB1 | YEL060C | s | 3, 8, 9 | vacuole | X | 3 | |
| SEL1 | YML013W | s | 3, 8, 9 | ubiquitin ligase complex, ER, mitochondrion | 3 | ||
| TPS2 | YDR074W | s | 3,5,7,8,10 | mitochondrion | X | 4 | |
| ITR1 | YDR497C | s | 4, 5, 7, 9 | plasma membrane | X | 9 | |
| NEW1 | YPL226W | s | 4, 5, 9, 8 | cytoplasm, mitochondrion | 8 | ||
| AQR2 | YBR043C | s | 4, 7 | plasma membrane | 1 | ||
| GTR1 | YML121W | s | 4, 9 | cytoplasm, endosome, nucleus, vacuole | T, E, R | 5 | |
| VAM10 | YOR068C | SL | 4, 9 | vacuolar membrane | 11 | ||
| SER1 | YOR184W | s | 5 | cytoplasm | 1 | ||
| SER2 | YGR208W | s | 5 | cytoplasm, nucleus | 5 | ||
| GCR2 | YNL199C | SL | 5, 10, 3 | nucleus | E | 30 | |
| ARO1 | YDR127W | SL | 5, 6 | cytoplasm | E | 0 | |
| ARO2 | YGL148W | SL | 5, 6 | cytoplasm | 1 | ||
| ETR1 | YBR026C | s | 5, 6 | mitochondrion | 0 | ||
| LIP2 | YLR239C | s | 5, 6 | mitochondrion | 7 | ||
| LIP5 | YOR196C | S | 5, 6 | mitochondrion | 2 | ||
| OAR1 | YKL055C | s | 5, 6 | mitochondrion | 0 | ||
| ERG3 | YLR056W | s | 5, 9 | ER | 19 | ||
| MDM34 | YGL219C | S | 6 | mitochondrial outer membrane | E, R | 10 | |
| KGD2 | YDR148C | S | 6, 5 | mitochondrion | 0 | ||
| MDM10 | YAL010C | s | 6, 9 | Mdm10/Mdm12/Mmm1 complex, mitochondrial outer membrane | T, E | 11 | |
| BEM1 | YBR200W | S | 7, 3, 1 | bud neck | E | 33 | |
| BEM4 | YPL161C | s | 7, 3, 1 | cytoplasm, nucleus | T, E | X | 29 |
| Smi1 | YGR229C | s | 7, 3, 1 | bud tip, nucleus | T, E, CW, R | 111 | |
| ROM2 | YLR371W | SL | 7, 3, 9 | bud tip | X | 16 | |
| CSF1 | YLR087C | s | 7, 5, 6 | mitochondrion | CW | 24 | |
| ECM30 | YLR436C | s | 7, 9 | cytoplasm | CW | 1 | |
| KRE1 | YNL322C | s | 7, 9 | cell wall | 64 | ||
| VIP1 | YLR410W | S | 7, 9 | cytoplasm | 15 | ||
| TPM1 | YNL079C | S | 7,9,3 | actin cable | E, CW, R | 21 | |
| RPS27B | YHR021C | S | 8 | ribosome | T | 1 | |
| RPN4 | YDL020C | S | 8, 10, 1, 3 | nucleus, proteasome | T, E | X | 242 |
| YDJ1 | YNL064C | S | 8, 9, 6 | cytosol, microsome | R | X | 27 |
| CCZ1 | YBR131W | s | 9 | late endosome, membrane | E, R | X | 6 |
| MON1 | YGL124C | s | 9 | cytosol, vacuolar membrane | 12 | ||
| VAM3 | YOR106W | s | 9 | vacuolar membrane | R | 9 | |
| VAM7 | YGL212W | s | 9 | vacuolar membrane | T, E, CW, R | 8 | |
| PIB2 | YGL023C | s | 9, 1, 7 | late endosome, mitochondrion | R | 0 | |
| GSG1 | YDR108W | s | 9, 3 | TRAPP complex | X | 11 | |
| YDR049W | s | 9, 3, 8 | cytoplasm, mitochondrion | E, R | |||
| MEH1 | YKR007W | S | 9, 4, 5 | cytosol, vacuolar membrane | X | 2 | |
| PMR1 | YGL167C | s | 9, 4, 7 | Golgi membrane | E | 41 | |
| AUT7 | YBL078C | s | 9, 5 | autophagic vacuole, cytosol | X | 2 | |
| CCW12 | YLR110C | s | 9, 7 | cell wall | CW | 13 | |
| COG5 | YNL051W | S | 9, 7 | Golgi | 14 | ||
| COG6 | YNL041C | S | 9, 7 | Golgi | 17 | ||
| COG7 | YGL005C | S | 9, 7 | Golgi | 12 | ||
| COG8 | YML071C | S | 9, 7 | Golgi | 17 | ||
| DID2 | YKR035W | s | 9, 7 | cytoplasm, late endosome | 2 | ||
| GYP1 | YOR070C | S | 9, 7 | Golgi, mitochondrion | R | 48 | |
| MNN10 | YDR245W | s | 9, 7 | α-1,6-mannosyltransferase complex | E, R, CW | 19 | |
| MON2 | YNL297C | s | 9, 7 | cytosol, endosome | R | 40 | |
| RIC1 | YLR039C | S | 9, 7 | Golgi, nucleus | E | 159 | |
| RUD3 | YOR216C | s | 9, 7 | Golgi | 26 | ||
| SEC22 | YLR268W | s | 9, 7 | ER, Golgi | CW | 44 | |
| TLG2 | YOL018C | s | 9, 7 | early endosome, trans Golgi | R | 12 | |
| VPS1 | YKR001C | S | 9, 7 | cytoplasm, membrane fraction | 28 | ||
| VPS17 | YOR132W | s | 9, 7 | endosome, retromer complex | 11 | ||
| VPS38 | YLR360W | s | 9, 7 | Vps34 complex | R | 12 | |
| VPS41 | YDR080W | s | 9, 7 | HOPS complex, vacuolar membrane, endosome | R | 7 | |
