Abstract
The discovery of cyanobacterial phytochrome histidine kinases, together with the evidence that phytochromes from higher plants display protein kinase activity, bind ATP analogs, and possess C-terminal domains similar to bacterial histidine kinases, has fueled the controversial hypothesis that the eukaryotic phytochrome family of photoreceptors are light-regulated enzymes. Here we demonstrate that purified recombinant phytochromes from a higher plant and a green alga exhibit serine/threonine kinase activity similar to that of phytochrome isolated from dark grown seedlings. Phosphorylation of recombinant oat phytochrome is a light- and chromophore-regulated intramolecular process. Based on comparative protein sequence alignments and biochemical cross-talk experiments with the response regulator substrate of the cyanobacterial phytochrome Cph1, we propose that eukaryotic phytochromes are histidine kinase paralogs with serine/threonine specificity whose enzymatic activity diverged from that of a prokaryotic ancestor after duplication of the transmitter module.
Keywords: plant photoreceptor/protein phosphorylation/molecular evolution/two-component signal transduction/PAS domain
The phytochromes, a family of biliprotein photoreceptors in higher plants, cryptophytes, and cyanobacteria, provide photosynthetic organisms with a means to detect suboptimal light conditions and to appropriately respond via changes in growth and development (1–5). Primarily responsible for red and far-red light perception, phytochromes exist in two photointerconvertible species, designated Pr and Pfr, respectively. Since phytochrome’s discovery nearly 40 years ago (6), it has been implicitly assumed that the Pr and Pfr forms possess distinct regulatory roles, the nature of which has eluded biochemical characterization.
The recent discovery that the cyanobacterial phytochrome Cph1 is a light-regulated histidine kinase (7) implicated that prokaryotic phytochrome predecessors were light-modulated enzymes. As summarized in two recent reviews (8, 9), evidence has been accumulating that suggests eukaryotic phytochromes are atypical protein kinases. In this regard, biochemical analysis of purified oat phytochrome A in the mid-1980s (10–13) revealed an associated polycation-stimulated, pyrophosphate-inhibited serine/threonine (Ser/Thr) protein kinase activity implicating phytochrome itself to be the enzyme responsible. More recently, Biermann et al. (14) reported that immunoprecipitated maize phytochrome also possessed protein kinase activity (14). Owing to the isolation of enzymatically inactive oat phytochrome A preparations (15, 16) and the observation that eukaryotic phytochromes lack the recognizable ATP binding motif of the Ser/Thr/Tyr protein kinase superfamily (17), the kinase hypothesis has been met with a great deal of skepticism.
The protein kinase hypothesis resurfaced in 1991, when Schneider-Poestsch and colleagues (18) noted that the C-terminal region of phytochromes is similar to histidine kinase transmitter modules found on bacterial sensor proteins. The functional significance of this sequence similarity has been questioned by the observation that many phytochromes lack the conserved histidine autophosphorylation site and the demonstration that other conserved residues within this domain are not required for photoregulatory activity in transgenic plants (19). Given the evidence that Cph1 is a histidine kinase (7), we chose to re-examine the hypothesis that eukaryotic phytochromes are protein kinases. The ability to express, reconstitute, and purify recombinant eukaryotic phytochromes (20) has enabled us to establish that higher plant and algal phytochromes are Ser/Thr protein kinases, thus implicating phytochrome-mediated protein phosphorylation in the transduction of the light signal in plants.
MATERIALS AND METHODS
Recombinant Oat and Green Algal Phytochrome Preparations.
Yeast expression, bilin attachment, and purification of Strep-Tagged (ST) recombinant phytochromes from oat Avena sativa L. (AsphyA-ST) and the green alga Mesotaenium caldariorum (Mcphy1b-ST) were performed as described (20, 21). In some experiments, bilin treatment was omitted to permit isolation of apophytochrome. After concentration, purified recombinant phytochromes were diluted with TEGE buffer (25 mM Tris⋅HCl, pH 8.0/25% ethylene glycol/1 mM EDTA) to a final concentration of 0.2–0.5 μg/μl and stored at −80°C. The predicted subunit molecular masses for AsphyA-ST and Mcphy1b-ST are both 126.6 kDa.
Protein Kinase Assays.
