Abstract
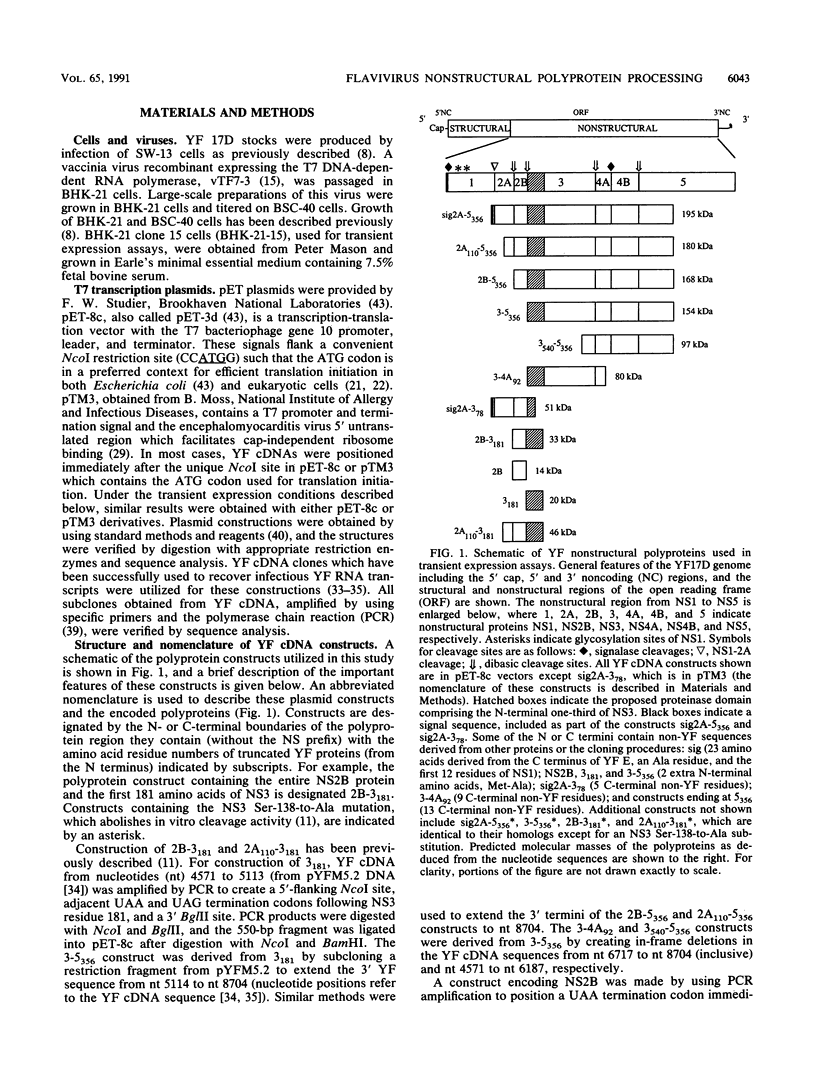
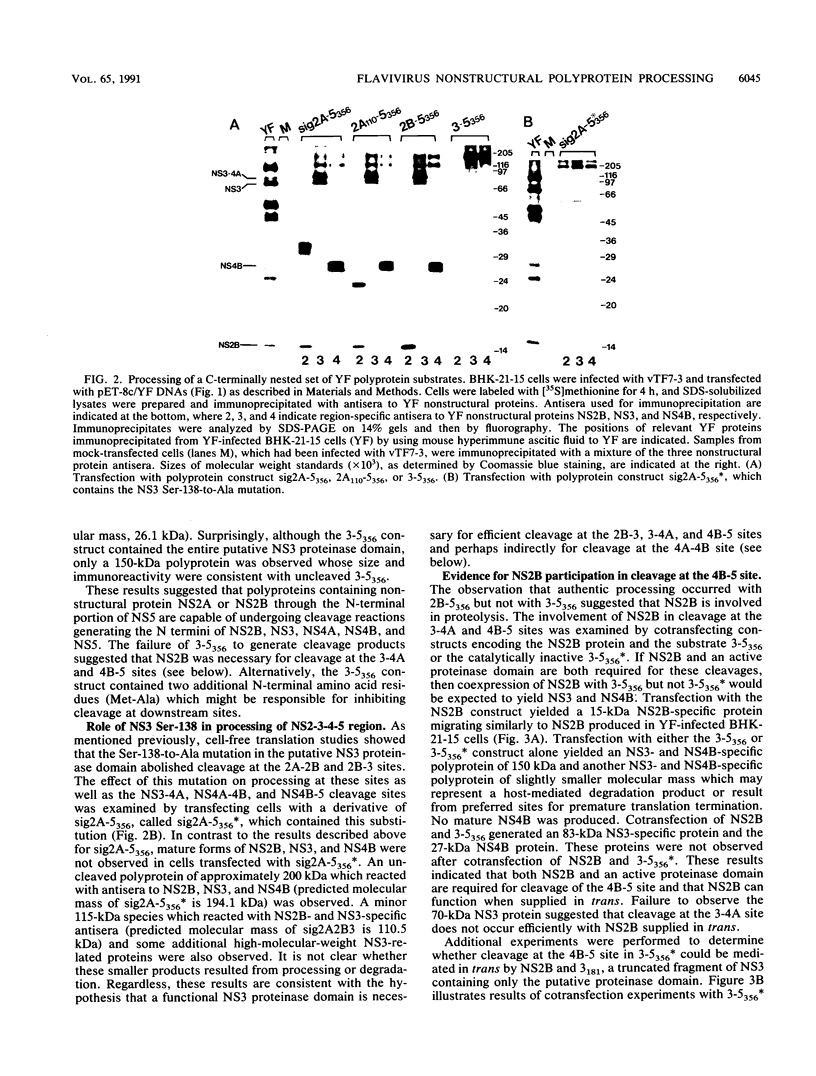
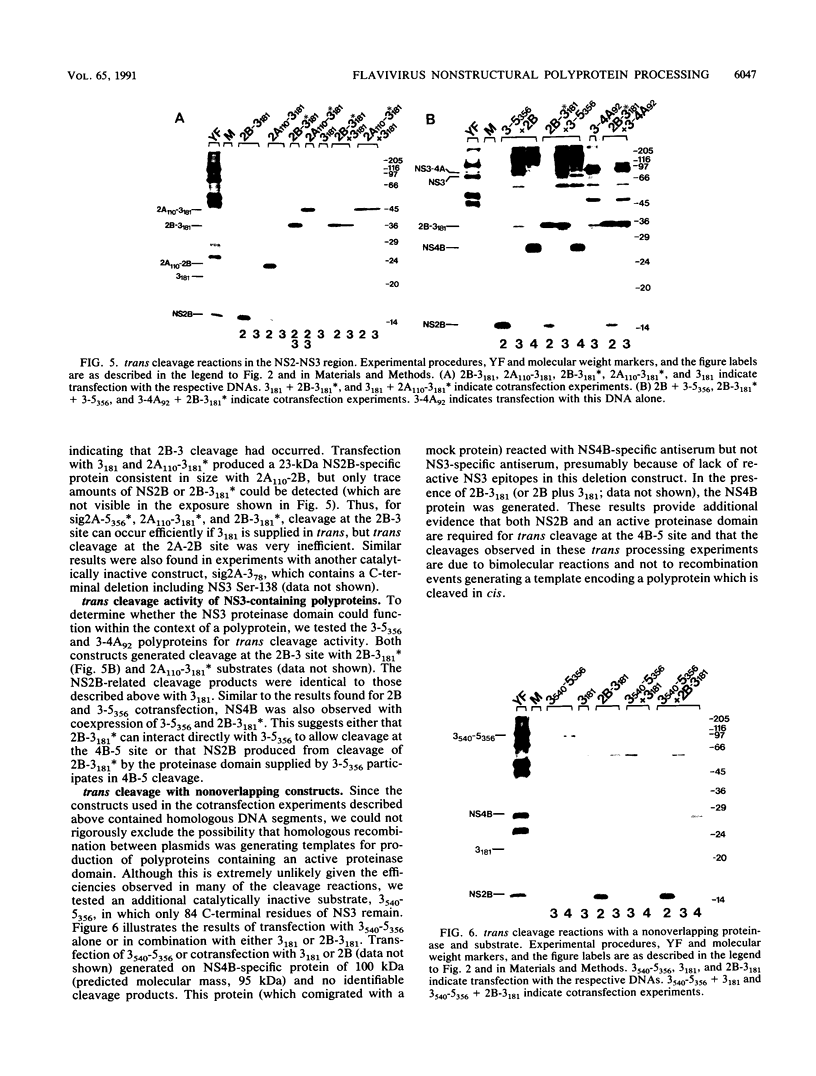
The vaccinia virus-T7 transient expression system was used to further examine the role of the NS3 proteinase in processing of the yellow fever (YF) virus nonstructural polyprotein in BHK cells. YF virus-specific polyproteins and cleavage products were identified by immunoprecipitation with region-specific antisera, by size, and by comparison with authentic YF virus polypeptides. A YF virus polyprotein initiating with a signal sequence derived from the E protein fused to the N terminus of NS2A and extending through the N-terminal 356 amino acids of NS5 exhibited processing at the 2A-2B, 2B-3, 3-4A, 4A-4B, and 4B-5 cleavage sites. Similar results were obtained with polyproteins whose N termini began within NS2A (position 110) or with NS2B. When the NS3 proteinase domain was inactivated by replacing the proposed catalytic Ser-138 with Ala, processing at all sites was abolished. The results suggest that an active NS3 proteinase domain is necessary for cleavage at the diabasic nonstructural cleavage sites and that cleavage at the proposed 4A-4B signalase site requires prior cleavage at the 4B-5 site. Cleavages were not observed with a polyprotein whose N terminus began with NS3, but cleavage at the 4B-5 site could be restored by supplying the the NS2B protein in trans. Several experimental results suggested that trans cleavage at the 4B-5 site requires association of NS2B and the NS3 proteinase domain. Coexpression of different proteinases and catalytically inactive polyprotein substrates revealed that trans cleavage at the 2B-3 and 4B-5 sites was relatively efficient when compared with trans cleavage at the 2A-2B and 3-4A sites.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bazan J. F., Fletterick R. J. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology. 1989 Aug;171(2):637–639. doi: 10.1016/0042-6822(89)90639-9. [DOI] [PubMed] [Google Scholar]