| VPS53 | YJL029C | SL | 9, 7 | GARP complex, Golgi, cytoplasm | T, R | 10 | |
| YPT6 | YLR262C | s | 9, 7 | Golgi | E, R | 181 | |
| YPT7 | YML001W | s | 9, 7 | vacuole, mitochondrion | T, R | 6 | |
| YLR111W | s | 9, 7 | CW | 0 | |||
| GET1 | YGL020C | S | 9, 8 | GET complex, ER, mitochondrion | R | X | 284 |
| SSE1 | YPL106C | SL | 9, 8 | cytoplasm | T | 15 | |
| BTS1 | YPL069C | S | 9,5,6 | mitochondrion | CW | 16 | |
| UBP3 | YER151C | s | 9,7,10,1,3 | cytoplasm | R | 32 | |
| YDL180W | s | vacuole | 0 | ||||
| YEL033W | s | T | 0 | ||||
| YOR235W | s | 0 | |||||
1 Genes are placed according to related GO terms with relevant groupings separated by lines.
2 Phenotype of double mutants. SL, synthetic lethal; S, severe slow growth; s, slow growth (in SC media at 33.5°C). For those underlined the phenotype is most easily detected on YPD at 30C as the single mutant shows reduced growth on SC media
3 Gene ontology: (1) DNA replication/repair and chromosome stability, (2) RNA processing, (3) cell cycle/microtubule, (4) general transport, (5) metabolism, (6) mitochondria, (7) polarity/cell wall, (8) protein/ribosome biosynthesis, (9) secretion/protein trafficking, (10) transcription, (11) unknown
4 Cellular compartments and/or components as indicated in the Saccharomyces Genome Database
5 Relevant phenotypes of single mutants as described in the Saccharomyces Genome Database. E, sensitivity to ethanol; CW, sensitivity to calcofluor white; T, shortened telomeres; R, sensitivity to rapamycin
6 Annotated in the Saccharomyces Genome Database as involved in stress response or autophagy
7 Number of previously identified SSL interactions in the Saccharomyces Genome Database
Before pursuing further analysis of the genes identified in the SGA screen, we wanted to eliminate those that may have arisen through indirect effects on neighboring genes. For instance, YLR111W is a dubious ORF located adjacent to CCW12 that encodes a cell wall component. Since disruption of YLR111W may simply act by affecting expression of CCW12, it was not considered in further analyses. Several other dubious ORFs were eliminated because they overlapped a second identified gene. Other pairs of adjacent genes were kgd2 and num1, spt8 and erg3, and tpm1 and eos1, though these were not removed from the analysis since potentially both could be involved in SSL interactions with tra1SRR3413.
Also indicated in Table 1 is the total number of additional SSL interactions listed for each of the genes in the Saccharomyces Genome Database. It has been argued that the number of interactions may be a measure of the importance of a gene for cellular fitness [11]. The relatively large number of SSL interactions for tra1SRR3413 may also reflect its involvement in both SAGA/SLIK and NuA4 complexes. As with any synthetic lethal analysis we can not eliminate the possibility that some of the apparent interactions are due to additive growth defects rather than a demonstration that the genes act in the same or related pathways.
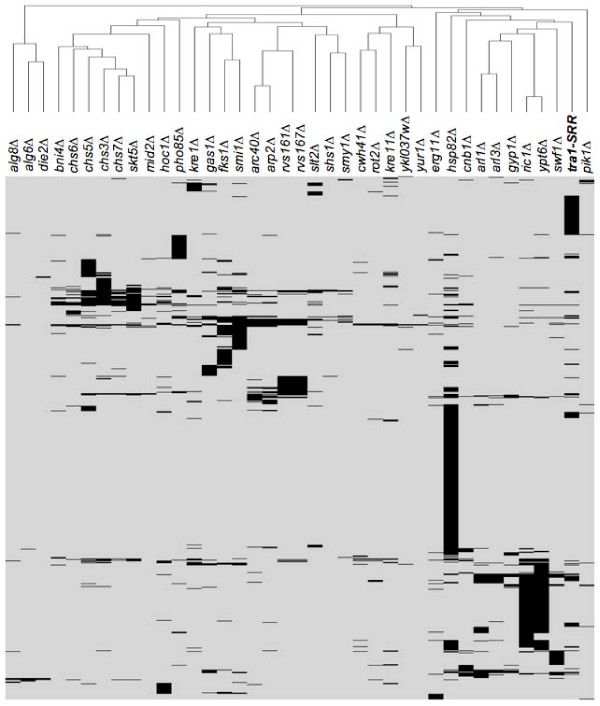
To identify functional relationships for tra1SRR3413, a hierarchal cluster analysis of SSL tra1SRR3413 interactions was performed with the data sets of Tong et al. [11], Measday et al. [12], Reguly et al. [13], Pan et al. [14] and Mitchell et al. [15] plus the SSL interactions of additional SAGA/SLIK and NuA4 components as listed in the Saccharomyces Genome Database (Figure 1). tra1SRR3413 clustered most closely to a group including arl1Δ0, arl3Δ0, gyp1Δ0, ric1Δ0, ypt6Δ0 and swf1Δ0. Three of these (ric1, ypt6 and gyp1) show synthetic interactions with tra1SRR3413 (Table 1). All encode key regulatory molecules in the processes of membrane sorting and protein trafficking [16], and with the exception of SWF1 are GTPases of the Ras-family or regulatory proteins of these molecules. Interestingly, ric1 was first identified as a temperature sensitive allele that affected transcription of both ribosomal protein genes and rRNA [17]. Also closely associated with this group was cnb1Δ0, which is SSL with tra1SRR3413 and encodes a Ca2+/calmodulin dependent protein phosphatase required for cell cycle regulation, stress induced gene expression and cell wall synthesis. To gain additional confidence in the apparent association of tra1SRR3413 with the family of GTPases, the cluster analysis was repeated in the absence of these genes. In this case, tra1SRR3413 clustered with a group containing SAGA/SLIK and NuA4 components (not shown).
