Abstract
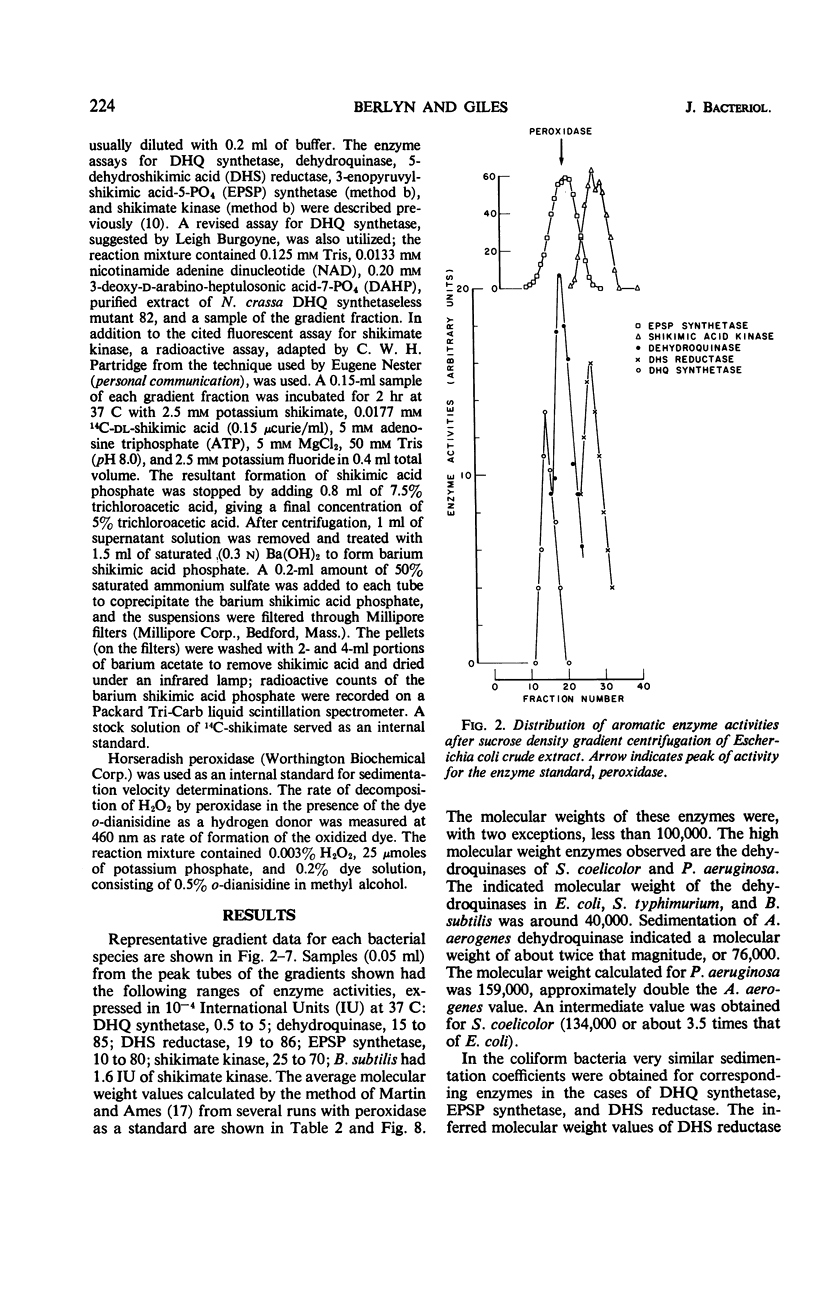
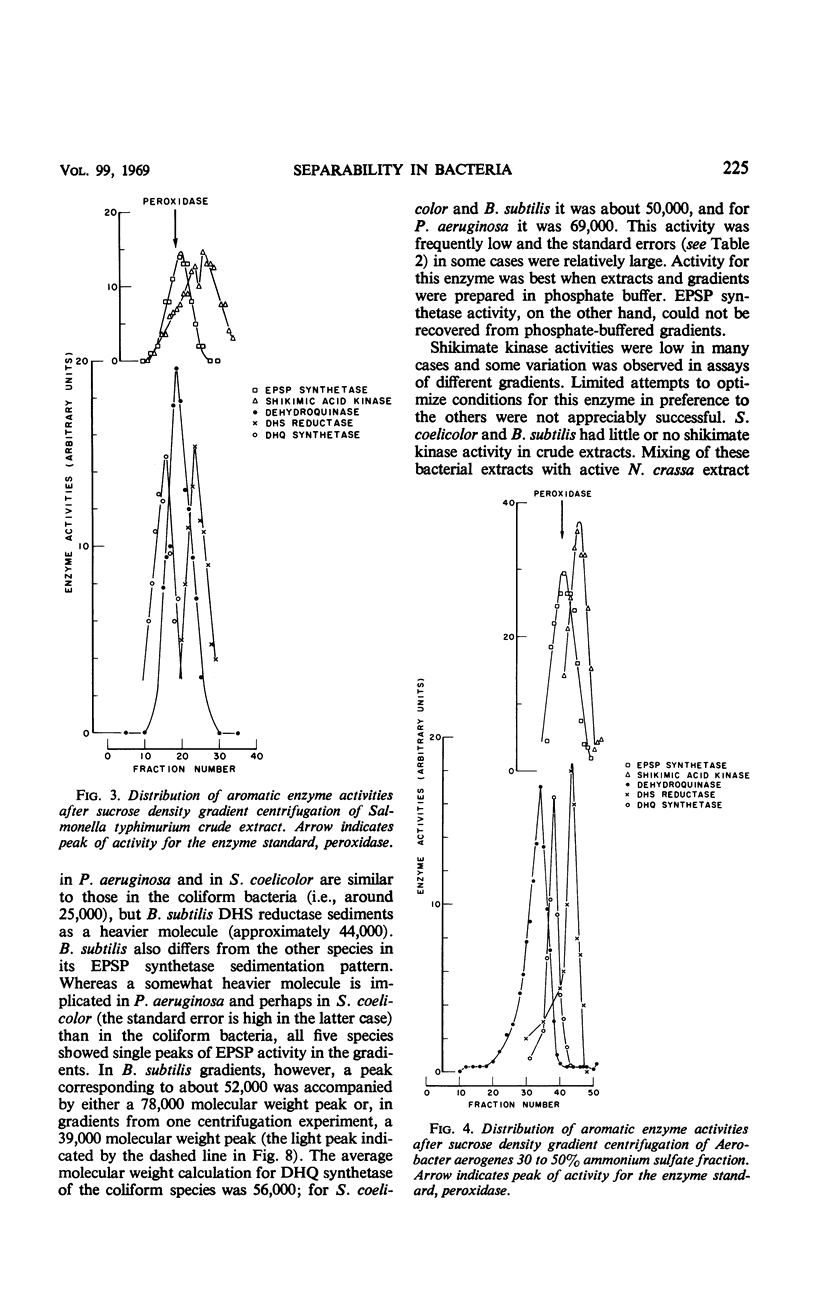
Ultracentrifugation in sucrose density gradients was employed to estimate the molecular weights and to determine possible physical aggregation of the five enzymes catalyzing steps two to six in the prechorismic acid portion of the polyaromatic synthetic pathway in six species of bacteria: Escherichia coli, Salmonella typhimurium, Aerobacter aerogenes, Bacillus subtilis, Pseudomonas aeruginosa, and Streptomyces coelicolor. The five enzymes were not aggregated in extracts of any of the species examined, nor are the genes encoding these enzymes clustered in those bacterial species for which genetic evidence exists. (An initial examination of the blue-green alga Anabaena variabilis indicates nonaggregation in this species also.) This situation in bacteria is in marked contrast to that found in Neurospora crassa and other fungi in which the same five enzymes are associated as an aggregate encoded (at least in the case of N. crassa) by a cluster of five genes. In addition, also in contrast to N. crassa, no evidence was obtained for more than one kind of dehydroquinase activity in any of the bacteria examined. These comparative results are discussed in relation to the origin, evolution, and functional significance of the gene-enzyme relationships existing in the early steps of aromatic biosynthesis in bacteria and fungi.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AHMED A., CASE M. E., GILES N. H. THE NATURE OF COMPLEMENTATION AMONG MUTANTS IN THE HISTIDINE-3 REGION OF NEUROSPORA CRASSA. Brookhaven Symp Biol. 1964 Dec;17:53–65. [PubMed] [Google Scholar]
- Ahmed S. I., Giles N. H. Organization of enzymes in the common aromatic synthetic pathway: evidence for aggregation in fungi. J Bacteriol. 1969 Jul;99(1):231–237. doi: 10.1128/jb.99.1.231-237.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUTTIN G. M'ECANISMES R'EGULATEURS DANS LA BIOSYNTH'ESE DES ENZYMES DU M'ETABOLISME DU GALACTOSE CHEZ ESCHERICHIA COLI K12. II. LE D'ETERMINISME G'EN'ETIQUE DE LA R'EGULATION. J Mol Biol. 1963 Aug;7:183–205. doi: 10.1016/s0022-2836(63)80045-5. [DOI] [PubMed] [Google Scholar]
