Abstract
Inhibition of DNA replication and physical DNA damage induce checkpoint responses that arrest cell cycle progression at two different stages. In Saccharomyces cerevisiae, the execution of both checkpoint responses requires the Mec1 and Rad53 proteins. This observation led to the suggestion that these checkpoint responses are mediated through a common signal transduction pathway. However, because the checkpoint-induced arrests occur at different cell cycle stages, the downstream effectors mediating these arrests are likely to be distinct. We have previously shown that the S. cerevisiae protein Pds1p is an anaphase inhibitor and is essential for cell cycle arrest in mitosis in the presence DNA damage. Herein we show that DNA damage, but not inhibition of DNA replication, induces the phosphorylation of Pds1p. Analyses of Pds1p phosphorylation in different checkpoint mutants reveal that in the presence of DNA damage, Pds1p is phosphorylated in a Mec1p- and Rad9p-dependent but Rad53p-independent manner. Our data place Pds1p and Rad53p on parallel branches of the DNA damage checkpoint pathway. We suggest that Pds1p is a downstream target of the DNA damage checkpoint pathway and that it is involved in implementing the DNA damage checkpoint arrest specifically in mitosis.
In response to certain types of impairments to normal DNA metabolism, the activity of surveillance mechanisms known as checkpoints is induced (1, 2). As a result, cell cycle progression is halted at specific points of the cell cycle, depending on the type of perturbation that initiated the checkpoint signal. The checkpoint arrest prevents the potentially lethal consequences of cell cycle progression in the presence of abnormal DNA structures (for review, see ref. 3).
In Saccharomyces cerevisiae, as well as in other organisms, the types of impairments to DNA metabolism that lead to a checkpoint arrest can be roughly classified into two major categories, DNA damage and inhibition of DNA replication. In both cases, the checkpoint arrest inhibits mitosis from taking place in the presence of damaged or unreplicated DNA. The arresting points, however, differ. In the budding yeast, inhibition of DNA replication results in an S phase arrest, whereas DNA damage leads to a mitotic arrest and G1 and S phase delays (4–7). The induction of these checkpoint signals requires two different sets of proteins: Polɛ, Rfc5p, and Dpb11p for generating the inhibition of DNA replication signal and Rad9p, Rad17p, Rad24p, and Mec3p for generating the DNA damage signal (for review, see ref. 2).
Unlike the generation of the checkpoint signals, the propagation of these signals in both the DNA damage and the replication checkpoint pathways requires the activity of the same proteins: Mec1p and Rad53p. Mec1p belongs to a superfamily of lipid kinases that includes the protein defective in Ataxia telangiectasia patients, ATM (for review, see refs. 8 and 9). The Schizosaccharomyces pombe Mec1p homolog Rad3p was also shown to be required for both the replication and the DNA damage checkpoints (10, 11). Rad3p and the ATM protein were shown to have protein kinase activity (12–14), suggesting that Mec1p may also be a protein kinase. Rad53p is a protein kinase that is required for the transcriptional activation of DNA-damage-inducible genes, although this may not be its only function (15). In the presence of DNA damage, or when DNA replication is inhibited, Rad53p is phosphorylated in a Mec1p-dependent manner, suggesting that Rad53p is downstream of Mec1p (16, 17). Whether Rad53p is a direct substrate of Mec1p is currently unknown. However, these and other observations prompted the idea that in S. cerevisiae different types of abnormal DNA structures induce checkpoint signals that merge into a common pathway in which the signal is transduced via the Mec1p-dependent phosphorylation of Rad53p (2, 16, 17).
Although the DNA replication and DNA damage checkpoint pathways appear to use a common signal transduction pathway, these checkpoint pathways result in arrest at different stages of the cell cycle, suggesting that these pathways mediate arrest by using distinct downstream effectors. We have recently demonstrated that in budding yeast, Pds1p is a mitotic inhibitor whose inactivation via protein degradation is required for anaphase initiation (18, 19). Unlike wild-type cells, γ-irradiation of pds1 mutant cells does not result in the inhibition of mitosis, as evidenced by sister chromatid separation, spindle elongation, and cytokinesis (18). In contrast, treatment of pds1 mutant cells with replication inhibitors leads to a complete inhibition of mitosis (ref. 18 and unpublished results). These observations suggest that the mechanisms that govern the inhibition of mitosis in the presence of replication inhibitors or DNA damage are distinct. Moreover, these findings suggest that Pds1p is involved in mitotic regulation not only under normal growth conditions but also as part of the DNA damage checkpoint pathway. Herein, we establish the position of Pds1p in the DNA damage checkpoint pathway and suggest an alternative model for the pathways by which the checkpoint signals are transduced.