Protein phosphorylation experiments were performed in 25-μl reaction mixtures containing kinase buffer (25 mM Tris⋅HCl, pH 7.5/0.2 mM EDTA/5 mM MgCl2/4 mM 2-mercaptoethanol), 0.1 mM [γ-32P]ATP (2,000–5,000 cpm/pmol), and 0.5–1 μg of purified recombinant phytochrome (10). Histone H1 (a gift of Joyce Hamaguchi, University of California, Davis) purified by method 1 of Johns (22), maltose binding protein (MBP; expressed in pMALc2-containing Escherichia coli cells and purified according to New England BioLabs instructions), MBP-Rcp1, the maltose binding protein fusion of the response regulator for Cph1 (7), and/or sodium pyrophosphate were included in some assays as specified. Phosphorylation reactions were initiated by adding [γ-32P]ATP, incubated at 30°C for 30 min and stopped by adding 10 μl of 4× SDS sample buffer under dim green safe light (10). Proteins were resolved on 10% SDS/polyacrylamide gels (23) and transblotted to poly(vinylidene difluoride) membranes (Immobilon P, Millipore) for autoradiographic analysis using a Storm 860 PhosphorImager (Molecular Dynamics) and for subsequent zinc blot analysis and Coomassie blue protein staining to verify protein loading (7). Acid and base hydrolytic stability of phosphorylated proteins were performed as described previously (7). For kinetic assays, reactions were performed at 30°C for 20 min and terminated by spotting samples onto Whatman GF/C glass fiber filter disks and plunging into ice-cold 10% (wt/vol) trichloroacetic acid (TCA) containing 1% (wt/vol) sodium pyrophosphate (24). After incubation for 30 min, the filter disks were washed three times for 30 min each in ice-cold 5% (wt/vol) TCA containing 1% (wt/vol) sodium pyrophosphate. The disks were rinsed briefly in ethanol, dried, and placed into scintillation fluid to determine the amount of 32P incorporation (24).
RESULTS
Recombinant Oat and Algal Phytochromes Possess Serine/Threonine Protein Kinase Activity.
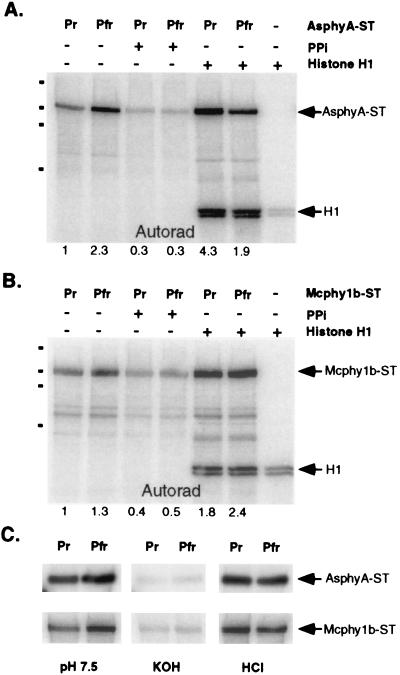
Purified affinity-peptide tagged versions of recombinant oat (AsphyA-ST) and green algal (Mcphy1b-ST) phytochromes, expressed in the yeasts Saccharomyces cerevisiae and Pichia pastoris, respectively, and preassembled with phycocyanobilin (PCB), were analyzed for protein kinase activity. Fig. 1 A and B shows that both proteins were phosphorylated upon incubation with [γ-32P]ATP and that phosphorylation of both proteins was light modulated. For these experiments, the Pfr forms of both oat and algal phytochromes were 2.3-fold and 1.3-fold more labeled than their respective Pr forms, with mol percent incorporations for the Pfr form of AsphyA-ST being approximately 8% in a 20-min reaction. Because the rates of 32P labeling were linear during a 40-min assay period (data not shown), these results indicated that the Pfr forms were either more active and/or were better substrates for the protein kinase. Fig. 1 also shows that the phosphorylations of both AsphyA-ST and Mcphy1b-ST were inhibited by pyrophosphate and stimulated by addition of histone H1 (particularly the Pr form), similar to that described for purified oat phytochrome A from dark grown seedlings (11). As was observed in previous studies (10, 11), histone H1 was partially “labeled” in the absence of phytochrome, which we attribute to nonspecific [32P]ATP/phosphate binding. Histone H1 phosphorylation increased 20-fold upon addition of AsphyA-ST, indicating that the histone H1 kinase activity was phytochrome associated. Unlike the phosphorylation of AsphyA-ST, histone H1 phosphorylation was not light regulated. Fig. 1C shows that both AsphyA-ST and Mcphy1b-ST phosphorylations were acid stable and base labile, as is characteristic of phosphoserine or phosphothreonine. Phosphoamino analyses revealed serine to be the predominant residue phosphorylated in both Pr and Pfr forms of AsphyA-ST (data not shown).
Figure 1.
Recombinant oat and algal phytochromes possess protein kinase activity. (A) Autoradiograph of Pr and Pfr forms of PCB adducts of recombinant oat phytochrome A (AsphyA-ST; 0.5 μg) after incubation with radiolabeled ATP under standard kinase conditions in the presence (+) or absence (−) of 2 μg histone H1 and/or 2 mM pyrophosphate (PPi) was obtained as described in Materials and Methods. The relative amount of phosphorylation indicated below the autoradiographs was obtained by using imagequant software (Molecular Dynamics). Dots on the left side of the gel represent molecular mass standards with 197 kDa, 117 kDa, 89 kDa, and 52 kDa (top to bottom). (B) An autoradiograph of algal phytochrome (Mcphy1b-ST; 1 μg) assayed identically as AsphyA-ST (above). (C) Hydrolytic stability of autophosphorylated phytochromes examined by treatment of poly(vinylidene difluoride) membranes with neutral (50 mM Tris⋅HCl, pH 7.5), basic (3 M KOH), or acidic (1 M HCl) solutions for 2.5 hr at room temperature with gentle shaking.
