Abstract
After landing on a wet stigma, pollen grains hydrate and germination generally occurs. However, there is no certainty of the pollen tube growth through the style to reach the ovary. The pistil is a gatekeeper that evolved in many species to recognize and reject the self-pollen, avoiding endogamy and encouraging cross-pollination. However, recognition is a complex process, and specific factors are needed. Here the isolation and characterization of a stigma-specific protein from N. alata, NaStEP (N. alata Stigma Expressed Protein), that is homologous to Kunitz-type proteinase inhibitors, are reported. Activity gel assays showed that NaStEP is not a functional serine proteinase inhibitor. Immunohistochemical and protein blot analyses revealed that NaStEP is detectable in stigmas of self-incompatible (SI) species N. alata, N. forgetiana, and N. bonariensis, but not in self-compatible (SC) species N. tabacum, N. plumbaginifolia, N. benthamiana, N. longiflora, and N. glauca. NaStEP contains the vacuolar targeting sequence NPIVL, and immunocytochemistry experiments showed vacuolar localization in unpollinated stigmas. After self-pollination or pollination with pollen from the SC species N. tabacum or N. plumbaginifolia, NaStEP was also found in the stigmatic exudate. The synthesis and presence in the stigmatic exudate of this protein was strongly induced in N. alata following incompatible pollination with N. tabacum pollen. The transfer of NaStEP to the stigmatic exudate was accompanied by perforation of the stigmatic cell wall, which appeared to release the vacuolar contents to the apoplastic space. The increase in NaStEP synthesis after pollination and its presence in the stigmatic exudates suggest that this protein may play a role in the early pollen–stigma interactions that regulate pollen tube growth in Nicotiana.
Keywords: Kunitz proteinase inhibitor homologue, Nicotiana, pollen–pistil interactions, pollination
Introduction
Once a pollen grain lands on the stigma, several recognition cascades are activated in the pistil. Although many types of pollen may land on the stigmatic surface, usually only pollen of the same or closely related species will be accepted. Compatible pollen grains germinate on the stigma surface, and the emerging pollen tubes grow through the extracellular matrix (ECM) of the transmitting tissue of the style toward the ovary. The ECM is rich in lipids, carbohydrates, amino acids, glycoproteins, and polysaccharides (Clarke et al., 1979; Gleeson and Clarke, 1979; Herrero and Dickinson, 1979; Gell et al., 1986; Wu et al., 1995; Cheung et al., 1995). The role of the pistil is multifunctional. It is thought to provide factors that act as directional cues, as well as providing physical and nutritional support for pollen tube growth. The pistil also produces factors required for recognition and rejection of incompatible pollen (Lord and Sanders, 1992). In general, all the pistil side factors involved in pollen–pistil interactions need to be secreted to the ECM. Arabinogalactan proteins (AGPs) involved in pollen tube growth, such as pistil extensin-like protein III (PELPIII) and transmitting tract-specific glycoprotein (TTS), comprise an abundant component of the pistil ECM (Goldman et al., 1992; Cheung et al., 1995; Wu et al., 1995; Schultz et al., 1997; Graaf et al., 2004; Cruz-García et al., 2005). The S-RNases required in self-incompatibility (SI), one of the best characterized pollen–pistil interactions (Kao and Tsukamoto, 2004; Takayama and Isogai, 2005), are major protein components of this matrix as well. In Nicotiana, besides S-RNases, the other stylar proteins required for the biochemical pathway of pollen rejection in Solanaceae, such as HT-B (McClure et al., 1999; O'Brien et al., 2002) and the 120 kDa glycoprotein (120K) (Hancock et al., 2005; Goldraij et al., 2006), are also secreted to the ECM.
In addition to those proteins present in the ECM, several proteins that may have implications in pollen–pistil interactions have been identified in stigmatic exudates. In fact, in wet-type stigma species (i.e. Nicotiana sp.), stigma germination and pollen tube growth depend on exudate components produced by stigmatic cells. Several of these identified proteins have homology to various classes of proteins but do not possess the implied activity. For example, pistil pollen allergen-like (PPAL) and lipid transfer protein (LTP) are two proteins present in the stigmatic exudate of N. tabacum (Nieuwland et al., 2005). Although PPAL has homology to β-expansins suggesting it may be involved in cell wall loosening (Pezzotti et al., 2002), no cell wall-loosening activity has been demonstrated. Instead, LTP, a cysteine-rich protein, has cell wall-loosening activity even though it shares no homology to β-expansins (Nieuwland et al., 2005). Proteins related to pathogen resistance proteins such as thaumatin-like proteins, β-1-3 glucanases, and chitinases have also been identified in stigmatic exudates, but their expression characteristics suggest that they may have a role in plant sexual reproduction as well as plant defence (Leung, 1992; Kuboyama et al., 1997; Kuboyama, 1998). In particular, proteinase inhibitors, known to have a role in plant defence, have been identified in stigma exudates. The processing of the N. alata serine proteinase inhibitors (Na-PI II and Na-PI IV) from precursor protein to mature peptide is correlated with floral development, suggesting an alternative role (Atkinson et al., 1993; Miller et al., 2000).
How and if some of these proteins in both the stigmatic exudates and the ECM contribute to either pollen tube growth, pollen tube recognition by the pistil (implying acceptance or rejection of pollen tubes), or both is still not clear. While attempting to identify genes that could be involved in pollen–pistil interactions, NaStEP (N. alata Stigma Expressed Protein), a Kunitz proteinase inhibitor homologue specifically expressed in N. alata stigmas, was isolated. Activity gel assays show that NaStEP is not a serine proteinase inhibitor, implying minimal involvement in plant defense. Protein blot and immunohistochemical analysis showed that NaStEP has a restricted localization defense in mature stigmas of SI Nicotiana and is deposited into the stigmatic cell vacuoles. When N. alata stigmas were pollinated with N. alata or N. tabacum pollen, NaStEP synthesis was stimulated and released onto the stigma exudate by a novel mechanism involving cell rupture. Taken together, the results suggest that NaStEP may play a role in the early pollen–stigma interactions that regulate pollen tube growth in Nicotiana.