Figure 1.
Hierarchal cluster analysis of tra1SRR3413 SSL interactions. Agglomerative hierarchical clustering based on the average linkage of uncentered correlations using CLUSTER 3.0 software [50] was performed on the profile obtained with the tra1SRR3413 strain and the combined data sets of Tong et al. [11], Measday et al. [12], Reguly et al. [13], Pan et al. [14], Mitchell et al. [15] and including SSL interactions of additional SAGA/SLIK and NuA4 components as listed in the Saccharomyces Genome Database. Only the profiles clustering close to tra1SRR3413 are shown.
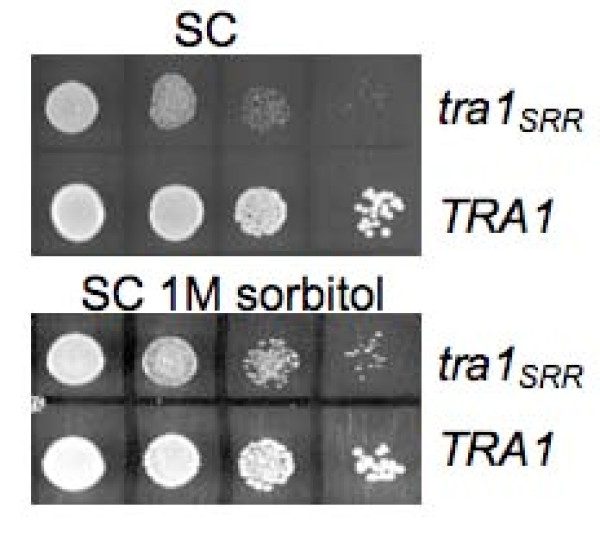
Sorbitol partially suppresses defects due to tra1SRR3413
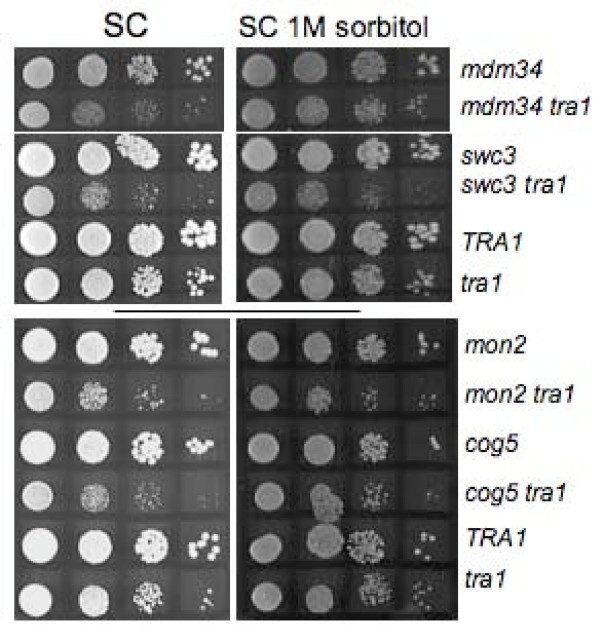
SSL interactions with a number of genes (CCW12, CNB1, ECM30, KRE1, MNN10, ROM2, SMI1 and TOR1) that have functional links to cell wall organization and biogenesis was consistent with the calcofluor white and ethanol sensitivity of the tra1SRR3413 strain [9]. We thus investigated the extent to which the temperature sensitivity of the tra1SRR3413 strain results from cell wall destabilization by examining if it is suppressed by growth in media containing 1 M sorbitol. As shown in Figure 2, sorbitol partially, but not completely, suppressed the temperature sensitive growth at 37°C of this strain (compare relative growth of tra1SRR and TRA1 in SC with sorbitol, lower panel, and without sorbitol, upper panel), indicating that defects in cell wall function contribute to but are not exclusively responsible for the temperature sensitivity. We also examined if some of the SSL interactions were largely due to cell wall instability. mdm34Δ0 tra1SRR3413, swc3Δ0 tra1SRR3413, mon2Δ0 tra1SRR3413 and cog5Δ0 tra1SRR3413 were examined as representative of mitochondrial, chromosomal and membrane sorting groups. As shown in Figure 3, growth inhibition of the mdm34Δ0 tra1SRR3413 strain (at 33.5°C for this experiment in contrast to 37°C for Figure 2) was partially rescued by 1 M sorbitol. Slight suppression was seen for the cog5Δ0 tra1SRR3413 strain, while growth of the swc3Δ0 tra1SRR3413 and mon2Δ0 tra1SRR3413 strains was relatively unchanged by sorbitol. The strain dependent differences in the ability of sorbitol to suppress SSL effects further establish that Tra1 has roles in multiple processes.
Figure 2.
Sorbitol partially suppresses slow growth at 37°C due to tra1SRR3413. Yeast strains CY2222 (tra1SRR3413) and BY7092 (TRA1) were grown overnight to saturation in YPD at 30°C. Cells were diluted to approximately 1000 cells per μl and 10 μl of 10-fold serial dilutions spotted on synthetic complete media containing 2% glucose without (SC) or with 1.0 M sorbitol. Cells were grown for 4 days at 37°C.
Figure 3.
Growth of tra1SRR3413 double mutant strains in sorbitol. Strains containing the indicated single disruptions (mdm34, swc3, cog5 and mon2) and combinations with tra1SRR3413 (tra1) were generated by sporulation of diploids obtained from SGA analysis. These strains plus CY2222 (tra1SRR3413) and BY7092 (TRA1) were grown overnight to saturation in YPD at 30°C. Cells were diluted to approximately 2000 cells per μl and 5 μl of 10-fold serial dilutions spotted on synthetic complete media containing 2% glucose without (SC) or with 1.0 M sorbitol. Cells were grown for 3 days (upper grouping) or 2 days (lower grouping) at 33.5°C.