AsphyA-ST Phosphorylation Is Concentration Independent.
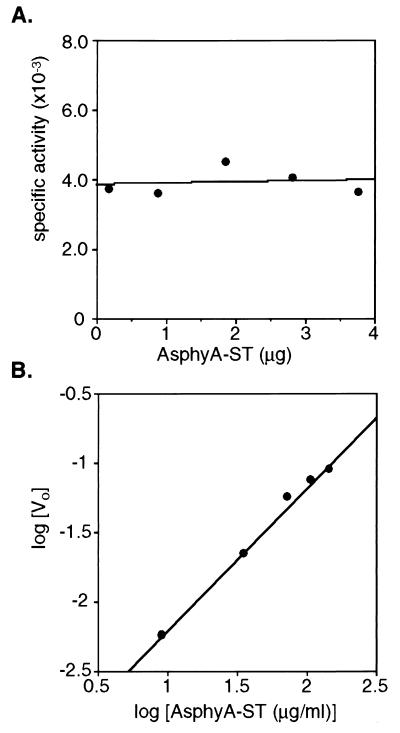
To address the hypothesis that the protein kinase activity reflected an intrinsic property of the phytochrome molecule, we tested whether the ATP-dependent phosphorylation of AsphyA-ST was concentration dependent. These experiments used the Pfr form of AsphyA-ST (PCB adduct) because of its higher molar 32P incorporation compared with the Pr form. For an intramolecular phosphotransfer, the initial velocity at a fixed ATP level is expected to increase linearly with protein concentration, whereas the concentration dependence for an intermolecular phosphotransfer should be parabolic (25). Consistent with the former, the rate of AsphyA-ST phosphorylation was proportional to the concentration of AsphyA-ST within a range of 40 to 620 nM. This data is depicted in Fig. 2A, which illustrates that the kinase-specific activity was independent of AsphyA-ST concentration, as expected for intramolecular phosphotransfer (26). The van’t Hoff plot shown in Fig. 2B yielded a slope equal to 1.02. A slope of one predicts an intramolecular mechanism whereas a slope of two indicates that the phosphotransfer is a bimolecular reaction (25–27). Taken together, these experiments indicate that either phytochrome is itself a protein kinase or it is associated with a tightly bound enzyme that cannot phosphorylate another phytochrome molecule. Because AsphyA-ST is a homodimer (20), intersubunit phosphotransfer is also consistent with these results.
Figure 2.
ATP-dependent phosphorylation of the PCB adduct of recombinant oat phytochrome is an intramolecular process. (A) Specific activity of AsphyA-ST autophosphorylation (dpm/μg AsphyA-ST; Pfr form) plotted versus μg AsphyA-ST analyzed. (B) A van’t Hoff plot of the same data obtained by plotting the logarithm of the initial rate (Vo) of AsphyA-ST phosphorylation, determined by converting the phosphorylation level (dpm) to pmol incorporation per minute, versus the logarithm of AsphyA-ST concentration (μg/ml). The slope of this line was 1.02 (r2 = 0.994).
AsphyA-ST Autophosphorylation Is Both Chromophore and Light Regulated.
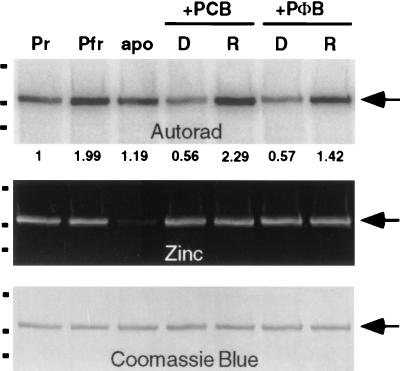
Expression of AsphyA-ST in the yeast S. cerevisiae yields a phytochrome species that lacks a bilin chromophore (20, 28). This observation enabled us to examine the effect of chromophore attachment on the protein kinase activity of AsphyA-ST. Because bilin attachment in the absence of light produces the Pr form of the photoreceptor (29), assembly was performed in darkness. Fig. 3 (Top) shows that both PCB and phytochromobilin (PΦB) attachment to AsphyA-ST inhibited phytochrome autophosphorylation. In this regard, the AsphyA-ST apoprotein (labeled apo in lane 3) was 2-fold more phosphorylated than either bilin adduct (labeled D in lanes 4 and 6). Photoconversion of both bilin adducts to their Pfr forms with red light irradiation stimulated AsphyA-ST autophosphorylation activity by roughly 3- to 4-fold (compare lanes labeled D and R). The stimulatory effect of red light also was observed in preassembled samples that had been separated from unbound PCB pigment (compare lanes labeled Pr and Pfr). This result demonstrated that the presence of free bilin pigment was not responsible for the light stimulation. In addition, the apparent autophosphorylation activity of a newly dark-synthesized Pr sample of AsphyA-ST (PCB adduct) was invariably less than that of a photocycled Pr sample (compare lanes labeled D and Pr; data not shown). The molecular basis of this interesting photocycling effect, which might reflect “irreversible” activation of AsphyA-ST’s kinase activity or dephosphorylation of sites originally labeled, is presently under investigation. Fig. 3 (Middle) shows the corresponding zinc blot in which the bilin-linked polypeptides were visualized by their fluorescence in the presence of zinc ion (30). The presence of fluorescent bands of similar intensity in dark and red light-treated samples shows that the extent of bilin binding was not responsible for the light modulation of AsphyA-ST’s kinase activity. These results demonstrate that AsphyA-ST Ser/Thr kinase activity is regulated by bilin binding and also by light.