MATERIALS AND METHODS
Plasmids and Strains.
Strains used were as follows: OCF1522 Mata bar1 PDS1-HA∷URA3 leu2 trp1 his3 ade2; OCF1523, like OCF1522 but mec1–1; OCF1524, like OCF1522 but rad53–21; OCF1525, like OCF1522 but pds1Δ∷LEU2, ura3; 1544, Mata rad9∷LEU2 PDS1-HA∷URA3 trp1 his7. Plasmids used were as follows: pRS313 and pRS314 are as described (20), pOC75 contains the MEC1 gene (on an ApaI–SacI fragment) inserted into the ApaI and SacI sites of pRS313, and pOC81 contains a SalI–BamHI fragment that includes the RAD9 gene.
Media and Reagents.
YEPD medium contained 1% yeast extract, 2% bactopeptone, adenine (2.5 mg/liter), and 2% glucose. Hydroxyurea (HU), α mating pheromone, Pronase, and β-glycerophosphate were from Sigma. Nocodazole was from Aldrich. Calf intestine alkaline phosphatase was from Boehringer Mannheim.
Western Blot Analysis.
Protein extract preparation, SDS/PAGE, and Western blot analysis were carried out as described (19), except that the acrylamide/bisacrylamide ratio in the SDS/PAGE gels used was 30:0.2. This was done to improve the separation between the DNA damage induced and noninduced forms of Pds1p-HA (where HA is hemagglutinin).
Irradiation.
Cell were grown to logarithmic phase in YEPD and treated with nocodazole (final concentration, 15 μg/ml), α mating factor (final concentration, 10−7 M), or HU (final concentration, 0.1 M). In all cases, cells appeared morphologically to be arrested after 2 h when grown at 30°C or 3.5 h when grown at 23°C. After cell cycle arrest, the cells were concentrated, plated on YEPD plates, and irradiated. When UV-irradiating, the cell density was about 2 × 107 cells per 100-mm plate. When γ-irradiating, the cell density was about 1 × 108 cells per 60-mm plate. After irradiation, the cells were resuspended in YEPD to their original cell density. When cells were released form a G1 phase arrest, they were washed by centrifugation at least three times in YEPD containing Pronase (0.1 mg/ml) and then resuspended in YEPD with Pronase. Irradiation was performed by using a UV Stratalinker (Stratagene; wavelength = 254 nm) or a cesium source at 220 rad/min (1 rad = 0.1 Gy).
Phosphatase Treatment.
UV-irradiation of nocodazole-arrested cells was carried out as described above. Protein extracts were prepared as followed: Cells were concentrated 50-fold and resuspended in 50 mM Tris⋅HCl, pH 7.4/100 mM NaCl/2 mM EDTA/1% SDS/1 mM phenylmethylsulfonyl fluoride. Extracts were prepared by bead breaking and boiling, after which the lysed cells were centrifuged, and the clarified protein extract was collected. The protein concentration was approximately 1 mg/ml. The 200-μl phosphatase reactions included 50 μl of protein extract, 35 mM Tris⋅HCl (pH 8.0), 6 mM MgCl2, calf intestine alkaline phosphatase (0, 40, or 80 units), β-glycerophosphate (0 or 200 mM), and 1 mM phenylmethylsulfonyl fluoride. Reaction were carried out for 30 min at 37°C and were terminated by adding 30 μl of ice-cold 100% trichloroacetic acid. The proteins were pelleted by centrifugation, resuspended in sample buffer (50 mM Tris⋅HCl, pH 6.8/2% 2-mercaptoethanol/2% SDS/0.01% bromophenol blue/10% glycerol), and used in Western blot analysis as described above.
Densitometry Scans.
Autoradiographs were scanned by using a Molecular Dynamics computing densitometer. Graphs were created by using Molecular Dynamics imagequant, version 1.11.
RESULTS
Pds1p Is Phosphorylated in the Presence of DNA Damage but Not When DNA Replication Is Inhibited.