Materials and methods
Plant material
Nicotiana alata (S105S105, SC10SC10, and SA2SA2 genotypes), N. glauca, and N. tabacum cv Praecox have been previously described (Murfett et al., 1994, 1996; Beecher and McClure, 2001). Nicotiana plumbaginifolia (inventory number TW107, accession 43B), N. longiflora (inventory number TW79, accession 30A), N. forgetiana (inventory number TW50, 21A), and N. bonariensis (inventory number TW28, accession 11) were obtained from the US Tobacco Germplasm Collection (Crops Research Laboratory, USDA-ARS, Oxford, NC, USA). Nicotiana benthamiana has been previously described (Juárez-Díaz et al., 2006). Nicotinan alata cv Breakthrough was obtained from Thompson and Morgan, Jackson, NJ, USA. Nicotiana alata (SC10S105, and SA2S105 genotypes) have been previously described (Goldraij et al., 2006). All plant material was grown under greenhouse conditions.
cDNA library construction and screening
A cDNA library was constructed with the SMART cDNA library construction kit (BD Biosciencies, Palo Alto, CA, USA) using a poly(A)+ RNA from SI N. alata pistils and cloned in λTriplEx2™ according to the manufacturer's instructions. S-RNase, HT-B, and 120K cDNA probes were used to identify and discard some previously identified sequences implicated in SI. A total of ∼31 000 colonies were screened. RNA corresponding to some of the clones was determined to be strongly expressed in mature SI N. alata pistils and undetectable in mature SC N. plumbaginifolia pistils. Partial sequencing and RNA blot analyses were used to identify nine pistil-specific sequences. Eight sequences were identical, corresponding to NaStEP, and one was slightly different, NaSoEP (N. alata Sexual Organs Expressed Protein).
Sequence analysis
The nucleotide sequences of NaStEP and NaSoEP are accessible at GenBank under the accession numbers EU253563 and EU253564, respectively. For homology searches, the NCBI GenBank BLAST server 2.2.12 was used (Altschul et al., 1997), through the BLASTX program provided by the San Diego Supercomputer Center (http://workbench.sdsc.edu). Protein sequences were aligned using the ClustalX program (Thompson et al., 1997). Protein identities were calculated with ALIGN (http://workbench.sdsc.edu; Myers and Miller, 1988). The signalP program (http://us.expasy.org/) was used for signal peptide and cleavage site predictions using neural networks (Bendtsen et al., 2004) and hidden Markov models (Nielsen and Krogh, 1998) trained on eukaryotes. Protein subcellular localization was predicted with PA-SUB (Lu et al., 2004; www.cs.ualberta.ca/∼bioinfo/PA/Sub/) or with the LOCSVMPSI 1.3 server (Xie et al., 2005; http://bioinformatics.ustc.edu.cn).
RNA blot analysis
Leaves, stems, sepals, petals, anthers, stigmas, styles, and stigmas plus styles (pistils excluding the ovary) were ground under liquid nitrogen, and the total RNA was prepared and separated in 2% agarose formaldehyde gels as described by McClure et al. (1990). RNA was blotted onto Hybond N+ (Amersham) and methylene blue stained to check for equal loading. Filters were blocked in hybridization solution (0.3 M sodium phosphate buffer pH 7.2, 7% SDS, and 1 mM EDTA pH 8) for at least 1 h. The blots were then hybridized overnight with [32P]cDNAs probes at 68 °C, and stringent washes were performed in 0.1× SSC, 0.1% SDS at 68 °C. Hybridization was detected by autoradiography.
Cloning and expression of NaStEP
The coding region from the NaStEP cDNA was amplified with NaStEPBamHI-F (5′CTCTGGATCCATGCCCACTACTACTGATGATGA3′) and NaStEPEcoRI-R (5′CGCGGAATTCCTAGGTTACCACAAACCTAA3′) primers and ligated in-frame with the glutathione S-transferase (GST) gene in pGEX-4T-2 (Amersham Pharmacia Biotech, Piscataway, NJ, USA). After transformation into Escherichia coli BL21-CodonPlus (RIL) (Stratagene), cells were induced with isopropyl-β-D-1-thiogalactopyranoside (IPTG) and fusion proteins purified by batch affinity chromatography with glutathione–Sepharose 4B (Amersham Pharmacia Biotech) as recommended by the manufacturer.
Protein isolation
Roots, stems, leaves, flowers, sepals, petals, pistils, styles, stigmas, ovaries, and anthers were ground under liquid nitrogen and homogenized in 50 mM NaCl, 50 mM TRIS-HCl pH 8, and 1% β-mercaptoethanol. Extracts were clarified by centrifugation for 10 min at 13 000 g at 4 °C and supernatants were kept at –80 °C until use. Protein concentration was estimated by the method of Bradford (1976) using bovine serum albumin (BSA) as the standard.
High-performance liquid chromatography (HPLC) and ion exchange chromatography
For NaStEP purification, stigma crude extracts from SI N. alata were prepared as indicated above, and then incubated with a quaternary ammonium Q–Sepharose fast flow anion exchange resin (Sigma) equilibrated in 50 mM TRIS-HCl pH 8.0 [equilibration buffer (EB)] for 1 h at room temperature. The resin was then washed 10 times with five bed volumes of EB. Elution was carried out using an increasing NaCl gradient (50, 100, 150, 200, 250, 300, 400, 500, 600, 700, 800, 900, and 1000 mM prepared in EB). Fractions containing most of the specifically bound NaStEP were pooled, and the EB plus the salt was changed to a new buffer [50 mM CH3COONa pH 5.5 (NB)]. Partially purified NaStEP in NB was bound again to the same resin and fractions were eluted as described above using 50 mM CH3COONa pH 5.5 plus the same increasing NaCl series. NaStEP-enriched fractions were pooled and analysed by HPLC using a Superdex 200 molecular exclusion column (Amersham-Biosciences). The column was eluted with NB at a flow rate of 0.5 ml min−1. Protein fractions were separated by SDS–PAGE, and gels were stained with silver nitrate or blotted to nitrocellulose for immunoanalysis.