Figure 3.
Phytochrome autophosphorylation is both chromophore and light regulated. Autoradiograph (Top), zinc blot (Middle), and Coomassie blue-stained blot (Bottom) of an AsphyA-ST apoprotein sample divided into three fractions. One fraction was treated with dimethyl sulfoxide only (apo), and the other two were treated with 8 μM PCB and 8 μM phytochromobilin (PΦB) and incubated 30 min in complete darkness, and assayed for kinase activity as described in Materials and Methods. Apophytochrome (apo), newly assembled bilin-adducts (D), and red-light irradiated bilin adducts (R) are shown (0.5 μg). Preassembled Pr and Pfr forms of AsphyA-ST (PCB adduct; 0.5 μg) were included in the left two lanes for comparison. Relative phosphorylation and molecular mass markers were determined as in Fig. 1.
Molecular Evolution of Phytochromes: Duplication of a Transmitter Module of a Prokaryotic Phytochrome Ancestor.
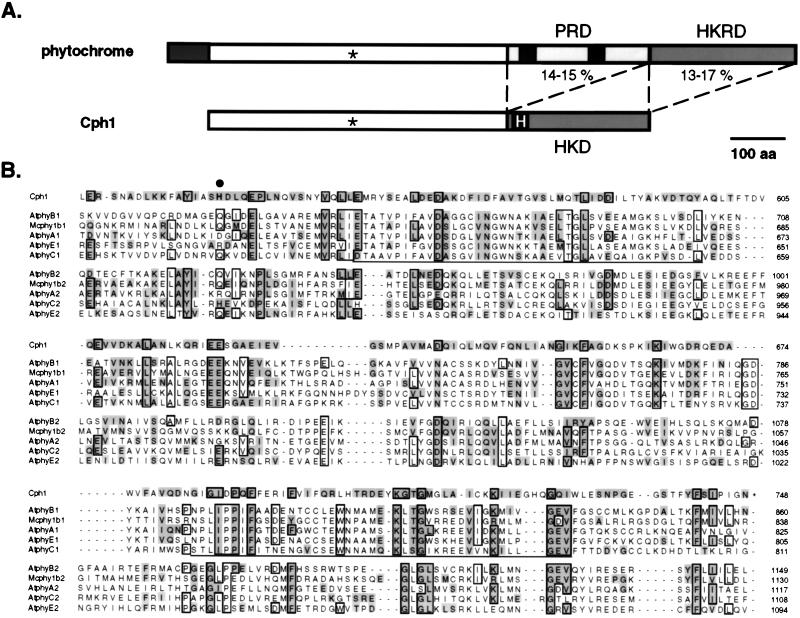
In view of the observed protein kinase activity of AsphyA-ST and the relatedness of its C-terminal region with histidine kinases of bacterial sensors (18), we began to consider the possibility that molecular evolution of a histidine kinase domain on a prokaryotic phytochrome ancestor similar to Cph1 could have produced a new type of ATP-binding domain with Ser/Thr protein kinase activity. When protein sequence alignments of Cph1 and 20 eukaryotic phytochromes were computed by using the Wisconsin Genetics Computer Group program pileup, we observed that Cph1’s histidine kinase domain (HKD) aligned with a region of eukarotic phytochromes adjacent to their histidine kinase-related domain (HKRD; see Fig. 4A for phytochrome domain model). We will refer to this region of phytochrome as the PAS-related domain (PRD), because of the presence of the 40-aa PAS motif found on the growing family of regulatory proteins typified by the Drosophila period clock protein PER, the mammalian aromatic hydrocarbon receptor nuclear translocator ARNT, and the Drosophila single-minded protein SIM (31–33). This unexpected alignment could be rationalized by the large penalty required to insert a 250-aa gap to align the eukaryotic phytochrome HKRD with the HKD of Cph1. Closer examination of this alignment suggested that both eukaryotic phytochrome PRD and HKRD were related to Cph1’s HKD (Fig. 4A). To assess this hypothesis, we divided the C-terminal region of 20 eukaryotic phytochromes into PRD and HKRD regions, and a multiple sequence alignment of the 40 sequences was computed by using pileup and saga (34). By using separate alignments of the Cph1 HKD with both PRD and HKRD regions of eukaryotic phytochromes to adjust this comparison, the consensus multiple sequence alignment shown in Fig. 4B was obtained. This alignment revealed that PRD and HKRD domains of eukaryotic phytochromes are equally related to the HKD of Cph1, with respective pair-wise equivalence values of 14–15% and 13–17% (see Fig. 4A and Table 1). Because the observed similarity is distributed throughout a large region approximately 250 amino acids in length, this result is consistent with the hypothesis that eukaryotic phytochrome PRD and HKRD regions share common ancestry and are likely to adopt a similar overall protein fold. Because eukaryotic phytochrome PRD and HKRD domains are more similar to Cph1’s HKD than they are to each other (Table 1), we speculate that the phytochrome ancestor possessed a histidine kinase domain similar to Cph1 whose duplication and molecular divergence accounted for the production of PRD and HKRD found on eukaryotic phytochromes.