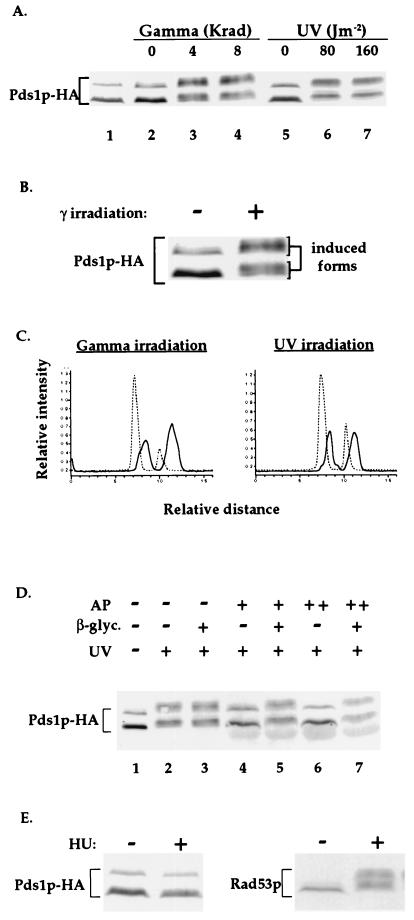
We have previously shown that Pds1p is a nuclear protein that is present in the cell from late G1 or early S phases to the time of anaphase initiation (19). Western blot analysis of a functional HA-tagged derivative of Pds1p (Pds1p-HA; ref. 19) from cycling cells revealed that the protein appeared as a doublet, with a minor form migrating slower than the major form (Fig. 1A, lane 1). To examine the effect of DNA damage on the mobility of Pds1p-HA, wild-type cells were arrested in mitosis with the drug nocodazole and exposed to either γ- or UV-irradiation (Fig. 1A). The nocodazole treatment itself did not alter Pds1p-HA’s mobility (Fig. 1A, compare lanes 1 with lanes 2 and 5), indicating that a mitotic arrest per se does not result in an apparent modification of Pds1-HA. However, exposure of the nocodazole-arrested cells to either UV- or γ-radiation resulted in a change in mobility and relative distribution of the various Pds1p-HA forms (Figs. 1 A–C). It is presently unknown what is the exact precursor–product relationship between the noninduced and the DNA-damage-induced Pds1p forms.
Figure 1.
Pds1p-HA is phosphorylated in the presence of DNA damage but not when DNA replication is inhibited. (A) Wild-type OCF1522 cells grown at 30°C were arrested in mitosis with nocodazole (lanes 2–7) and exposed to either UV- or γ-radiation at the indicated doses. Protein samples were prepared 20 min after irradiation and processed for Western blot analysis. The sample from the cycling cells (lane 1) was prepared from cells of the same culture taken prior to the addition of nocodazole. The protein bands corresponding to Pds1p-HA are indicated. Occasionally, a fast-migrating HA-cross-reacting band appears (data not shown). (B) An enlargement of the Pds1-HA bands from lanes 2 and 3 of A. (C) Densitometry scan of lanes 2, 4 (Left), 5, and 7 (Right) of A. The dashed and solid lines are of Pds1p-HA from nonirradiated and irradiated cells, respectively. Scans were preformed from bottom to top. (D) Protein extracts were prepared from nocodazole-arrested wild-type OCF1522 cells that were UV-irradiated at 80 J/m2 as described above. Reactions were carried out in the absence (−) or presence of calf intestine alkaline phosphatase (AP; +, 40 units per reaction; ++, 80 units per reaction) and in the absence (−) or presence (+) of the alkaline phosphatase inhibitor β-glycerophosphate (β-glyc.). (E) Wild-type OCF1522 cells grown at 30°C were not treated (−) or treated with 0.1 M HU for 2 h, after which protein extracts were prepared and examined by Western blot analysis for Pds1p-HA (Left) and Rad53p (Right). Identical results were obtained when cells were arrested with 0.2 M HU (data not shown). The S phase arrest after the HU treatment was verified by flow cytometry analysis (data not shown).
Treatment of protein extracts from UV-irradiated cells with alkaline phosphatase resulted in the reversion of the damage-induced modified Pds1p-HA forms to the forms seen in the absence of induced DNA damage (Fig. 1D). This was also observed when extracts were prepared from γ-irradiated cells (data not shown). This reversion was prevented when an inhibitor of alkaline phosphatase was present (Fig. 1D), demonstrating that the DNA damage induced modification of Pds1p-HA was due to phosphorylation.