Connection of TRA1 to cellular stress
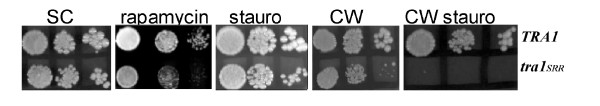
We were intrigued by the finding that amongst the genes showing SSL interactions with tra1SRR3413, 30 are annotated to stress response or autophagy (see Table 1). This is an underestimate as, for example, Swi4 [18], Rps27b [19], Caf40 [20], Vps1 [21] and Stp1 [22] have been implicated in stress response but are not directly annotated as such. Some of the stress response genes include key signaling and regulatory molecules such as Tor1, Msk1, Cnb1, and Rom2; others have functions in protein turnover (Ubi4 and Rpn4), organization of the cytoskeleton (Tpm1 and Bem4) and DNA repair (Rad52, Ckb2, Doa1). To further examine the link between Tra1 and cellular stress response we tested the sensitivity of the tra1SRR3413 strain to rapamycin and to staurosporine. As shown in Figure 4, the tra1SRR3413 strain was partially sensitive to rapamycin, and to a lesser extent staurosporine. As staurosporine results in cell wall instability through its action on Protein Kinase C [23,24], we also examined the tra1SRR3413 strain for sensitivity to the combination of calcofluor white and staurosporine. The tra1SRR3413 strain was extremely sensitive to calcofluor white in the presence of staurosporine, consistent with a role for Tra1 in events required for cell wall integrity.
Figure 4.
Growth of the tra1SRR3413 strain under conditions of cellular stress. Yeast strains BY7092 (TRA1) and CY2222 (tra1SRR3413) were grown overnight to saturation in YPD at 30°C. 5 μl of 10-fold serial dilutions were spotted onto synthetic complete media containing 2% glucose (SC) or with the addition of 2 nM rapamycin, 2 μg/ml staurosporine (stauro), 7.5 μg/ml calcofluor white (CW), or 2 μg/ml staurosporine plus 7.5 μg/ml calcofluor white (CW stauro). Cells were grown for 3 days on SC and 5 days on selective plates, at 30°C.
Relationship of tra1SRR3413 to other mutations in SAGA/SLIK and NuA4 components
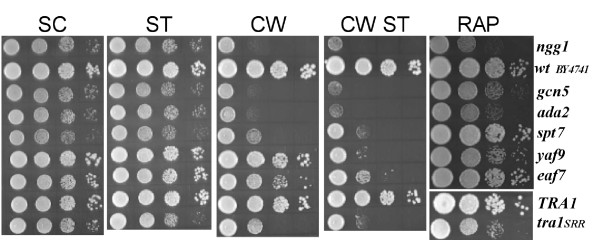
Bruno et al. [25] have shown that deletion of the Ada2 homologue in C. albicans results in sensitivity to cell wall destabilizing agents. In addition, both SAGA/SLIK and NuA4 have been suggested to be involved in stress response based upon the transcription profiles of deletion strains [26,27]. As some of the genetic interactions and phenotypes of the tra1SRR3413 strain may result from a SAGA/SLIK or NuA4-dependent inability to induce stress response genes, we analyzed staurosporine, calcofluor white, calcofluor white plus staurosporine and rapamycin sensitivity in strains deleted for additional components of these complexes (Figure 5). The effect of tra1SRR3413 under each of these conditions is similar to, though slightly less severe than, that seen upon deletion of the ada genes (ada2, ngg1/ada3 and gcn5/ada4) of the SAGA/SLIK complexes. Interestingly, despite the fact that deletion of spt7 results in disruption of the complexes, spt7Δ0 results in less of an effect on rapamycin and calcofluor white than deletion of the ada genes, suggesting a potential role for an independent Ada complex [28]. This similarity between the ada genes and tra1SRR3413 is also consistent with our previous finding that the expression profile in the tra1SRR3413 strain most closely resembled deletion of ada2 [9]. Less similarity was seen in the phenotypes with NuA4 components though deletion of yaf9 and eaf7 did lead to reduced growth in calcofluor white plus staurosporine containing media and deletion of yaf9 resulted in a slight reduction of growth in media containing rapamycin.
Figure 5.
Phenotypic comparison of tra1SRR3413 with deletions of SAGA/SLIK and NuA4 components. Yeast strains BY3534 (ngg1Δ0), BY4741 (wild-type background for deletion strains), BY7285 (gcn5Δ0), BY4282 (adaΔ0), BY3281 (spt7Δ0), BY4240 (yaf9Δ0), BY2940 (eaf7Δ0), BY7092 (TRA1) and CY2222 (tra1SRR3413) were grown overnight to saturation in YPD at 30°C. Cells were diluted to approximately 2000 cells per μl and 5 μl of 10-fold serial dilutions spotted on synthetic complete media containing 2% glucose (SC), with 2 μg/ml staurosporine (ST), 7.5 μg/ml calcofluor white (CW), or 2 μg/ml staurosporine plus 7.5 μg/ml calcofluor white (CW ST) or YPD with 2 nM rapamycin (RAP). SC and ST were grown for 2 days at 30°C, CW and CW ST for 3 days, and RAP for 4 days. For the rapamycin image, TRA1 and tra1SRR were grown on a separate plate.