Figure 4.
Duplication of a transmitter module of a prokaryotic phytochrome ancestor: A model for molecular evolution of eukaryotic phytochromes. (A) Structural comparison of eukaryotic phytochromes and Cph1. The conserved cysteine chromophore binding site is marked (∗), and the conserved histidine on the HKD of Cph1 is highlighted. The percent amino acid equivalence between the HKD of Cph1 and both PRD and HKRD of eukaryotic phytochromes is indicated (see Table 1 for details). (B) Multiple sequence alignment of the HKD of Cph1 with the PRD (sequences ending with 1) and HKRD (sequences ending with 2) regions of representative eukaryotic phytochromes including phytochromes from Arabidopsis thaliana (i.e., AtphyA, AtphyB, AtphyC, and AtphyE) and phytochrome from the green alga M. caldariorum. (Mcphy1b). Residues that are identical in six or more sequences are boxed, whereas residues equivalent to those in Cph1 (where I = V, L = M, R = K, S = T = A and D = E = N = Q) are highlighted. The conserved histidine phosphorylation site (H538) of Cph1 is marked with a large dot. The two PAS motifs in eukaryotic phytochromes are underlined. GenBank accession numbers for phytochrome sequences are AtphyA, L21154; AtphyB, X17342; AtphyC, Z32538; AtphyE, X76610; and Mcphy1b, U31284.
Table 1.
Amino acid equivalence between the C-terminal PHD (1) and HKR (2) domains of eukaryotic phytochromes and the HKD of Cph1
| Cph1 | phyB2 | Mcphy1b2 | phyA2 | phyC2 | phyE2 | |
|---|---|---|---|---|---|---|
| Cph1 | 100 | 15 | 17 | 13 | 15 | 13 |
| phyB1 | 15 | 11 | 12 | 8 | 6 | 11 |
| Mcphy1b1 | 14 | 9 | 9 | 9 | 8 | 9 |
| phyA1 | 14 | 12 | 10 | 9 | 14 | 10 |
| phyE1 | 15 | 9 | 11 | 10 | 6 | 8 |
| phyC1 | 14 | 9 | 8 | 7 | 9 | 8 |
Subdomains 1 and 2 represent PHD and HKR, respectively. The numbers indicate percent identity where I = V, L = M, D = N = Q = E, R = K and A = S = T.
AsphyA-ST Interacts with a Cyanobacterial Phytochrome Response Regulator.
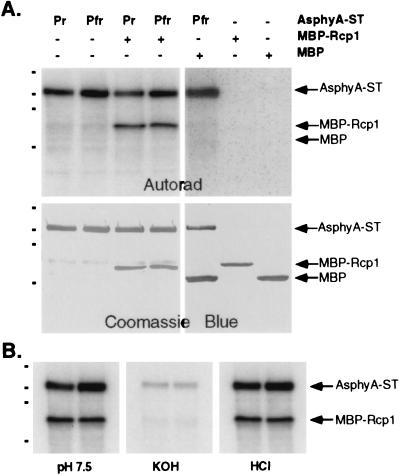
Because PRD and HKRD regulatory domains of AsphyA appear to be structurally related to bacterial histidine kinase domains, we reasoned that the natural substrates for eukaryotic phytochromes might resemble the response regulator substrates of bacterial sensors. In this regard, we tested whether AsphyA-ST could phosphorylate Rcp1, the response regulator substrate of Cph1 (7). Fig. 5A shows that Rcp1, as a MBP fusion, was a substrate for AsphyA-ST. Because MBP itself was not a substrate for AsphyA-ST and neither Rcp1 nor MBP possessed detectable kinase activity (Fig. 5A), these results document the interaction between AsphyA-ST and MBP-Rcp1. Like histone H1, however, MBP-Rcp1 phosphorylation by AsphyA-ST was not modulated by light as might be expected for a natural substrate for a eukaryotic phytochrome. Because the D68A mutant of Rcp1, which lacks the phospho-accepting aspartate residue for Cph1 (7), was as good a substrate as wild-type Rcp1 (data not shown), a different residue on Rcp1 must have been phosphorylated. Phosphoamino acid analysis indicated that residue(s) phosphorylated on MBP-Rcp1 was acid stable and base labile (Fig. 5B), indicating that MBP-Rcp1 was phosphorylated on a Ser/Thr residue(s).