No change in Pds1p-HA mobility was observed when cells were treated with HU (Fig. 1E Left), a DNA replication inhibitor that induces an S phase DNA replication checkpoint arrest. As has been observed (17), this treatment does lead to Rad53p phosphorylation (Fig. 1E Right). Thus, Pds1p-HA is specifically phosphorylated in response to types of DNA damage that elicit a mitotic checkpoint arrest but not in response to reagents that cause a DNA replication checkpoint arrest.
The DNA Damage-Dependent Phosphorylation of Pds1p Requires Mec1p and Rad9p.
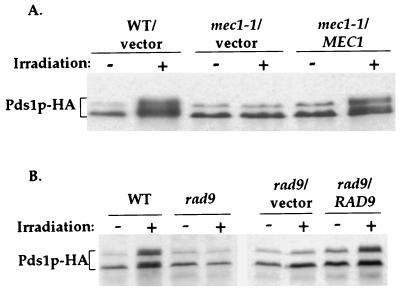
The phosphorylation of Pds1p in the presence of DNA damage raised the possibility that Pds1p is phosphorylated via the DNA damage checkpoint pathway. Therefore, strains that are defective in components of this pathway may fail to promote the DNA damage-dependent phosphorylation of Pds1p-HA. This was tested in haploid mec1, rad9, and rad53 mutant strains. The comparison of wild-type and checkpoint-defective mutant strains is complicated by the fact that checkpoint mutants do not arrest cell cycle progression in the presence of DNA damage and they may have a diminished DNA repair capability (21), resulting in the accumulation of more lesions per cells compared with wild-type strains. To minimize the contribution of these differences, cells were arrested in G1 phase with the α mating pheromone, irradiated, and then released into medium containing nocodazole. With this approach, the DNA damage induced in G1 phase was largely irreparable in both wild-type and mutant strains due to the absence of sister chromatids or homologous chromosomes that are used as undamaged templates for recombinational repair (22–24). In addition, the release into nocodazole ensured that all strains were at the same stage of the cell cycle when the phosphorylation of Pds1p was assayed. As can be seen in Fig. 2, mec1 and rad9 mutant strains were unable to promote Pds1p-HA phosphorylation after γ-irradiation, and their ability to do so was restored by introducing a copy of the respective wild-type gene. These results indicate that the DNA-damage-dependent phosphorylation of Pds1p was mediated by the DNA damage checkpoint machinery and that Pds1p is downstream of both Rad9p and Mec1p.
Figure 2.
DNA-damage-dependent phosphorylation of Pds1p-HA is Mec1p- and Rad9p-dependent. (A) Wild-type (OCF1522), mec1–1 (OCF1523), and mec1–1 cells carrying a centromere-based plasmid (pRS313, indicated as vector) or a pRS313 derivative carrying a copy of the wild-type MEC1 gene (pOC75, indicated as MEC1) were grown at 30°C and were arrested in G1 phase with the α mating pheromone. When more than 85% of the cells appeared arrested morphologically (shmooed), the cultures were either left untreated (−) or exposed to γ-radiation (+, 4 krad). After the irradiation, the cultures were released into medium containing Pronase and nocodazole. Samples for protein extracts were taken 2 h after the release from G1 phase and analyzed by Western blot analysis. At the time when the sample were taken, approximately 90% of the cells in all cultures had a mitotic arrest phenotype (large budded cell with a single nucleus) as determined by 4′,6-diamidino-2-phenylindole (DAPI) staining. (B) Wild-type cells (OCF1522), rad9 cells (OCF1544), and rad9 cells carrying a centromere-based plasmid (pRS314, indicated as vector) or a pRS314-derived plasmid containing a copy of the RAD9 wild-type gene (pOC81, indicated as RAD9) were treated as described in A.
Pds1p and Rad53p Are on Parallel Branches of the DNA Damage Checkpoint Pathway.