GFP-tagged Tra1 localizes to nucleus
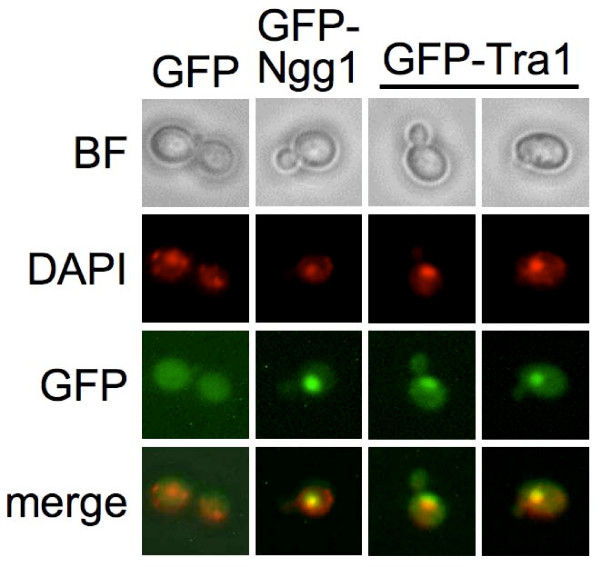
The requirement for an intact C-terminal FATC domain [1] has meant that Tra1 localization has not been tested in genome-wide screens. Localization of Tra1 to sites outside the nucleus potentially could account for the broad range of genetic interactions seen for the molecule. To examine cellular localization, we engineered an N-terminally GFP-tagged allele of TRA1 that is functional as determined by plasmid shuffling assays (not shown). As shown in Figure 6, the pattern of GFP-Tra1 distribution was very similar to that of the SAGA/SLIK component Ngg1/Ada3 and coincided with DAPI staining within the nucleus (Figure 6). A low level of fluorescence was apparent throughout the cell, making it difficult to fully exclude that lesser amounts exist in other compartments; however, GFP-Tra1 was not in foci other than the nucleus.
Figure 6.
GFP-Tra1 is found predominately in the nucleus. Yeast strain BY4741, expressing either GFP, GFP-Ngg1 or GFP-Tra1 from a URA3-centromeric plasmid was grown in synthetic complete media, stained with DAPI and examined by fluorescence microscopy. BF, bright field.
Discussion
Identification of the SSL interactions for the tra1SRR3413 allele has provided insights into the function of this molecule. The genes identified link Tra1 to many cellular functions including membrane sorting/protein trafficking, cell wall biogenesis/function, RNA processing, gene expression and mitochondrial function. The number of genetic interactions identified for tra1SRR3413 is likely due to its function in two key cellular regulatory complexes, SAGA/SLIK and NuA4.
The finding that the SSL profile for tra1SRR3413 does not share greater similarity with mutations of other SAGA/SLIK and NuA4 components agrees with our previous observations that these strains differ in their transcription profiles, effects on telomere length and sensitivity to the DNA damaging agent methylmethanesulfonate [9]. These differences may arise from the integrative effects of disturbing both complexes and/or the possibility that Tra1 has a unique function. While differences are apparent, we do note that if the cluster analysis is performed in the absence of the arl1Δ0, arl3Δ0, gyp1Δ0, ric1Δ0, ypt6Δ0 and swf1Δ0 group, the SSL profile for tra1SRR3413 clusters most closely with SAGA/SLIK and NuA4 components (not shown). In addition, as seen by the common sensitivity to ethanol and calcofluor white plus staurosporine, several of the tra1SRR3413 phenotypes are similar to deletions of the ada genes.
Nuclear function
SAGA/SLIK and NuA4 are nuclear complexes and indeed we have found Tra1 to be predominately, if not exclusively, localized in the nucleus. As an essential component of two histone-modifying complexes, it is not surprising that approximately 40 of the SSL interactions are with genes annotated principally as having roles in nuclear function or gene expression. Some of these genes (for example: pub1, pml39 and sky1) are involved with RNA processing, export or stability perhaps reflecting a role for the SAGA complex at the nuclear pore [29]. Another clear subset of SSL interactions includes genes of the SWR1 complex and htz1. In this case the relationship with tra1SRR3413 almost certainly reflects their combined importance in determining chromatin structure and their resulting influence on transcription and/or DNA repair. Supporting this argument, SSL interactions have been observed between many components of SAGA and NuA4 with htz1 and SWR1 complex genes [30]. Similarly the interaction of tra1SRR3413 with rad52 may relate to the function of the NuA4 complex in DNA repair.
Membrane processes
The association of SAGA/SLIK and NuA4 components with membrane processes has been noted previously. Gcn5 and Spt20 are required for the unfolded protein response [31,32]. A strain with deletion of eaf1/vid21 (Vacuolar Import Degradation) was identified in a screen for defects in sorting of carboxypeptidase Y to the vacuole [33] and results in sensitivity to ethanol [34]. In addition, Eaf1 interacts with Vac8, which is required for several aspects of vacuolar function [35]. eaf3, eaf5, eaf7, eaf1/vid21 and yaf9 were identified as Opi- mutants that result in overproduction of inositol, though not through direct repression of INO1 [36]. Mitchell et al. [15] have observed SSL interactions of nonessential NuA4 components with proteins involved in membrane sorting/protein trafficking. They find that deletion the NuA4 component genes leads to defects in vacuolar morphology and similar to what we suggest for the phenotypic effects of tra1SRR3413, predict that the vacuolar defects arise from changes in gene expression.
Mitochondrial function
Fifteen genes annotated to have links to mitochondrial function were identified in the screen. This may relate to the possible involvement of the SLIK complex in the retrograde response pathway [37]. Interestingly this pathway links mitochondrial metabolism with stress response, signaling the expression of genes that provide required biosynthetic precursors during mitochondrial dysfunction, which results in the loss of tricarboxylic acid cycle activity [38].
Stress response
Included in the tra1SRR3413 SSL genes are greater than 30 related to cellular stress or stress response. Genetic interactions for tra1SRR3413 with molecules related to cellular stress is consistent with the transcription profiles of strains containing deletions of SAGA/SLIK and NuA4 components [26,27] as well as the calcofluor white, ethanol, rapamycin and temperature sensitivity of the tra1SRR3413 strain. An aspect of the involvement of Tra1 in stress likely includes a role in the cell wall integrity response pathway as indicated by the calcofluor white sensitivity and interactions of the tra1SRR3413 SSL profile with molecules required for membrane sorting/protein trafficking and cell wall components (for example, Kre1 and Ccw12). The processes identified with Tra1 in many ways resemble those seen with Tor1, which as well as its initially defined role in integrating nutrient status with growth, has been genetically and functionally linked to membrane sorting/protein trafficking, cytoskeletal events, microautophagy and to cell well integrity [39-43].