Two-dimensional gel electrophoresis
Two-dimensional PAGE [isoelectric focusing (IEF) in the first dimension using Immobiline™ drystrip (pH 3–10) (Amersham Biosciences) and SDS–PAGE in the second dimension] was performed by using the Multiphor II system (Amersham). Protein samples were extracted as described above, precipitated with 10% trichloroacetic acid (TCA) in acetone, and dissolved in 125 μl of hydration buffer {4% CHAPS (3-[3-cholamidopropyl) dimethylammonio]-1-propane-sulphonate, 7 M urea, 2% IPG buffer, 10 mM dithiotreitol (DTT) and 0.002% bromophenol blue}. Protein samples were applied to dry polyacrylamide gel strips formulated with immobilized pH gradients and the gel strips were allowed to hydrate at room temperature for at least 16 h. IEF was performed in a Multiphor II system at 500 V for 1 h for the first phase, and for phase two the voltage was raised to 3500 V for 90 min. The voltage remained at 3500 V throughout phase three for 90 min, with an upper voltage limit of 5000 V. The gel strips were then dipped in SDS equilibration solution (50 mM TRIS-HCl pH 8.8, 6 M urea, 30% glycerol, 2% SDS, and a few traces of bromophenol blue) plus DTT (100 mg per 10 ml SDS equilibration buffer) for 15 min. The first equilibration solution was decanted, and iodoacetamide-containing equilibration solution was added to the strips (250 mg per 10 ml of SDS equilibration buffer) for 15 min. Second dimension SDS–PAGE was performed in a 15% gel (Laemmli, 1970), which was subsequently silver stained or blotted to nitrocellulose membranes (Millipore) for immunoanalysis.
Protein gel blot analysis
The affinity-purified polyclonal antiserum was prepared against the NaStEP peptide WEGSKDGMPVKFFT (Sigma). Equal amounts of protein were separated by 12% or 15% SDS–PAGE and blotted to nitrocellulose. Immunoblotting was performed essentially as described (Cruz-Garcia et al., 2005). Primary antibody dilutions were: anti-NaStEP 1:15 000 and anti-SA2-RNase 1:100 000.
Proteinase inhibitory activity assays
Proteinase inhibitory activity was assayed on SDS–polyacrylamide gels using a modified version of the method of Hou and Lin (1998). The protein samples were dissolved in the sample buffer without either DTT or boiling. After electrophoresis, gels were soaked twice in 25% isopropanol in 20 mM TRIS-HCl pH 8.0 for 10 min each prior to any additional treatment. The gel was then dipped in 20 mM TRIS-HCl buffer pH 8.0 for 30 min. Next the gel was placed in a trypsin solution [50 units of bovine trypsin (Sigma) in 1 ml of 50 mM TRIS-HCl pH 8.0, 50 mM CaCl2] for 30 min at 37 °C. The gel was rinsed in the same buffer before incubation in a fresh substrate–dye solution and then incubated for 50 min in the dark at 37 °C with 80 ml of the substrate–dye solution. The substrate–dye solution consisted of 20 mg of N-acetyl-DL-phenylalanine β-naphthyl ester (APNE, Sigma) in 8 ml of N,N-dimethylformamide that was brought to 80 ml with 72 ml of 50 mM TRIS-HCl pH 8.0, 50 mM CaCl2 in which 40 mg of tetrazotized O-dianisidine was dissolved. The gel was destained with 10% acetic acid for 30 min. The soybean trypsin inhibitor (BioChemika) was used as a positive control.
Sequential protein extractions
Nicotiana alata cv Breakthrough stigmas plus styles were treated as described previously (Wu et al., 2000; Juárez-Díaz et al., 2006), with the following modifications. Nicotiana alata cv Breakthrough stigmas plus styles were submerged sequentially in two different salt concentration buffers for 3 h at 4 °C: low-salt buffer (100 mM TRIS-HCl, pH 8.0) and high-salt buffer (400 mM NaCl, 40 mM TRIS-HCl, pH 8.0). Finally, the stigmas plus styles were ground in liquid N2, homogenized in 400 mM NaCl, 40 mM TRIS-HCl, pH 8.0, 1% Triton X-100, and then clarified at 13 000 g for 30 min.
Microscopy and immunolocalization
Mature pistils from SI N. alata SC10SC10, SI N. alata SA2SA2, SI N. alata S105S105, SI N. forgetiana, SC N. alata Breakthrough, SC N. longiflora, SC N. plumbaginifolia, SC N. tabacum, SC N. glauca, and SC N. benthamiana were harvested and fixed in 4% (v/v) formaldehyde in phsophate-buffered saline (PBS), dehydrated in an ethanol series, and embedded in Paraplast Plus (Polysciences Inc., Warrington, PA, USA). Sections of 6–7 μm were blocked with PBS plus 3% BSA, 0.01% sodium azide, and 0.1% Triton X-100 for 4 h at 4 °C. Sections were incubated with either primary rabbit anti-NaStEP antibody (1:3000 or 1:6000 dilutions) or the primary anti-HT-B antibody (1:3000 dilution) at 4 °C overnight. Sections were then incubated with either secondary goat anti-rabbit Alexa 568-fluorochrome- (HT-B) conjugated or goat anti-rabbit fluorescein isothiocyanate (FITC)-fluorochrome- (NaStEP) conjugated antibodies for 4 h at 4 °C. Sections were observed using a confocal fluorescence microscope.
Immunogold and transmission electron microscopy
Nicotiana alata flowers were emasculated 48 h before anthesis. When the flowers matured, pollinations were performed with N. tabacum, N. plumbaginifolia, or N. alata Breakthrough pollen. Unpollinated and pollinated stigmas were collected after 10 h and then fixed in 3% formaldehyde and 0.5% glutaraldehyde in PBS for 2 h at room temperature. Tissues were then dehydrated in a series of ethanol concentrations (30, 40, 50, 60, 70, 80, 90, 96, and 100%) and infiltrated with LR White resin by gradually increasing its concentration (25, 50, 75, and 100%) then rinsed in PBS and post-fixed with 1% osmium tetroxide. For immunolocalization, 800 nm sections were hydrated and blocked in TBS (20 mM TRIS pH 7.6, 150 mM NaCl, 20 mM sodium azide, 1% Tween-20, 5% BSA, and 5% normal goat serum) for 1 h at room temperature and incubated with anti-NaStEP antibody (1:30) at 4 °C overnight. The sections were washed four times in TBS and incubated for 1 h at room temperature with 20 nm gold particle conjugates of rabbit IgG (Zymed) at a dilution of 1:20 in TBS. Before observation, sections were stained with aqueous 5% uranyl acetate followed by 3% lead citrate, and then observed with a transmission electron microscope (JEOL) at 60 kV.