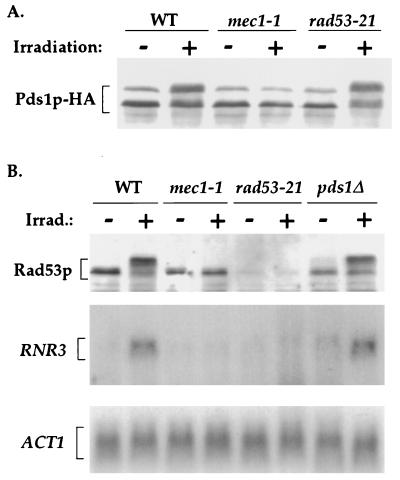
The DNA-damage-dependent phosphorylation of Pds1p did not require Rad53p (Fig. 3A). This was also true when a rad53Δ strain were used (data not shown). Because Rad53p is known to be required for the mitotic DNA damage checkpoint arrest (15, 25), its dispensability in the DNA-damage-dependent phosphorylation of Pds1p could be explained in one of two ways, (i) Pds1p is upstream of Rad53p in the DNA damage checkpoint pathway or (ii) Pds1p and Rad53p are on parallel branches of the pathway. To determine which of these possibilities is correct, we examined whether the DNA-damage-dependent transcriptional activation of RNR3 (which is Rad53p-dependent, ref. 15) and the DNA-damage-dependent phosphorylation of Rad53p (16, 17) occurred in the absence of Pds1p. If Pds1p is upstream of Rad53p, one would expect that the deletion of PDS1 (pds1Δ) would abrogate Rad53p-dependent processes. If, on the other hand, Pds1p and Rad53p are on parallel branches, pds1Δ would not affect Rad53p-dependent processes. As can be seen in Fig. 3B, both the damage-dependent phosphorylation of Rad53p and the transcriptional activation of RNR3 were independent of Pds1p, suggesting that Pds1p and Rad53p are on parallel branches of the DNA damage checkpoint pathway, both of which are controlled by Mec1p.
Figure 3.
DNA-damage-dependent phosphorylation of Pds1p is independent of Rad53p function. (A) Wild-type (OCF1522), mec1–1 (OCF1523), and rad53–21 (OCF1524) strains were arrested in G1 phase, irradiated, and released into medium containing nocodazole as described in Fig. 2A. (B) Wild-type (OCF1522), mec1–1 (OCF1523), rad53–21 (OCF1524), and pds1Δ (OCF1525) strains were arrested in G1 phase, irradiated, and released into medium containing nocodazole as described in Fig. 2A, except that samples were taken for both Western blot analysis (Top) and Northern blot analysis (Middle and Bottom), which were carried out as described (19).
The DNA-Damage-Dependent Phosphorylation of Pds1p Can Occur in S Phase-Arrested Cells.
As mentioned above, the S phase checkpoint arrest caused by inhibition of DNA replication and the mitotic checkpoint arrest caused by DNA damage both require the activities of the Mec1 and Rad53 proteins. This led to the notion that in both cases a common signal transduction pathway is being used. Indeed, Rad53p is phosphorylated in a Mec1p-dependent manner in the presence of DNA damage and in the presence of DNA replication inhibitors, albeit to a different extent (ref. 17 and unpublished observation). However, Pds1p is only phosphorylated in the presence of DNA damage but not when DNA replication is inhibited by HU (Fig. 1D). Formally, this differential phosphorylation of Rad53p and Pds1p could have resulted from the requirement for a cell cycle-specific factor in the phosphorylation of Pds1p but not Rad53p. In this case, a hypothetical factor would be absent is S phase but present in M phase, resulting in the phosphorylation of only Rad53p in the S phase checkpoint arrest and in the phosphorylation of both Rad53p and Pds1p in the mitotic checkpoint arrest.
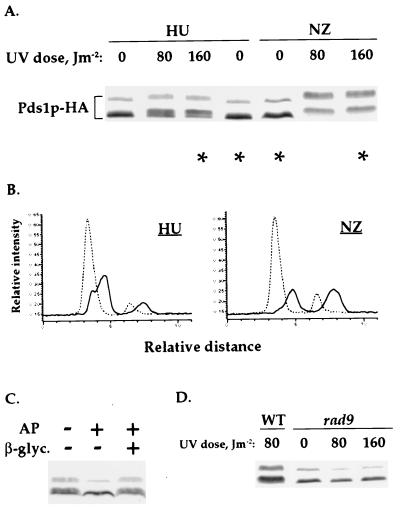
To test this possibility, we examined whether Pds1p-HA was phosphorylated in UV-irradiated cells that were arrested in S phase by HU. As a control we UV-irradiated cells arrested in mitosis by nocodazole. The arrests in S phase and M phase were verified by flow cytometry analysis (data not shown). As has been described above for cells arrested in mitosis, the UV-irradiation of S phase-arrested cells also lead to Pds1p-HA phosphorylation (Fig. 4 A–C). The S phase phosphorylation was dependent on Rad9p function (Fig. 4D), suggesting that it occurred via the DNA damage checkpoint pathway. These results suggest that the ability to phosphorylate Pds1p via the DNA damage checkpoint pathway is not restricted to mitosis.