The simplest interpretation of a possible link between Tra1 and stress is to suggest that Tra1 is involved in the expression of stress response genes or genes whose lack of expression results in stress. Consistent with this, in YPD media seventeen genes with an annotation or description indicative of an involvement in stress response have decreased expression of two-fold or greater in a tra1SRR3413 background ([9], Table 2). Many of these genes function in the response to oxidative stress but their exact role in determining phenotype is likely complex given that the tra1SRR3413 strain is not sensitive to the oxidizing agent tert-butylhydroperoxide and genes typically elevated in response to stress are not found induced in the tra1SRR3413 strain.
Table 2.
Stress related genes down regulated in a tra1SRR3413 background.
| Gene | Fold-decrease1 | Description2 |
| AAD4 | 11 | Putative aryl-alcohol dehydrogenase. Involved in oxidative stress response. |
| TSA2 | 6.5 | Stress induced thioredoxin peroxidase. |
| CCS1 | 5.5 | Copper chaperone for superoxide dismutase Sod1. Involved in oxidative stress protection. |
| AAD6 | 4.4 | Putative aryl-alcohol dehydrogenase. Involved in oxidative stress response. |
| GPX2 | 3.6 | Phospholipid hydroperoxide glutathione peroxidase. Protects cells from peroxides. |
| SSA4 | 3.3 | Heat shock protein that is highly induced upon stress. Role in SRP-dependent cotranslational protein-membrane targeting. |
| GRX2 | 3.1 | Glutaredoxin, thioltransferase, glutathione-dependent disulfide oxidoreductase. Maintains redox state of target proteins. |
| RHR2 | 3.0 | DL-glycerol-3-phosphatase. Induced by anaerobic and osmotic stress. |
| ALD2 | 2.4 | Aldehyde dehydrogenase. Stress induced. |
| TRX2 | 2.4 | Thioredoxin isoenzyme. Protects cells against oxidative stress. |
| GRE2 | 2.4 | 3-methylbutanal reductase and methylglyoxal reductase. Stress induced. |
| SRX1 | 2.4 | Sulfiredoxin. Contributes to oxidative stress resistance. |
| ALD3 | 2.3 | Cytoplasmic aldehyde dehydrogenase. Stress induced. |
| LSP1 | 2.3 | Component of eisosomes. Null mutants show activation of Pkc1p/Ypk1p stress pathways. |
| PST2 | 2.2 | Similarity to flavodoxin-like proteins. Induced by oxidative stress. |
| CMK1 | 2.2 | Calmodulin-dependent protein kinase. May play a role in stress response. |
| DAK1 | 2.0 | Dihydroxyacetone kinase. Involved in stress adaptation. |
1 Decrease in gene expression from microarray analysis [9], tra1SRR3413 versus TRA1
2 Modified from the Saccharomyces Genome Database
We previously demonstrated that tra1SRR3413 results in a generation dependent telomere shortening that is not characteristic of other SAGA/SLIK or NuA4 components. Fifteen of the genes with SSL interactions with tra1SRR3413 also show telomere shortening [44]. In many cases direct telomeric functions for these molecules have not been described but like tra1SRR3413, they display slow growth in response to ethanol, calcofluor white or rapamycin. This suggests the possibility that shortened telomeres in the tra1SRR3413 strain is the result of a similar indirect mechanism rather than direct action at the telomere.
Conclusion
Through the identification of synthetic sick/lethal interactions with tra1SRR3413 we have demonstrated a genetic association of Tra1 not only with nuclear processes but also with membrane events and mitochondrial function. The identity of the SSL genes also connects Tra1 with cellular stress, a result confirmed by the sensitivity of the tra1SRR3413 strain to a variety of conditions that result in a stress response. The transcription profile and SSL interactions indicate that the functions of Tra1 can not simply be ascribed individually to either SAGA/SLIK or NuA4 complexes. However, the finding that many patterns of the tra1SRR3413 phenotype resemble those seen with deletions of the Ada components of the SAGA/SLIK complex points toward a role for the PI3K domain of Tra1 in regulating the activity of the Ada molecules.
Methods
Yeast strains and growth
Yeast strains BY3534 (ngg1Δ0), BY7285 (gcn5Δ0), BY4282 (adaΔ0), BY3281 (spt7Δ0), BY4240 (yaf9Δ0), BY2940 (eaf7Δ0) are derivatives of BY4741 (MATa his3Δ0, leu2Δ0 met1Δ0 ura3Δ0; [45]) and were purchased from Open Biosystems. Yeast strain CY2222 is a derivative of BY7092 (MATα can1Δ::STE2pr-SpHIS5 lyp1Δhis3Δ1 leu2Δ0 ura3Δ0 met10 LYS2+) that has been gene replaced with tra1SRR3413 and selected for through the placement of Tn10LUK at the downstream BstBI site. To ensure that this integration did not hamper expression of YHR100C, a 2035 base pair EcoRI-HindIII fragment encompassing this gene was integrated after cloning into the LEU2 integrating vector YIplac128 [46] and digestion with MscI. Strains containing disruptions of individual genes and double mutants with tra1SRR3413 were obtained by tetrad dissection of diploids generated from the SGA analysis. Strains were spotted in ten-fold serial dilutions on synthetic complete media containing 2 nM rapamycin (LC Laboratories, Woburn Ma), 7.5 μg/ml calcofluor white (Sigma-Aldrich Canada, Oakville Ontario), 2 μg/ml staurosporine (LC Laboratories), calcofluor white plus staurosporine, or 1.0 M sorbitol. Growth was also compared on synthetic complete media containing 1% potassium acetate and 0.05% glucose as the carbon source.