Results
Identification of pistil-specific genes in SI Nicotiana alata
To identify factors that may contribute to early specific pollen–pistil interactions in SI Nicotiana, a cDNA library from SI N. alata stigma plus style was prepared and screened with S-RNase, HT-B, and 120K probes. After discarding the known sequences implicated in SI, clones that did not hybridize were sequenced and the ones that appeared more frequently were selected as good candidates. The screen identified one major sequence that met the necessary criteria, which was designated NaStEP. Figure 1a shows that NaStEP is highly expressed in N. alata pistils but is undetectable in SC N. plumbaginifolia, SC N. longiflora, and N. tabacum. Interestingly, all these species show DNA hybridization with a NaStEP probe in DNA blot assays (Supplementary Fig. S1 available at JXB online).
Fig. 1.
Species-specific and developmental expression of NaStEP and NaSoEP. (a) Detection of NaStEP in different genetic backgrounds. Total RNA (10 μg) from mature pistils of SI N. alata (Na) and SC N. plumbaginifolia (Np), N. longiflora (Nl), and N. tabacum (Nt) was blotted and hybridized with 32P-labelled NaStEP. (b) NaStEP expression in developing anthers and pistils of N. alata SC10SC10. Total RNA (10 μg) was loaded in each lane, blotted, and probed with 32P-labelled NaStEP. (c) NaSoEP expression in developing anthers and pistils of N. alata SC10SC10. Total RNA (10 μg) was loaded in each lane, blotted, and probed with 32P-labelled NaSoEP. (d) Densitometric analysis of NaStEP and NaSoEP expression during the development of styles plus stigmas. Numbers 1–5 represents the developmental stages of stigmas plus styles: 1, 0.5–1.0 cm; 2, 1–2 cm; 3, 2–3.5 cm; 4, 3.5–6 cm; and 5, 6 cm. (e) NaStEP and NaSoEP expression in non-sexual organs of N. alata. Total RNA (10 μg) was loaded in each lane, blotted, and probed with a 32P-labelled NaStEP (upper) or NaSoEP (lower). The same amount of stigma RNA was loaded as hybridization control. To ascertain equal RNA loading, blots were stained with methylene blue (lower sections of a–c and e).
The compatibility phenotypes of Nicotiana species used in this work are shown in Supplementary Table S1 at JXB online. Figure 1b shows that NaStEP displays stigma-specific expression. NaStEP transcripts were expressed in styles plus stigmas from early developing stages but not in developing anthers. NaStEP expression tends to increase towards maturity, since the highest expression was detected in mature pistils (Fig. 1b–d, 6 cm). RNA blot analysis with other tissues (stems, leaves, sepals, and petals) showed no expression of NaStEP (Fig. 1e, upper panel). Interestingly, a similar sequence isolated in the screening, NaSoEP (i.e. one of nine clones), was expressed in early pistil developmental stages, with a marked decrease in expression as the pistil matures (Fig. 1c, d). However, those very low levels of NaSoEP were still detectable from the 2–3.5 cm to 6 cm stages (Fig. 1c) in contrast to the complete lack of transcript detected in non-sexual organs (Fig. 1e, lower panel).
Fig. 6.
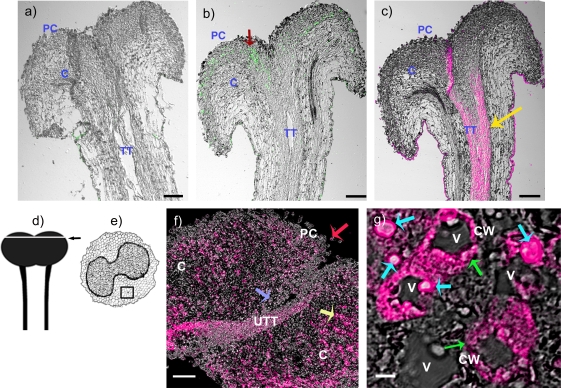
Immunohistochemical localization of NaStEP in stigmatic tissues of SC N. alata Breakthrough. (a) Stigma and style treated with pre-immune serum. (b) Stigma and style treated with anti-NaStEP antibody (green). (c) Stigma and style treated with anti-HT-B antibody (magenta). (d) and (e) Diagrams of stigma plus style of N. alata and a transverse section of a stigma. The arrow and square show zones where images were taken. (f) Stigma cross-sections showing NaStEP (magenta) in papillary cells (red arrow), secretory cells (yellow arrow), basal cells (blue arrow), and in a portion of the upper transmitting tract. (g) High magnification view of stigmatic secretory cells shows that NaStEP is in small bodies (blue arrows) and in the proximity of the cell wall (green arrows). All figures represent merges of immunostained and phase contrast images of stigmas plus styles. CW, cell wall; V, vacuole; TT, transmitting tissue; UTT, upper transmitting track; C, cortex; PC, papillae cell. Bars, 50 μm for (a), (b), (c) and 20 μm for (f) and 5 μm for (g).
Sequence analysis suggests that NaStEP and NaSoEP are vacuolar proteins. NaStEP and NaSoEP encode very similar proteins of 243 and 242 residues, respectively, with 70.9% sequence identity (Fig. 2). Although the predicted polypeptides are similar, NaStEP is acidic (pI 4.68), while NaSoEP is basic (pI 8.01). Both deduced polypeptides possess six cysteine residues that may form three disulphide bridges, putative N-glycosylation sites, and a predicted signal peptide (SP) (probability 0.994; Nielsen and Krogh, 1998; Bendtsen et al., 2004). Cleavage at the predicted site (Nielsen and Krogh, 1998; Bendtsen et al., 2004) shown in Fig. 2 would generate proteins of 24.37 kDa for NaStEP and 24.16 kDa for NaSoEP. The SP targets proteins for translocation across the endoplasmic reticulum (ER) membrane in eukaryotes (Von Heijne, 1990). Proteins translocated across the ER membrane devoid of specific retention signals in their sequence are by default secreted. NaStEP and NaSoEP both include a sequence-specific vacuolar sorting signal (ssVSS) NPIVL similar to the degenerate motif [N/L]-[P/L]-[I/S]-[R/P] [L/P/M] (Holwerda et al., 1992; Saalbach et al., 1996; Matsuoka and Nakamura, 1999; Frigerio et al., 2001). Although the LOCSVMpsi algorithm (Xie et al., 2005) predicts that both proteins are secreted to the extracellular space, the PA–SUB program (Lu et al., 2004) predicts that NaStEP and NaSoEP are likely to be either secreted to the extracellular space (100%) or sorted to the vacuole (99.9% and 99.75%, respectively). The signal peptide sequence and the putative vacuolar sorting signal are completely conserved in both proteins.