Figure 4.
Pds1p-HA is phosphorylated after the UV-irradiation of S phase-arrested cells. (A) Wild-type (OCF1522) cells were arrested in S phase with HU (0.1 M) or in mitosis with nocodazole (NZ, 15 μg/ml) and UV-irradiated at the indicated doses. Twenty minutes after irradiation, the cells were harvested and analyzed by Western blot analysis for Pds1p-HA. (B) Densitometry scans of the lanes indicated by asterisks in A: HU-treated cells, 0 and 160 J/m2; NZ-treated cells, 0 and 160 J/m2. Scans were done as described in Fig. 1B. The dashed and solid lines are of Pds1p-HA from nonirradiated and irradiated cells, respectively. (C) Wild-type (OCF1522) cells were arrested in S phase with HU (0.2 M) and UV-irradiated at 160 J/m2. The irradiated cells were harvested 20 min after irradiation and processed for phosphatase treatment. The phosphatase reaction were carried out in the presence (+) or absence (−) of 40 units of calf intestine alkaline phosphatase (AP) and 0.2 M β-glycerophosphate. (D) Wild-type (OCF1522) and rad9 (OCF1544) cells were arrested in S phase with HU (0.1 M) and UV-irradiated at the indicated UV doses as described in A.
Densitometry scanning of Pds1p-HA from irradiated and nonirradiated cells (Fig. 4A, lanes indicated by asterisks) revealed that the relative distribution of the UV-induced phosphorylated forms of Pds1p differed in cells that were irradiated in S phase vs. mitosis (Fig. 4B). The reason for this difference is currently unknown. It should be noted that under these experimental conditions, Mec1p is involved in transducing checkpoint signals that result from both replication arrest and DNA damage, and this may affect the efficiency of Pds1p phosphorylation. Alternatively, Pds1p may be a better substrate for modification during M phase, when the DNA is fully replicated.
DISCUSSION
DNA damage induced by UV- or γ-irradiation led to the phosphorylation of Pds1p. This phosphorylation was Mec1p- and Rad9p-dependent. Thus, with the earlier finding that cells lacking Pds1p fail to arrest in mitosis in response to DNA damage (18), these results strongly suggest that Pds1p is a downstream target of the DNA damage checkpoint pathway.
The DNA-damage-dependent phosphorylation of Pds1p was Rad53p-independent, as has also been observed for Rpa2 (26). Because Rad53p-related processes took place in the absence of Pds1p, we propose that Pds1p and Rad53p are on parallel branches of the DNA damage checkpoint pathway, both of which are controlled by Mec1p. At this point, however, we cannot rule out the possibility that Pds1p plays an additional role downstream of Rad53p. Whether or not Rad53p and Pds1p are directly modified by Mec1p is yet to be determined. Interestingly, pds1 and rad53 mutant strains are both less sensitive to UV- and γ-radiation than mec1 mutant strains (ref. 17 and unpublished observation). Furthermore, in the presence of DNA damage, pds1 and rad53 mutants slightly delay in mitosis, but the double pds1 rad53 mutant doesn’t (R. Gardner and T. Weinert, personal communication). Therefore, we propose that the Pds1p and Rad53p branches of the checkpoint pathway are not functionally redundant but that a complete mitotic cell cycle arrest in response to DNA damage requires the Mec1p-regulated activity of both branches of the pathway.