GFP fusion protein and microscopy
A URA3 centromeric plasmid that allowed expression of GFP fusions was engineered by inserting a BamHI-NotI fragment of the PCR product synthesized using oligonucleotides 5'-ATGCGGATCCACAATGGTGAGCAAGGGCGAGG-3 and 5'-TTTTCCTTTTCCGGCCGCCTTGTACAGCTCGTCCATG-3' as primers and pEGFP-N1 (Clontech Laboratories, Inc.) as template to replace the tags of YCpDed-TAP-Flag [9]. TRA1 and NGG1 were inserted into this vector as NotI-SstI fragments from YCpDed-TAP-Flag-TRA1 (9) and YCpDed-myc-NGG1 [47], respectively. Fluorescence microscopy was performed as described by Brachat et al. [48] with minor modification. Yeast expressing GFP-tagged proteins were grown in SC media lacking uracil at 30°C. At A600~0.8, 4',6-diamidino-2-phenylindole (DAPI) was added to a final concentration of 5 μg/ml and the cells incubated at 30°C for an additional 1–2 hr. Cells from 1 mL of the culture were pelleted, resuspended in 100 μL SC media, immobilized between a microscope slide and cover slip in SC media containing 0.7% agarose and observed using a Zeiss Axiovert 25 fluorescent microscope under 400× magnification. Images were captured, auto-equalized, colorized and merged using Northern Elite software (Empix Imaging, Inc.).
Systematic genetic-array analysis
SGA analysis with yeast strain CY2222 was performed as described by Tong and Boone [49]. Each strain was pinned in quadruplicate with strains analyzed on synthetic media at 26°C, 34°C and 36°C. Diploids of those strains showing slow growth at all temperatures were sporulated and subjected to tetrad dissection on YPD plates. When viable, the growth of single and double disruptions of the relevant gene were compared to CY2222 on YPD plates at 30°C and synthetic complete plates at 33.5°C. Agglomerative hierarchical clustering based on the average linkage of uncentered correlations using CLUSTER 3.0 software [50] was performed on the profile obtained with the data sets of Tong et al. [11], Measday et al. [12], Reguly et al. [13], Pan et al. [14] and Mitchell et al. [15], as compiled by Mitchell et al. [15]. SSL interactions, as listed in the Saccharomyces Genome Database (March 2008), of additional components of SAGA/SLIK and NuA4 complexes were also included in the analysis.
Authors' contributions
SMTH carried out microscopy, data analysis and co-wrote the manuscript with CJB. JG performed the SGA analysis under the supervision of BA. CJB performed the analysis of individual strains. All authors have read and approved the final manuscript.
Acknowledgments
Acknowledgements
We would like to thank Irina Mutiu, Megan Davey and David Edgell for comments on this manuscript, David Litchfield for use of microscope facilities, Julie Genereaux for technical assistance and Esther Loney for constructing the GFP molecule. This work was supported by a Canadian Institutes of Health Research grant to CB. SH is supported by Schulich Graduate and Ontario Graduate Scholarships.
Contributor Information
Stephen MT Hoke, Email: shoke@uwo.ca.
Julie Guzzo, Email: julie.guzzo@utoronto.ca.
Brenda Andrews, Email: brenda.andrews@utoronto.ca.
Christopher J Brandl, Email: cbrandl@uwo.ca.
References
- Bosotti R, Isacchi A, Sonnhammer EL. FAT: a novel domain in PIK-related kinases. Trends Biochem Sci. 2000;25:225–227. doi: 10.1016/S0968-0004(00)01563-2. [DOI] [PubMed] [Google Scholar]
- Saleh A, Schieltz D, Ting N, McMahon SB, Litchfield DW, Yates JR, 3rd, Lees-Miller SP, Cole MD, Brandl CJ. Tra1 is a component of the yeast Ada-Spt transcriptional regulatory complexes. J Biol Chem. 1998;273:26559–26565. doi: 10.1074/jbc.273.41.26559. [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Yates JR, 3rd, Workman JL. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 1998;2:863–867. doi: 10.1016/S1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, JWorkman JL, Côté J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishburn J, Mohibullah N, Hahn S. Function of a eukaryotic transcription activator during the transcription cycle. Mol Cell. 2005;18:369–378. doi: 10.1016/j.molcel.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Reeves WM, Hahn S. Targets of the Gal4 transcription activator in functional transcription complexes. Mol Cell Biol. 2005;25:9092–9102. doi: 10.1128/MCB.25.20.9092-9102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutiu AI, Hoke SMT, Genereaux J, Hannam C, MacKenzie , Jobin-Robitaille O, Guzzo J, Côté J, Andrews B, Haniford DB, Brandl CJ. Structure/function analysis of the phosphatidylinositol-3-kinase domain of yeast Tra1. Genetics. 2007;177:151–166. doi: 10.1534/genetics.107.074476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Krogan N, Li Z, Levinson JN, Lu H, Menard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burg CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Measday V, Baetz K, Guzzo J, Yuen K, Kwok T, Sheikh B, Ding H, Ueta R, Hoac T, Cheng B, Pot I, Tong A, Yamaguchi-Iwai Y, Boone C, Hieter P, Andrews B. Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc Natl Acad Sci USA. 2005;102:13956–13961. doi: 10.1073/pnas.0503504102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hon GC, Myers CL, Parsons A, Friesen H, Oughtred R, Tong A, Stark C, Ho Y, Botstein D, Andrews B, Boone C, Troyanskya OG, Ideker T, Dolinski K, Batada NN, Tyers M. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;10:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Mitchell L, Lambert JP, Gerdes M, Al-Madhoun AS, Skerjanc IS, Figeys D, Baetz K. Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 is essential for complex integrity. Mol Cell Biol. 2008;7:2244–2256. doi: 10.1128/MCB.01653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N. Ypt and Rab GTPases: insight into functions through novel interactions. Curr Opin Cell Biol. 2001;13:500–511. doi: 10.1016/S0955-0674(00)00242-8. [DOI] [PubMed] [Google Scholar]
- Mizuta K, Park JS, Sugiyama M, Nishiyama M, Warner JR. RIC1, a novel gene required for ribosome synthesis in Saccharomyces cerevisiae. Gene. 1997;187:171–178. doi: 10.1016/S0378-1119(96)00740-8. [DOI] [PubMed] [Google Scholar]