Fig. 2.
Alignment of the amino acid sequences of the cDNA encoding NaStEP and NaSoEP proteins. The black boxes under the alignment indicate the degree of sequence consensus. Arrowheads indicate the potential signal peptide cleavage sites. Cysteine residues are marked with an asterisk. Putative glycosylation sites are single underlined. Presumed vacuolar sorting signals are boxed. The antigenic region is double underlined. A dash in the sequence indicates a gap introduced to maintain a good alignment.
NaStEP and NaSoEP are most similar in sequence to Kunitz-type proteinase inhibitors. Alignment with a variety of serine, aspartic, and cysteine protease inhibitors showed identities ranging from 24.8% to 79.1%. Supplementary Fig. S2a at JXB online presents a phylogenetic tree showing six main clades of plant proteinase inhibitors or homologues, designated I–VI. According to the proteinase inhibitors classification of Rawlings et al. (2004), all these proteins belong to the I3 family (the plant Kunitz-type proteinase inhibitor). NaStEP and NaSoEP cluster with clade V together with NgPI, a putative proteinase inhibitor from N. glutinosa (KS Park et al., 2000), and double-headed cysteine and aspartic protease inhibitors from Solanum tuberosum and Solanum lycopersicum (tomato, formerly Lycopersicon esculentum). Supplementary Fig. S2b at JXB online shows an alignment of the proteins in clade V. All the proteins have a predicted signal peptide (Supplementary Fig. S2b) and an ssVSS in their N-terminal propeptides (NTPPs). The putative binding sites for trypsin-like (Arg99Phe100) and chymotrypsin-like (Leu144Leu145) inhibitors are conserved in the five aspartic proteinase inhibitors (Supplementary Fig. S2b, red boxes and arrowheads). Only three of the proteins shown in the alignment (PDI, PCPI, and S. tuberosum cathepsin D inhibitor) have been demonstrated to exhibit protease inhibitor activity. The percentage identities of NaStEP and NaSoEP with proteins that fall in clade V are shown in Supplementary Table S2 at JXB online.
NaStEP is a stigma-specific protein
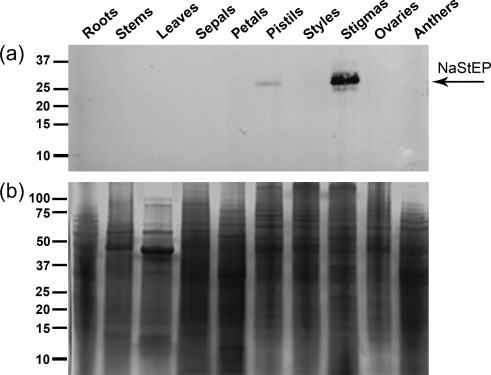
Because NaStEP was strongly expressed in mature stigmas, it seemed to be the most promising candidate for pollen–stigma interactions deserving further characterization. For these studies, a NaStEP-specific antibody was raised against the sequence WEGSKDGMPVKFFT, which shares only seven identical positions with NaSoEP. Antibody specificity was proved against a GST:NaStEP fusion protein. No binding was observed to GST alone or other E. coli proteins (Supplementary Fig. S3b, d at JXB online). The antibody reacted specifically with a GST:NaStEP fusion protein expressed in E. coli, confirming that the antibody recognized the NaStEP epitope (Supplementary Fig. S3a, c). Figure 3 shows antibody detection of a 32 kDa protein in SC N. alata cv Breakthrough, a natural mutant that does not express any S-RNase (Murfett et al., 1996; Beecher and McClure, 2001), but keeps the genetic background of an SI plant. Because of this, no differences are expected in the NaStEP cell biology with respect to the wild-type SI N. alata. The protein was evident in the mature stigma and pistils but not in any other sexual or non-sexual organ. Protein blots of stigma polypeptides separated by 2D PAGE were used to test whether the antibody detects only NaStEP and does not cross-react with NaSoEP. Since mature NaStEP has a predicted pI of 4.68 and mature NaSoEP a predicted pI of 8.01, an anti-SA2-RNase (i.e. pI 8.016) antibody was used as a control. The results showed that the anti-NaStEP antibody detects a small family of acidic polypeptides in extracts of SI N. alata SA2SA2, but shows no cross-reaction with any basic polypeptide (Fig. 4). Deglycosylation assays performed with peptide-N-glycosidase F (PNGase-F) (Supplementary Fig. S4c at JXB online), and concanavalin A (Con A) binding experiments (Supplementary Fig. S4a) revealed that some of these acidic polypeptides are glycosylated forms of NaStEP.
Fig. 3.
Immunoanalysis of NaStEP in N. alata organs. Equal amounts of total protein (10 μg) of roots, stems, leaves, sepals, petals, pistils, styles, stigmas, ovaries, and anthers were separated by SDS–PAGE. (a) Proteins transferred onto nitrocellulose and immunostained with anti-NaStEP antibody. (b) Coomassie blue-stained proteins.
Fig. 4.