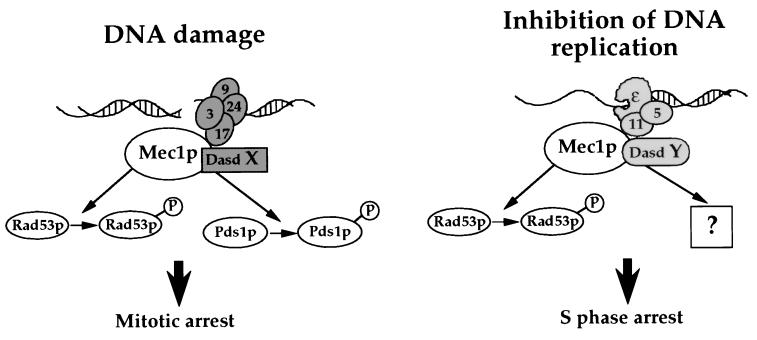
Mec1p and Rad53p are involved in transducing both the DNA replication and the DNA damage checkpoint signals. A common signal transduction pathway could have resulted in the loss of identity of the DNA alteration from which the checkpoint signal had originated. However, Pds1p is phosphorylated in the presence of DNA damage but not when DNA replication is inhibited by HU. In addition, Pds1p phosphorylation can be induced in HU-arrested cells if they are exposed to DNA damaging agents. These results suggest that the identity of the initiating signal is maintained through the signal transduction pathway. Therefore, Mec1p can apparently discriminate between incoming signals that originate from different types of DNA alterations and activate only those downstream effectors that are required for the appropriate checkpoint response. Thus, we propose that the link between the type of DNA alteration and the activation of specific checkpoint effectors is mediated through DNA-alteration-specific determinants (Dasds) that recognize specific types of altered DNA structures caused by DNA damaging agents or inhibitors of DNA replication (Fig. 5). These Dasds interact with Mec1p and/or its downstream targets, thereby determining which effectors will be activated. It is conceivable that the Dasd function is carried out by proteins that are already known to be involved in damage recognition and/or processing. Because Mec1p may associate with different factors when participating in either the DNA damage checkpoint or the replication checkpoint, we propose that these checkpoint responses do not share a common signal transduction pathway. Rather, they are mediated by two distinct pathways that use common components. We predict that it should be possible to isolate mec1 mutants that are defective in only one of the two checkpoint pathways. This is consistent with observations in S. pombe, in which certain mutant forms of proteins that are required for both checkpoint responses are defective in either the replication or the DNA damage checkpoint (27–29).
Figure 5.
Model for the signal transduction pathways of the DNA damage and replication checkpoint. DNA damage is represented by a double-strand break. In the presence of DNA damage, Mec1 and/or its downstream target associate with DNA-alteration-specific determinant X (Dasd X), leading to the phosphorylation of Pds1p and Rad53p. Inhibition of DNA replication activates a different Dasd (Dasd Y) that will not promote the phosphorylation of Pds1p but perhaps the phosphorylation of an S phase-specific mitotic inhibitor. The proximity of different proteins indicates their possible localization relative to the DNA and does not imply physical interactions between the proteins. Symbols are as follows: 9, Rad9p; 17, Rad17p; 24, Rad24p; 3, Mec3p; ɛ, polɛ; 5, Rfc5p; 11, Dpb11p.
The existence of specific effectors that mediate specific cell cycle checkpoint arrests has been suggested previously for the fission yeast S. pombe (30, 31). Unlike in S. cerevisiae, in which DNA damage appears to cause a mitotic arrest (as indicated by the high Clb2p-associated Cdc28 kinase activity; ref. 5), in S. pombe, as well as in most other organisms for which a DNA damage checkpoint has been demonstrated, the DNA damage checkpoint arrest occurs in G2 phase. Although the arrest point differs, the signal transduction pathway involves damage-dependent phosphorylation that is mediated by a conserved element (i.e., Mec1p in S. cerevisiae, Rad3p in S. pombe, and perhaps ATM in human cells). Moreover, both S. cerevisiae and S. pombe appear to use the same strategy in which the DNA replication and DNA damage checkpoints are mediated through a shared component. Therefore, the determinants that allow the specific activation of downstream effectors may also be conserved.
Although Pds1p is clearly an essential component of the DNA damage checkpoint pathway (18), it is still unknown whether its phosphorylation is required for the checkpoint arrest. The DNA-damage-dependent phosphorylation of Pds1p in S phase-arrested cells suggests that this modification is not a consequence of the mitotic checkpoint arrest but could be involved in implementing it. Because under normal conditions Pds1p must be degraded for cells to initiate anaphase (19), an attractive model would be that the DNA-damage-dependent phosphorylation of Pds1p renders it resistant to proteolysis, thereby preventing progression through mitosis. Alternatively, phosphorylated Pds1p may serve as a reporter that indicates to the cell that its DNA had been damaged and that it must induce an appropriate response. Future studies should determine the role of phosphorylation in the checkpoint function of Pds1p.