- Berlyn M. B. Gene-enzyme relationships in histidine biosynthesis in Aspergillus nidulans. Genetics. 1967 Nov;57(3):561–570. doi: 10.1093/genetics/57.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COTTON R. G., GIBSON F. THE BIOSYNTHESIS OF PHENYLALANINE AND TYROSINE; ENZYMES CONVERTING CHORISMIC ACID INTO PREPHENIC ACID AND THEIR RELATIONSHIPS TO PREPHENATE DEHYDRATASE AND PREPHENATE DEHYDROGENASE. Biochim Biophys Acta. 1965 Apr 12;100:76–88. doi: 10.1016/0304-4165(65)90429-0. [DOI] [PubMed] [Google Scholar]
- DEMEREC M. CLUSTERING OF FUNCTIONALLY RELATED GENES IN SALMONELLA TYPHIMURIUM. Proc Natl Acad Sci U S A. 1964 Jun;51:1057–1060. doi: 10.1073/pnas.51.6.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas H. C., Hawthorne D. C. Regulation of genes controlling synthesis of the galactose pathway enzymes in yeast. Genetics. 1966 Sep;54(3):911–916. doi: 10.1093/genetics/54.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G. R. A cluster of genes controlling three enzymes in histidine biosynthesis in Saccharomyces cerevisiae. Genetics. 1966 Mar;53(3):445–459. doi: 10.1093/genetics/53.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles N. H., Case M. E., Partridge C. W., Ahmed S. I. A gene cluster in Nuerospora crassa coding for an aggregate of five aromatic synthetic enzymes. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1453–1460. doi: 10.1073/pnas.58.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles N. H., Partridge C. W., Ahmed S. I., Case M. E. The occurrence of two dehydroquinases in Neurospora crassa, one constitutive and one inducible. Proc Natl Acad Sci U S A. 1967 Nov;58(5):1930–1937. doi: 10.1073/pnas.58.5.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollub E., Zalkin H., Sprinson D. B. Correlation of genes and enzymes, and studies on regulation of the aromatic pathway in Salmonella. J Biol Chem. 1967 Nov 25;242(22):5323–5328. [PubMed] [Google Scholar]