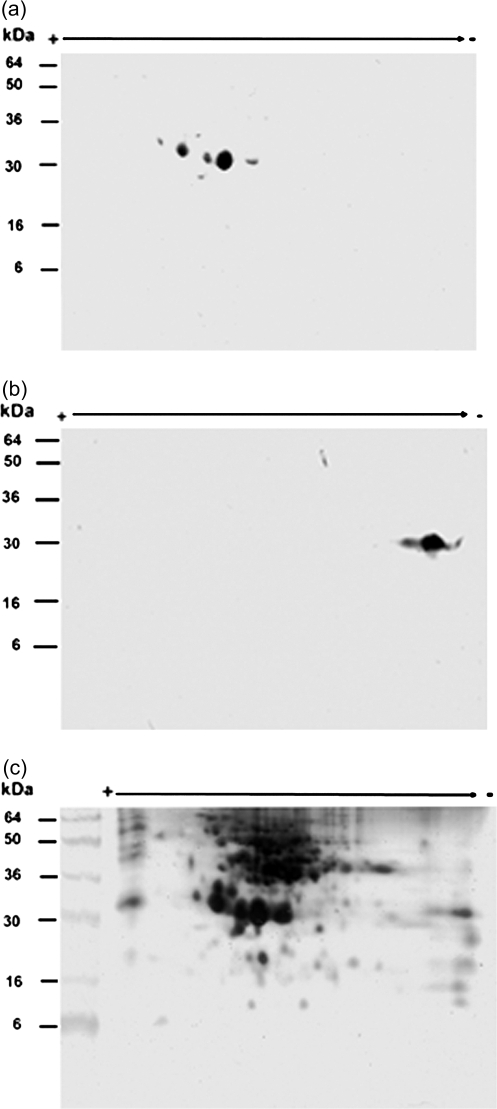
Two-dimensional gel analysis of NaStEP in stigmatic extracts. Total protein (50 μg) from matures styles plus stigmas of SI N. alata SA2SA2 were fractionated by two-dimensional electrophoresis. (a) Proteins transferred onto nitrocellulose and immunodetected with anti-NaStEP antibody. (b) Proteins blotted and immunodetected with the anti-SA2-RNase antibody. (c) Mature proteins silver stained.
Since NaStEP is a homologue of Kunitz-type proteinase inhibitors, we determined whether NaStEP is an active proteinase inhibitor. The native NaStEP protein was purified to homogeneity by ionic and gel filtration chromatography (Fig. 5a, b) and tested for proteinase inhibitory activity (Fig. 5c, d). In activity gel assays, the catalytic activity of the trypsin is visualized for the whole stained gel surface, except those areas where the inhibitor is present. Figure 5c shows that NaStEP does not exhibit inhibitory activity against trypsin as the soybean trypsin inhibitor does. Similar results were obtained when three NaStEP-enriched fractions from Con A affinity chromatography (fractions 3–5, Supplementary Fig. S4a, b at JXB online) were tested for proteinase inhibitory activity (data not shown).
Fig. 5.
Native NaStEP from N. alata stigmas and serine proteinase inhibitor activity assay. Purified HPLC fractions (29–36) of NaStEP were resolved by SDS–PAGE. (a) Proteins present in the HPLC fractions were transferred onto nitrocellulose and immunostained with the anti-NaStEP antibody. (b) Silver-stained proteins. (c) Inhibitory activity gel assay against trypsin. A 3 μg aliquot of the soybean trypsin inhibitor (STI) was run as a positive control. (d) A silver-stained replicate of the inhibitory activity assay gel.
To investigate further the tissue specificity of NaStEP localization, immunohistochemical analyses were performed in stigmatic tissues of SC N. alata cv Breakthrough. Longitudinal sections of unpollinated styles, including the stigma and the upper portion of the style, were immunostained with anti-NaStEP or anti-HT-B antibodies (Goldraij et al., 2006) and viewed by three-dimensional confocal microscopy. Figure 6 shows that NaStEP expression was restricted to the stigma and was especially conspicuous in papillary cells (Fig. 6b); while HT-B was restricted to the stylar transmitting tissue (Fig. 6c). A cross-section of the unpollinated stigma shows NaStEP immunostaining in the secretory zone (yellow arrow) and stigmatic basal cells (blue arrow), as well as in the papillary cells (red arrow) (Fig. 6f). In Fig. 6g, a higher magnification shows NaStEP localization in the periphery of the cells (green arrows) and inside subcellular vacuolated compartments (blue arrows).
NaStEP is predominantly expressed in SI Nicotiana species
We examined expression of NaStEP-like proteins in other Nicotiana species and N. alata accessions using the anti-NaStEP antibody. Figure 7a shows immunoblot results obtained with pistil extracts (i.e. stigma plus style) from SI N. forgetiana, N. bonariensis, and three SI and two SC accessions of N. alata. NaStEP-like proteins were readily detected in all cases, although the expression levels vary. As expected from the transcript level results shown in Fig. 1a, no NaStEP-like proteins were detected in the SC species N. plumbaginifolia, N. tabacum, N. longiflora, N. glauca, or Nicotiana alata×N. plumbaginifolia hybrids, which also express NaStEP. Similar results were obtained in immunolocalization experiments. Stigma cross-sections (including the upper portion of the transmitting tract) from two SI (N. alata and N. forgetiana) and four SC species (N. tabacum, N. plumbaginifolia, N. glauca, and N. benthamiana) show that NaStEP was strongly expressed in the stigmas of the SI species, but not in the SC species (Fig. 7b).
Fig. 7.
NaStEP-like proteins are expressed in stigmas of SI Nicotiana species but no in SC species. (a) Total proteins (10 μg) from stigmas and styles of SI N. bonariensis, SI N. forgetiana, SI N. alata SA2SA2, SI N. alata S105SC10, SI N. alata S105S105, SC N. alata S105SA2, SC N. alata Breakthrough, SI Hybrid 1 (SC N. plumbaginifolia×SI N. alata S105S105), SI Hybrid 2 (SC N. plumbaginifolia×SI N. alata SC10SC10), SC N. plumbaginifolia, SC N. tabacum, SC N. longiflora, and SC N. glauca were separated by SDS–PAGE and either Coomassie blue stained (left) or blotted to nitrocellulose for immunostaining with anti-NaStEP antibody (right). (b) Cross-sections of N. alata Breakthrough, SI N. alata SC10SC10, SI N. forgetiana, SC N. tabacum, SC N. plumbaginifolia, SC N. glauca, and SC N. benthamiana stigmas immunostained with anti-NaStEP. I and II: diagrams of a stigma plus style of N. alata and a transverse section of a stigma. The arrow and square show zones where images were taken. UTT, upper transmitting tissue; SC, secretory cells; PC, papillae cell. Bars, 50 mm.