- Bell J. R., Kinney R. M., Trent D. W., Lenches E. M., Dalgarno L., Strauss J. H. Amino-terminal amino acid sequences of structural proteins of three flaviviruses. Virology. 1985 May;143(1):224–229. doi: 10.1016/0042-6822(85)90110-2. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Cauchi M. R., Henchal E. A., Wright P. J. The sensitivity of cell-associated dengue virus proteins to trypsin and the detection of trypsin-resistant fragments of the nonstructural glycoprotein NS1. Virology. 1991 Feb;180(2):659–667. doi: 10.1016/0042-6822(91)90079-q. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., Hahn C. S., Galler R., Rice C. M. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., McCourt D. W., Rice C. M. Production of yellow fever virus proteins in infected cells: identification of discrete polyprotein species and analysis of cleavage kinetics using region-specific polyclonal antisera. Virology. 1990 Jul;177(1):159–174. doi: 10.1016/0042-6822(90)90470-c. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., McCourt D. W., Rice C. M. Yellow fever virus proteins NS2A, NS2B, and NS4B: identification and partial N-terminal amino acid sequence analysis. Virology. 1989 Mar;169(1):100–109. doi: 10.1016/0042-6822(89)90045-7. [DOI] [PubMed] [Google Scholar]
- Falgout B., Chanock R., Lai C. J. Proper processing of dengue virus nonstructural glycoprotein NS1 requires the N-terminal hydrophobic signal sequence and the downstream nonstructural protein NS2a. J Virol. 1989 May;63(5):1852–1860. doi: 10.1128/jvi.63.5.1852-1860.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B., Pethel M., Zhang Y. M., Lai C. J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991 May;65(5):2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst T. R., Niles E. G., Studier F. W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Donchenko A. P., Koonin E. V., Blinov V. M. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 1989 May 25;17(10):3889–3897. doi: 10.1093/nar/17.10.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989 Jun 26;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Lai C. J. Cleavage of dengue virus NS1-NS2A requires an octapeptide sequence at the C terminus of NS1. J Virol. 1990 Sep;64(9):4573–4577. doi: 10.1128/jvi.64.9.4573-4577.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore J., De Geus B., Jackson R. J., Pouwels P. H., Enger-Valk B. E. Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J Gen Virol. 1988 Jul;69(Pt 7):1627–1636. doi: 10.1099/0022-1317-69-7-1627. [DOI] [PubMed] [Google Scholar]