Acknowledgments
We are grateful to Ted Weinert for strains, plasmids, and communicating results before publication and to Steve Elledge and Yolanda Sanchez for anti-Rad53 antibodies. We also thank Ted Weinert, Steve Elledge, Paul Megee, Shikha Laloraya, and Kathleen Wilsbach for commenting on this manuscript. O.C.F. is supported by a National Institutes of Health grant (GM18382–02). D.K. is supported by a National Institutes of Health grant (GM1718) and is an Investigator with the Howard Hughes Medical Institute.
ABBREVIATIONS
- HU
hydroxyurea
- Dasd
DNA-alteration-specific determinant
References
- 1.Hartwell L H, Weinert T A. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 2.Elledge S. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 3.Paulovich A, Toczyski D, Hartwell L. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 4.Weinert T, Hartwell L. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 5.Amon S, Surana U, Muroff I, Nasmyth K. Nature (London) 1992;355:368–371. doi: 10.1038/355368a0. [DOI] [PubMed] [Google Scholar]
- 6.Siede W, Friedberg A S, Friedberg E C. Proc Natl Acad Sci USA. 1993;90:7985–7989. doi: 10.1073/pnas.90.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Deshaies R. Cell. 1993;74:223–226. doi: 10.1016/0092-8674(93)90413-k. [DOI] [PubMed] [Google Scholar]
- 8.Zakian V. Cell. 1995;82:685–687. doi: 10.1016/0092-8674(95)90463-8. [DOI] [PubMed] [Google Scholar]
- 9.Carr A. Curr Opin Genet Dev. 1997;7:93–98. doi: 10.1016/s0959-437x(97)80115-3. [DOI] [PubMed] [Google Scholar]
- 10.al-Khodairy F, Carr A. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jimenez G, Yucel J, Rowley R, Subramani S. Proc Natl Acad Sci USA. 1992;89:4952–4956. doi: 10.1073/pnas.89.11.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentley N, Holtzman D, Flaggs G, Keegan K, DeMaggio A, Ford J, Hoekstra M, Carr A. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- 13.Keegan K S, Holtzman D A, Plug A W, Christenson E R, Brainerd E E, Flaggs G, Bentley N J, Taylor E M, Meyn M S, Moss S B, Carr A M, Ashley T, Hoekstra M F. Genes Dev. 1996;10:2423–2437. doi: 10.1101/gad.10.19.2423. [DOI] [PubMed] [Google Scholar]
- 14.Jung M, Kondratyev A, Lee S, Dimtchev A, Dritschilo A. Cancer Res. 1997;57:24–27. [PubMed] [Google Scholar]
- 15.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. Genes Dev. 1994;8:2416–2428. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 16.Sun Z, Fay D S, Marini F, Foiani M, Stern D F. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto A, Guacci V, Koshland D. J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen-Fix O, Peters J, Kirschner M, Koshland D. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 20.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aboussekhra A, Vialard J, Morrison D, de la Torre-Ruiz M A, Cernakova L, Fabre F, Lowndes N. EMBO J. 1996;15:3912–3922. [PMC free article] [PubMed] [Google Scholar]
- 22.Brunborg G, Resnick M, Williamson D. Radiat Res. 1980;82:547–558. [PubMed] [Google Scholar]
- 23.Resnick M. J Theor Biol. 1976;59:97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- 24.Game J C. In: Yeast Genetics: Fundamental and Applied Aspects. Spencer J F T, Spencer D M, Smith A R W, editors. New York: Springer; 1983. pp. 109–137. [Google Scholar]
- 25.Weinert T A, Kiser G L, Hartwell L H. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 26.Brush G, Morrow D, Hieter P, Kelly T. Proc Natl Acad Sci USA. 1996;93:15075–15080. doi: 10.1073/pnas.93.26.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanter-Smoler G, Knudsen K, Jimenez G, Sunnerhagen P, Subramani S. Mol Biol Cell. 1995;6:1793–1805. doi: 10.1091/mbc.6.12.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchiyama M, Galli I, Griffiths D, Wang T. Mol Cell Biol. 1997;17:3103–3115. doi: 10.1128/mcb.17.6.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Francesconi S, Grenon M, Bouvier D, Baldacci G. EMBO J. 1997;16:1332–1341. doi: 10.1093/emboj/16.6.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart E, Enoch T. Curr Opin Cell Biol. 1996;8:781–787. doi: 10.1016/s0955-0674(96)80078-0. [DOI] [PubMed] [Google Scholar]
- 31.Carr A. Science. 1996;271:314–315. doi: 10.1126/science.271.5247.314. [DOI] [PubMed] [Google Scholar]