Pollination-induced discharge of NaStEP to stigmatic exudate
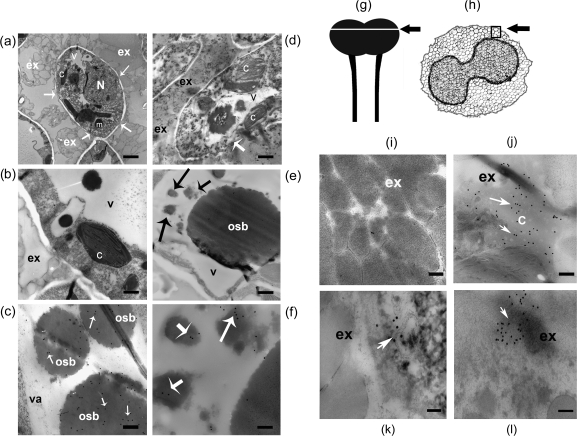
Electron microscopy and immunogold labelling experiments showed that NaStEP is discharged into the stigmatic exudate upon pollination (Fig. 8). Figure 8a shows a papillary cell cross-section from a mature/open flower, surrounded by copious masses of secretory droplets in the exudate. The cell wall shows discrete interruptions (arrows, Fig. 8a, b). The cytoplasm is displaced to the periphery of the papillary cells by large vacuoles containing osmiophilic bodies (electron-dense bodies, structures darkly stained with osmium tetroxide). These osmiophilic bodies were also present in smaller vesicles (Fig. 8b). Immunogold labelling with anti-NaStEP antibody showed that NaStEP accumulates mainly in these osmiophilic bodies (Fig. 8c). No NaStEP was detected in stigmatic exudate in unpollinated stigmas (Fig. 8i). However, after pollination, several changes were observed. Figure 8d shows that 10 h after pollination with N. tabacum pollen, the papillary cell cytoplasm appears different. Vacuoles and other organelles in these cells seem to degenerate and the osmiophilic bodies are smaller than in unpollinated stigmas. The papillary cell wall interruptions were more pronounced (arrow), and the large osmiophilic bodies did not label well with anti-NaStEP (Fig. 8e). Rather, labelling was localized in smaller vacuoles with smaller osmiophilic bodies (Fig. 8f).
Fig. 8.
NaStEP is discharged from the vacuole to the stigmatic exudate upon pollination. Unpollinated stigma from SC N. alata cv Breakthrough showing NaStEP localization in vacuoles and osmiophilic bodies. (a) Cross-section of mature papillae surrounded by copious exudate containing secretory droplets. Arrows show cell wall interruptions. (b) A papillary cell with a large vacuole and osmiophilic bodies. (c) Anti-NaStEP labelling of vacuolar electron-dense bodies. Arrows show immunogold secondary antibody labelling. Nicotiana alata cv Breakthrough stigma 10 h after pollination with N. tabacum pollen. (d) Degenerating papillary cells (arrow), showing abundant exudate and a disordered cytoplasm with abnormal organelles and disintegrated vacuoles. (e) Vacuole showing poor anti-NaStEP labelling of the osmiophilic bodies (arrows). (f) A close-up of the labelling shown in (e) (black arrows) showing anti-NaStEP labelling (white arrows) in smaller osmiophilic bodies. Stigmatic exudate of N. alata cv Breakthrough. (g) and (h) Diagrams of a stigma plus style of N. alata and a transverse section of a stigma. The arrow and square show zones where images were taken. (i) Unpollinated stigmas. (j) At 10 h after pollination with N. tabacum pollen. (k) At 10 h after pollination with N. plumbaginifolia pollen. (l) At 10 h after self-pollination. N, nucleus; m, mitochondria; c, chloroplast; v, vacuole; osb, osmiophilic bodies; ex, exudate. Bars, 500 nm for (a), 1 μm for (b) and (d), 240 nm for (c), 500 nm for (e), and 200 nm for (i)–(l).
After pollination, stigmatic exudate was more abundant. Cellular organelles were present in the exudate, indicating that some papillary cells discharged their contents (Fig. 8d). NaStEP labelling was abundant in the exudate after pollination with N. tabacum, N. plumbaginifolia, or N. alata cv Breakthrough pollen (Fig. 8j–l).
Synthesis and release of NaStEP onto the exudate is strongly stimulated by incompatible pollination
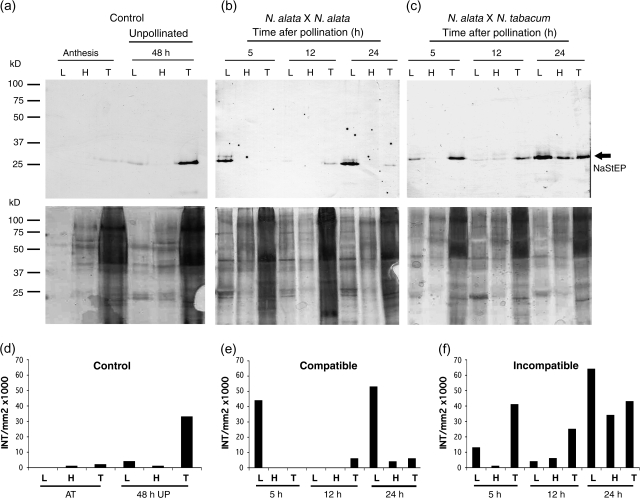
To investigate further the effect of compatible and incompatible pollination on NaStEP subcellular localization in the stigma, proteins present in the exudate were differentially extracted using sequential washes with low and high salt buffers as described (Wu et al., 2000; Juárez-Díaz et al., 2006). Under this procedure, the low salt buffer elutes soluble proteins, whereas more tightly bound proteins are released after the high salt buffer wash. The remaining cellular proteins were collected after N2 grinding. Sequential protein extractions were performed from N. alata cv Breakthrough stigmas, pollinated with either self or N. tabacum pollen. Soluble proteins were recovered at 5, 12, and 24 h after pollination (hap). Figure 9b and c shows that after pollination with either compatible or incompatible pollen, NaStEP was observed in the low salt fraction after 5 h, with a noteworthy increase at 24 hap. NaStEP was not detectable in the exudate from unpollinated stigmas 48 h after anthesis (Fig. 9a); however, the protein was clearly present in the non-soluble component (intracellular). Whether the levels of NaStEP before and after pollination are compared, it seems that the total NaStEP synthesis was increased 3.7-fold (24 h after incompatible pollination, compared with 48 h for the control) and 1.7-fold (24 h, compared with 48 h for the control) after compatible pollination (Fig. 9d–f). In addition, the deposition of NaStEP into the stigmatic exudate was stimulated 20-fold (24 h, compared with 48 h for the control) in incompatible pollination (Fig. 9f) and 12-fold (24 h, compared with 48 h for the control) in compatible pollinations (Fig. 9e). Based on these results, it was concluded that NaStEP synthesis and release onto the stigmatic exudates were both stimulated by pollination, with a stronger response after N. tabacum pollination.