Figure 5.
MBP-Rcp1 is a substrate for AsphyA-ST kinase activity. (A) Autoradiograph and Coomassie blue-stained blot of Pr and Pfr forms of AsphyA-ST (PCB adduct; 1 μg) incubated alone or with MBP-Rcp1 (1 μg) under standard kinase assay conditions (see Materials and Methods). As controls, a mixture of purified MBP (1.5 μg) and the Pfr form of AsphyA-ST (PCB adduct; 1 μg), purified MBP-RCP1 (1 μg) or purified MBP (1.5 μg) incubated under standard kinase assay conditions (see Materials and Methods) are shown in the right three lanes. (B) Acid and base hydrolytic stability of the mixture of AsphyA-ST (PCB adduct, 1 μg) and MBP-Rcp1 (1 μg) was determined as described in Fig. 1.
DISCUSSION
Eukaryotic Phytochromes Are Protein Kinases.
Our investigations support the conclusion that eukaryotic phytochromes are protein kinases. The alternative hypothesis, that the Ser/Thr kinase activity of phytochrome is caused by a copurifying contaminant, is less favored for several reasons. First, protein kinase activity is observed for highly purified recombinant phytochromes from both a higher plant and a green alga expressed in two different yeast systems. Second, the biochemical properties of this protein kinase (i.e., polycation stimulation, pyrophosphate inhibition, and Pr/Pfr differential phosphorylation) are strikingly similar to those of phytochrome isolated from plant tissue (12). Indeed, even the specific activity of AsphyA-ST is comparable to that of native oat phytochrome A (11). It is also noteworthy that the properties of the protein kinases separated from phytochrome by others (15, 16) are distinct from those described here and previously (see discussion in ref. 12). Third, ATP-dependent phytochrome phosphorylation is independent of phytochrome concentration in the range of 40 to 620 nM. Fourth, phytochrome (auto)phosphorylation is both chromophore and light modulated. Although these data do not categorically rule out the presence of a copurifying enzyme, this protein kinase would have to be produced in both S. cerevisiae and P. pastoris cells, to bind oat (and algal) phytochrome with an apparent (sub)nanomolar affinity, to be unable to phosphorylate another phytochrome molecule intermolecularly, and to exhibit an ability to distinguish between apophytochrome, Pr, and Pfr forms as substrates (or to be differentially regulated by these forms of phytochrome). Because we believe this scenario to be unlikely, we conclude that phytochrome is a Ser/Thr protein kinase that catalyzes intramolecular phosphotransfer between the subunits of the phytochrome homodimer, an autophosphorylation mechanism that is well documented for many protein kinases (17).
What Is the Biological Significance of Phytochrome’s Kinase Activity?
Based on the light-regulated phosphotransferase activities of Cph1 (7), it is reasonable that the primary signal transduction pathway for higher plant phytochrome also will use a light-modulated phosphotransfer mechanism. The ability of oat phytochrome A to phosphorylate MBP-Rcp1, the substrate for Cph1, suggests that the endogenous substrates for eukaryotic phytochromes may be response regulator homologs. Eukaryotic phytochromes possess Ser/Thr kinase activity, however, not histidine kinase activity, and an active site aspartate residue on Rcp1 is not required for phosphotransfer. The chemical mechanism for phosphotransfer thus appears to be quite dissimilar for prokaryotic and eukaryotic phytochromes. This conclusion is further supported by the observation that that key conserved residues within the HKRD of oat phytochrome are not required for its photoregulatory activity in transgenic plants (19). Considering the possible duplication of an ancestral HKD region of a prokaryotic phytochrome, we hypothesize that the paralogous PRD region of eukaryotic phytochromes is responsible for Ser/Thr kinase activity of these photoreceptors. Missense mutations that inhibit the biological function of phytochrome also heavily cluster within the PRD region, suggesting that this region is critical to the photoreceptor’s regulatory activity (35). We speculate that the duplication of the transmitter domain of a prokaryotic phytochrome led to the evolution of the eukaryotic HKRD, which retains the ability to interact with response regulator receiver domains, and the PRD region, that not only interacts with proteins containing the PAS motif (32, 33), but also possesses ATP-dependent Ser/Thr phosphotransferase activity. Thus, the HKRD might serve to position a substrate for PRD-mediated phosphotransfer. Alternatively, the PRD region might be responsible for positioning substrates for phosphotransfer because the PAS domains have been strongly implicated in protein-protein interactions in numerous regulatory proteins (36–38). Mutagenesis experiments to address the importance of conserved residues within PRD and HKRD domains to phytochrome’s protein kinase activity and biological activity in planta should shed light on these hypotheses.