- Henning U., Dennert G., Hertel R., Shipp W. S. Translation of the structural genes of the E. coli pyruvate dehydrogenase complex. Cold Spring Harb Symp Quant Biol. 1966;31:227–234. doi: 10.1101/sqb.1966.031.01.031. [DOI] [PubMed] [Google Scholar]
- Lorence J. H., Nester E. W. Multiple molecular forms of chorismate mutase in Bacillus subtillis. Biochemistry. 1967 May;6(5):1541–1553. doi: 10.1021/bi00857a041. [DOI] [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- MITSUHASHI S., DAVIS B. D. Aromatic biosynthesis. XIII. Conversion of quinic acid to 5-dehydroquinic acid by quinic dehydrogenase. Biochim Biophys Acta. 1954 Oct;15(2):268–280. doi: 10.1016/0006-3002(54)90069-4. [DOI] [PubMed] [Google Scholar]
- Morell H., Sprinson D. B. Shikimate kinase isoenzymes in Salmonella typhimurium. J Biol Chem. 1968 Feb 10;243(3):676–677. [PubMed] [Google Scholar]
- Nasser D., Nester E. W. Aromatic amino acid biosynthesis: gene-enzyme relationships in Bacillus subtilis. J Bacteriol. 1967 Nov;94(5):1706–1714. doi: 10.1128/jb.94.5.1706-1714.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nester E. W., Lorence J. H., Nasser D. S. An enzyme aggregate involved in the biosynthesis of aromatic amino acids in Bacillus subtilis. Its possible function in feedback regulation. Biochemistry. 1967 May;6(5):1553–1563. doi: 10.1021/bi00857a042. [DOI] [PubMed] [Google Scholar]
- Nishioka Y., Demerec M., Eisenstark A. Genetic analysis of aromatic mutants of Salmonella typhimurium. Genetics. 1967 Jun;56(2):341–351. doi: 10.1093/genetics/56.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittard J., Wallace B. J. Distribution and function of genes concerned with aromatic biosynthesis in Escherichia coli. J Bacteriol. 1966 Apr;91(4):1494–1508. doi: 10.1128/jb.91.4.1494-1508.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Wallace B. J., Pittard J. Genetic and biochemical analysis of the isoenzymes concerned in the first reaction of aromatic biosynthesis in Escherichia coli. J Bacteriol. 1967 Jan;93(1):237–244. doi: 10.1128/jb.93.1.237-244.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]