- Katoh I., Ikawa Y., Yoshinaka Y. Retrovirus protease characterized as a dimeric aspartic proteinase. J Virol. 1989 May;63(5):2226–2232. doi: 10.1128/jvi.63.5.2226-2232.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Markoff L. In vitro processing of dengue virus structural proteins: cleavage of the pre-membrane protein. J Virol. 1989 Aug;63(8):3345–3352. doi: 10.1128/jvi.63.8.3345-3352.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P. W. Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology. 1989 Apr;169(2):354–364. doi: 10.1016/0042-6822(89)90161-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P. W., McAda P. C., Dalrymple J. M., Fournier M. J., Mason T. L. Expression of Japanese encephalitis virus antigens in Escherichia coli. Virology. 1987 Jun;158(2):361–372. doi: 10.1016/0042-6822(87)90208-x. [DOI] [PubMed] [Google Scholar]
- Moss B., Elroy-Stein O., Mizukami T., Alexander W. A., Fuerst T. R. Product review. New mammalian expression vectors. Nature. 1990 Nov 1;348(6296):91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- Nowak T., Färber P. M., Wengler G., Wengler G. Analyses of the terminal sequences of West Nile virus structural proteins and of the in vitro translation of these proteins allow the proposal of a complete scheme of the proteolytic cleavages involved in their synthesis. Virology. 1989 Apr;169(2):365–376. doi: 10.1016/0042-6822(89)90162-1. [DOI] [PubMed] [Google Scholar]
- Preugschat F., Yao C. W., Strauss J. H. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J Virol. 1990 Sep;64(9):4364–4374. doi: 10.1128/jvi.64.9.4364-4374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Aebersold R., Teplow D. B., Pata J., Bell J. R., Vorndam A. V., Trent D. W., Brandriss M. W., Schlesinger J. J., Strauss J. H. Partial N-terminal amino acid sequences of three nonstructural proteins of two flaviviruses. Virology. 1986 May;151(1):1–9. doi: 10.1016/0042-6822(86)90098-x. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Grakoui A., Galler R., Chambers T. J. Transcription of infectious yellow fever RNA from full-length cDNA templates produced by in vitro ligation. New Biol. 1989 Dec;1(3):285–296. [PubMed] [Google Scholar]