With regard to the role of phytochrome autophosphorylation, mutagenesis of the serine-rich region near the N terminus of phytochrome has been shown to lead to enhanced biological activity in transgenic plants (39, 40). Because this region encompasses the serine site for both in vitro and in vivo phosphorylation of oat phytochrome A (41, 42), but is not essential for biological activity in transgenic plants (43, 44), autophosphorylation appears to serve a regulatory role or, as suggested recently, to modulate phytochrome’s association with other regulatory molecules (5, 9). This serine-rich extension has been established to undergo a significant light-dependent conformational change (reviewed in ref. 5); however, this region is not present on Cph1. Its apparently recent evolution thus may reflect its role to regulate phytochrome’s protein kinase activity toward other substrates (possibly via interaction with HKRD and/or PRD regions). Indeed, autophosphorylation of a regulatory or pseudosubstrate domain is a well-established mechanism for modulating the activity or specificity of protein kinases (45). Until bona fide phytochrome substrates are identified, the biological role of phytochrome’s kinase activity in the transmission of the light signal in plants remains hypothetical. We anticipate that the combination of genetic and biochemical approaches, such as yeast two-hybrid screens and cross talk with the growing families of plant response regulators (46–49) and PAS domain-containing proteins (32, 33), soon will identify molecules in the phytochrome signal transduction pathway whose activities are regulated by protein phosphorylation.
Acknowledgments
We thank Shu-Hsing Wu for the preparation of Mcphy1b-ST, Dan Buster for phosphoamino acid analysis, and Hugh Nicholas for advice with protein sequence alignments. We acknowledge grant support from the National Science Foundation (MCB 96-04511) to J.C.L. and from the National Institutes of Health National Center for Research Resources Resource Grant 2 P41 RR06009 to the Pittsburgh Supercomputing Center.
ABBREVIATIONS
- Cph1
cyanobacterial phytochrome 1
- AsphyA
Avena sativa phytochrome A
- HKD
histidine kinase domain
- HKRD
histidine kinase-related domain
- MBP
maltose binding protein
- Mcphy1b
Mesotaenium caldariorum phytochrome 1b
- PAS
motif found in the protein superfamily typified by the products of per, arnt, and sim
- PCB
phycocyanobilin
- PRD
PAS-related domain
- Pfr
far-red light absorbing form of phytochrome
- Pr
red light absorbing form of phytochrome
- Rcp1
response regulator for Cph1
- ST
Strep-Tag
Footnotes
A Commentary on this article begins on page 13358.
References
- 1.Sage L C. Pigment of the Imagination: A History of Phytochrome Research. San Diego: Academic; 1992. [Google Scholar]
- 2.Smith H. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:289–315. [Google Scholar]
- 3.Pratt L H. Photochem Photobiol. 1995;61:10–21. doi: 10.1111/j.1751-1097.1975.tb06635.x. [DOI] [PubMed] [Google Scholar]
- 4.Furuya M, Schafer E. Trends Plant Sci. 1996;1:301–307. [Google Scholar]
- 5.Quail P H. Plant Cell Environ. 1997;20:657–665. [Google Scholar]
- 6.Butler W L, Norris K H, Seigelman H W, Hendricks S B. Proc Natl Acad Sci USA. 1959;45:1703–1708. doi: 10.1073/pnas.45.12.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeh K-C, Wu S-H, Murphy J T, Lagarias J C. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- 8.Elich T D, Chory J. Cell. 1997;91:713–716. doi: 10.1016/s0092-8674(00)80458-4. [DOI] [PubMed] [Google Scholar]
- 9.Quail P H. BioEssays. 1997;19:571–579. doi: 10.1002/bies.950190708. [DOI] [PubMed] [Google Scholar]
- 10.Wong Y S, Cheng H C, Walsh D A, Lagarias J C. J Biol Chem. 1986;261:12089–12097. [PubMed] [Google Scholar]
- 11.Wong Y S, McMichael R W, Lagarias J C. Plant Physiol. 1989;91:709–718. doi: 10.1104/pp.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMichael R W, Lagarias J C. In: Current Topics in Plant Biochemistry and Physiology. Randall D D, Blevins D G, editors. Vol. 9. Columbia: University of Missouri; 1990. pp. 259–270. [Google Scholar]
- 13.Wong Y S, Lagarias J C. Proc Natl Acad Sci USA. 1989;86:3469–3473. doi: 10.1073/pnas.86.10.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biermann B J, Pao L I, Feldman L J. Plant Physiol. 1994;105:243–251. doi: 10.1104/pp.105.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimm R, Gast D, Rudiger W. Planta. 1989;178:199–206. doi: 10.1007/BF00393195. [DOI] [PubMed] [Google Scholar]