- Baetz K, Moffat J, Haynes J, Chang M, Andrews B. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol Cell Biol. 2001;21:6515–6528. doi: 10.1128/MCB.21.19.6515-6528.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesono Y, Toh-e A. Transient inhibition of translation initiation by osmotic stress. J Biol Chem. 2001;277:13848–13855. doi: 10.1074/jbc.M108848200. [DOI] [PubMed] [Google Scholar]
- Lenssen E, Azzouz N, Michel A, Landrieux E, Collart MA. The Ccr4-Not Complex Regulates Skn7 through Srb10 Kinase. Eukaryot Cell. 2007;6:2251–2259. doi: 10.1128/EC.00327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand JD, Grant CM. The thioredoxin system protects ribosomes against stress-induced aggregation. Mol Biol Cell. 2006;17:387–401. doi: 10.1091/mbc.E05-06-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson C, Ljungdahl PO. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 2002;16:3158–3172. doi: 10.1101/gad.239202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Ikeda E, Uno I, Mitsuzawa H. Characterization of a staurosporine- and temperature-sensitive mutant, stt1, of Saccharomyces cerevisiae. Mol Gen Genet. 1992;231:337–344. doi: 10.1007/BF00292700. [DOI] [PubMed] [Google Scholar]
- Helliwell SB, Howald I, Barbet N, Hall MN. TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics. 1998;148:99–112. doi: 10.1093/genetics/148.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno VM, Kalachikov S, Subaran R, Nobile CJ, Kyratsous C, Mitchell AP. Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog. 2006;2:e21. doi: 10.1371/journal.ppat.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 13:573–585. doi: 10.1016/S1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Lindstrom KC, Vary JC, Jr, Parthun MR, Delrow J, Tsukiyama T. Isw1 functions in parallel with the NuA4 and Swr1 complexes in stress-induced gene repression. Mol Cell Biol. 2006;26:6117–6129. doi: 10.1128/MCB.00642-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Lang V, Cook R, Brandl CJ. Identification of native complexes containing the yeast coactivator/repressor proteins Ngg1/Ada3 and Ada2. J Biol Chem. 1997;272:5571–5578. doi: 10.1074/jbc.272.9.5571. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/S0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger SL, Hieter P, Zhang Z, Brown GW, Ingles CJ, Emili A, Allis CD, Toczyski DP, Weissman JS, Greenblatt JF, Krogan NJ. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Welihinda AA, Tirasophon W, Green SR, Kaufman RJ. Gene induction in response to unfolded protein in the endoplasmic reticulum is mediated through Ire1p kinase interaction with a transcriptional coactivator complex containing Ada5p. Proc Natl Acad Sci USA. 1997;94:4289–4294. doi: 10.1073/pnas.94.9.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welihinda AA, Tirasophon W, Kaufman RJ. The transcriptional co-activator ADA5 is required for HAC1 mRNA processing in vivo. J Biol Chem. 2000;275:3377–3381. doi: 10.1074/jbc.275.5.3377. [DOI] [PubMed] [Google Scholar]
- Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K, Matsuyama A, Kobayashi Y, Iwahashi H. The genome-wide screening of yeast deletion mutants to identify the genes required for tolerance to ethanol and other alcohols. FEMS Yeast Res. 2006;6:744–750. doi: 10.1111/j.1567-1364.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- Tang F, Peng Y, Nau JJ, Kauffman EJ, Weisman LS. Vac8p, an armadillo repeat protein, coordinates vacuole inheritance with multiple vacuolar processes. Traffic. 2006;7:1368–1377. doi: 10.1111/j.1600-0854.2006.00458.x. [DOI] [PubMed] [Google Scholar]
- Hancock LC, Behta RP, Lopes JM. Genomic analysis of the Opi- phenotype. Genetics. 2006;173:621–634. doi: 10.1534/genetics.106.057489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, Workman JL, Grant PA. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol. 2002;22:8774–8786. doi: 10.1128/MCB.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski SM. The retrograde response links metabolism with stress responses, chromatin-dependent gene activation, and genome stability in yeast aging. Gene. 2005;34:22–27. doi: 10.1016/j.gene.2005.03.040. [DOI] [PubMed] [Google Scholar]
- Cutler NS, Pan X, Heitman J, Cardenas ME. The Tor signal transduction cascade controls cellular differentiation in response to nutrients. Mol Cell Biol. 2001;12:4103–4113. doi: 10.1091/mbc.12.12.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Uesono Y, Oguchi T, Toh-e A. LAS24/KOG1, a component of the TOR complex 1 (TORC1), is needed for resistance to local anesthetic tetracaine and normal distribution of actin cytoskeleton in yeast. Genes Genet Syst. 2005;80:325–343. doi: 10.1266/ggs.80.325. [DOI] [PubMed] [Google Scholar]
- Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Aronova S, Wedaman K, Anderson S, Yates J, III, Powers T. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2779–2794. doi: 10.1091/mbc.E07-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci USA. 2004;101:9515–9516. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast E. coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1998;74:255–269. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Brandl CJ, Furlanetto AM, Martens JA, Hamilton KS. Characterization of NGG1, a novel yeast gene required for glucose repression of GAL4p-regulated transcription. EMBO J. 1993;12:5255–5265. doi: 10.1002/j.1460-2075.1993.tb06221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachat A, Liebundguth N, Rebischung C, Lemire S, Schärer F, Hoepfner D, Demchyshyn V, Howald I, Düsterhöft A, Möstl D, Pöhlmann R, Kötter P, Hall MN, Wach A, Philippsen P. Analysis of deletion phenotypes and GFP fusions of 21 novel Saccharomyces cerevisiae open reading frames. Yeast. 2000;3:241–253. doi: 10.1002/(SICI)1097-0061(200002)16:3<241::AID-YEA517>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–148. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]