- Rice C. M., Lenches E. M., Eddy S. R., Shin S. J., Sheets R. L., Strauss J. H. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985 Aug 23;229(4715):726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Ruiz-Linares A., Cahour A., Després P., Girard M., Bouloy M. Processing of yellow fever virus polyprotein: role of cellular proteases in maturation of the structural proteins. J Virol. 1989 Oct;63(10):4199–4209. doi: 10.1128/jvi.63.10.4199-4209.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Speight G., Coia G., Parker M. D., Westaway E. G. Gene mapping and positive identification of the non-structural proteins NS2A, NS2B, NS3, NS4B and NS5 of the flavivirus Kunjin and their cleavage sites. J Gen Virol. 1988 Jan;69(Pt 1):23–34. doi: 10.1099/0022-1317-69-1-23. [DOI] [PubMed] [Google Scholar]
- Speight G., Westaway E. G. Carboxy-terminal analysis of nine proteins specified by the flavivirus Kunjin: evidence that only the intracellular core protein is truncated. J Gen Virol. 1989 Aug;70(Pt 8):2209–2214. doi: 10.1099/0022-1317-70-8-2209. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Vos P., Verver J., Jaegle M., Wellink J., van Kammen A., Goldbach R. Two viral proteins involved in the proteolytic processing of the cowpea mosaic virus polyproteins. Nucleic Acids Res. 1988 Mar 25;16(5):1967–1985. doi: 10.1093/nar/16.5.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G., Czaya G., Färber P. M., Hegemann J. H. In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J Gen Virol. 1991 Apr;72(Pt 4):851–858. doi: 10.1099/0022-1317-72-4-851. [DOI] [PubMed] [Google Scholar]
- Wengler G., Wengler G., Nowak T., Castle E. Description of a procedure which allows isolation of viral nonstructural proteins from BHK vertebrate cells infected with the West Nile flavivirus in a state which allows their direct chemical characterization. Virology. 1990 Aug;177(2):795–801. doi: 10.1016/0042-6822(90)90552-3. [DOI] [PubMed] [Google Scholar]
- Westaway E. G. Flavivirus replication strategy. Adv Virus Res. 1987;33:45–90. doi: 10.1016/s0065-3527(08)60316-4. [DOI] [PubMed] [Google Scholar]
- Ypma-Wong M. F., Dewalt P. G., Johnson V. H., Lamb J. G., Semler B. L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988 Sep;166(1):265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- de Groot R. J., Hardy W. R., Shirako Y., Strauss J. H. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 1990 Aug;9(8):2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]