- 16.Kim, I., Bai, U. & Song, P. (1989) Photochem. Photobiol. 319–323. [DOI] [PubMed]
- 17.Hanks S K, Hunter T. In: The Eukaryotic Protein Kinase Superfamily. Hardie G, Hanks S, editors. London: Academic; 1995. pp. 7–47. [Google Scholar]
- 18.Schneider-Poetsch H A W, Braun B, Marx S, Schaumburg A. FEBS Lett. 1991;281:245–249. doi: 10.1016/0014-5793(91)80403-p. [DOI] [PubMed] [Google Scholar]
- 19.Boylan M T, Quail P H. Protoplasma. 1996;195:12–17. [Google Scholar]
- 20.Murphy J T, Lagarias J C. Photochem Photobiol. 1997;65:750–758. doi: 10.1111/j.1751-1097.1997.tb01920.x. [DOI] [PubMed] [Google Scholar]
- 21.Wu S-H, Lagarias J C. Proc Natl Acad Sci USA. 1996;93:8989–8994. doi: 10.1073/pnas.93.17.8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johns E W. Biochem J. 1964;92:55–59. doi: 10.1042/bj0920055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Hess J F, Oosawa K, Matsumura P, Simon M I. Proc Natl Acad Sci USA. 1987;84:7609–7613. doi: 10.1073/pnas.84.21.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Todhunter J A, Purich D L. Biochim Biophys Acta. 1977;485:87–94. doi: 10.1016/0005-2744(77)90195-4. [DOI] [PubMed] [Google Scholar]
- 26.Huang K-P, Chan K-F J, Singh T J, Nakabayashi H, Huang F L. J Biol Chem. 1986;261:12134–12140. [PubMed] [Google Scholar]
- 27.van’t Hoff J H. Etudes de Dynamique Chimique. Amsterdam: Muller; 1884. p. 87. [Google Scholar]
- 28.Wahleithner J A, Li L, Lagarias J C. Proc Natl Acad Sci USA. 1991;88:10387–10391. doi: 10.1073/pnas.88.23.10387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Lagarias J C. J Biol Chem. 1992;267:19204–19210. [PubMed] [Google Scholar]
- 30.Berkelman T R, Lagarias J C. Anal Biochem. 1986;156:194–201. doi: 10.1016/0003-2697(86)90173-9. [DOI] [PubMed] [Google Scholar]
- 31.Lagarias D M, Wu S-H, Lagarias J C. Plant Mol Biol. 1995;29:1127–1142. doi: 10.1007/BF00020457. [DOI] [PubMed] [Google Scholar]
- 32.Ponting C P, Aravind L. Curr Biol. 1997;7:R674–R677. doi: 10.1016/s0960-9822(06)00352-6. [DOI] [PubMed] [Google Scholar]
- 33.Zhulin I B, Taylor B L. Trends Biochem Sci. 1997;22:331–333. doi: 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]
- 34.Notredame C, Higgins D G. Nucleic Acids Res. 1996;24:1515–1524. doi: 10.1093/nar/24.8.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quail P H, Boylan M T, Parks B M, Short T W, Xu Y, Wagner D. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- 36.Lindebro M C, Poellinger L, Whitelaw M L. EMBO J. 1995;14:3528–3539. doi: 10.1002/j.1460-2075.1995.tb07359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Z J, Edery I, Rosbash M. Nature (London) 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 38.Zelzer E, Wappner P, Shilo B Z. Genes Dev. 1997;11:2079–2089. doi: 10.1101/gad.11.16.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stockhaus J, Nagatani A, Halfter U, Kay S, Furuya M, Chua N H. Genes Dev. 1992;6:2364–2372. doi: 10.1101/gad.6.12a.2364. [DOI] [PubMed] [Google Scholar]
- 40.Jordan E T, Marita J M, Clough R C, Vierstra R D. Plant Physiol. 1997;115:693–704. doi: 10.1104/pp.115.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMichael R W, Lagarias J C. Biochemistry. 1990;29:3872–3878. doi: 10.1021/bi00468a011. [DOI] [PubMed] [Google Scholar]
- 42.Lapko V N, Jiang X Y, Smith D L, Song P S. Biochemistry. 1997;36:10595–10599. doi: 10.1021/bi970708z. [DOI] [PubMed] [Google Scholar]
- 43.Boylan M, Douglas N, Quail P H. Plant Cell. 1994;6:449–460. doi: 10.1105/tpc.6.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jordan E T, Cherry J R, Walker J M, Vierstra R D. Plant J. 1996;9:243–257. doi: 10.1046/j.1365-313x.1996.09020243.x. [DOI] [PubMed] [Google Scholar]
- 45.Johnson L N, Noble M E M, Owen D J. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 46.Chang C, Stewart R C. Plant Physiol. 1998;117:723–731. doi: 10.1104/pp.117.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imamura A, Hanaki N, Umeda H, Nakamura A, Suzuki T, Ueguchi C, Mizuno T. Proc Natl Acad Sci USA. 1998;95:2691–2696. doi: 10.1073/pnas.95.5.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakakibara H, Suzuki M, Takei K, Deji A, Taniguchi M, Sugiyama T. Plant J. 1998;14:337–344. doi: 10.1046/j.1365-313x.1998.00134.x. [DOI] [PubMed] [Google Scholar]
- 49.Brandstatter I, Kieber J J. Plant Cell. 1998;10:1009–1019. doi: 10.1105/tpc.10.6.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]