Fig. 9.
Pollination induces synthesis and release of NaStEP into the N. alata stigmatic exudate. Total protein from exudate of unpollinated and pollinated stigmas of N. alata cv Breakthrough were sequentially extracted, separated by SDS–PAGE, and blotted to nitrocellulose for immunostaining with anti-NaStEP (top) or silver nitrate stained (bottom). Equal volumes (30 μl) were loaded in each lane. (a) Total protein from exudate of unpollinated stigmas at anthesis and at 48 h old. (b) Total proteins from exudate after 5, 12, and 24 h of self-pollination. (c) Total proteins from exudate after 5, 12, and 24 h of pollination with N. tabacum pollen. (d), (e), and (f) Densitometric analysis of NaStEP expression and its presence in the stigmatic exudate of N. alata before and after pollination with self- and N. tabacum pollen. L, low-salt wash; H, high-salt wash; T, proteins remaining in the stigma after washes.
Discussion
NaStEP and NaSoEP cDNAs were isolated as part of an effort to identify N. alata factors with high potential to contribute to pollen recognition. Although central and N-terminal regions of both proteins show extensive sequence similarity to Kunitz homologue proteinase inhibitors (Supplementaryy Fig. S2a, b at JXB online), no serine proteinase inhibitor activity was associated with NaStEP. The present data show that NaStEP is stored in the vacuole of stigmatic papillae and is released upon pollination. Moreover, NaStEP is not expressed in a selection of species that accept N. tabacum but is expressed in several species showing N. tabacum pollen rejection. DNA blot analysis (Supplementary Fig. S1 and data not shown) showed that SC species such as N. plumbaginifolia, N. glauca, and N. tabacum present positive hybridization against a NaStEP probe, suggesting that these species encode a gene orthologous to NaStEP, but they have lost the ability to express it, since neither transcripts (Fig. 1a) nor proteins (Fig. 7) were detected in their stigmas.
Proteinase inhibitors are common in plants and have been previously identified in sexual tissues of solanaceous plants. Atkinson et al. (1993) described Na-PI II, a stigma-specific proteinase inhibitor from N. alata that inhibits the digestive proteases of several insect species (Heath et al., 1997). Na-PI II is a type II proteinase inhibitor that is delivered to the vacuole as a 40.2 kDa precursor and processed to release six mature 6 kDa trypsin or chymotrypsin inhibitors (Atkinson et al., 1993; Heath et al., 1995; Lee et al., 1999). A trypsin inhibitor from N. attenuata has been shown to function in defence against herbivore attack (Zavala et al., 2004). LAT52, a protein with similarity to Kunitz-type proteinase inhibitors (Twell et al., 1989; McCormick et al., 1991), has been directly implicated in pollination. LAT52 is highly expressed in mature pollen and is essential for normal pollen hydration and tube growth (Muschietti et al., 1994). It binds to the receptor kinase LePRK2 in yeast-two hybrid and co-immunoprecipitation experiments, and it has been proposed to be a ligand in vivo (Tang et al., 2002). This interaction may activate a signalling cascade that regulates pollen tube growth (Johnson and Preuss, 2003).
We focused attention on NaStEP because its expression and pollination-induced synthesis and deposition onto the stigma surface are consistent with a role in early pollen–pistil interactions (Figs 1b, d, 3, 6–9). Although NaSoEP is a similar gene, its expression pattern in early anther and pistil development (Fig. 1c) is different and is more likely to be involved in flower development. Nevertheless, since there is low NaSoEP expression in mature stigmas, a possible role in pollination cannot be totally excluded. Stigma proteins have been previously implicated in the regulation of pollen tube growth. For example, a lily lipid transfer-like protein, SCA (stigma/stylar cysteine-rich adhesin) is necessary for adhesion of pollen tubes in the style (SY Park et al., 2000). SCA, together with a chemocyanin, displays chemotropic in vitro activity in lily pollen tube growth (SY Park et al., 2000; Kim et al., 2003; Park and Lord, 2003). LeSTIG1, the Solanum lycopersicum orthologue of the stigma-specific protein STIG1 from N. tabacum (Goldman et al., 1994), interacts with the extracellular domain of the tomato pollen-specific receptor kinases LePRK1 and LePRK2 and promotes pollen tube growth in vitro (Tang et al., 2004). As already mentioned, LAT52, a pollen protein, also binds LePRK2. Therefore, stigmatic LeSTIG1 might displace LAT52 after germination and contribute to growth regulation through the pistil (Tang et al., 2004). A role for STIG1 in the regulation of stigmatic exudate secretion has also been proposed in Petunia hybrida and N. tabacum (Verhoeven et al., 2005). Because the expression of NaStEP-like proteins is induced by pollination and preferentially expressed in SI species (Fig. 7), general roles in defence, as with Na-PI (Heath et al., 1997), are unlikely.
NaStEP may play a role in specific pollen–pistil interactions like LaT52, which also has similarity to Kunitz-type proteinase inhibitors (Twell et al., 1989; McCormick et al., 1991; Muschietti et al., 1994), but it is unlikely to encode a functional proteinase inhibitor (Lee and Lee, 2003). Since NaStEP-like protein expression is correlated with SI among species and its expression is strongly induced by pollination with incompatible pollen, it is hypothesized that it may also contribute to unilateral incompatibility. In this process, interspecific pollen from SC species is frequently inhibited directly on the SI stigma or very soon after pollen tube penetration into the style (de Nettancourt, 2001; Mutschler and Leidl, 1994). As a consequence of the interaction between NaStEP and pollen factors, pollen tubes to be rejected would trigger signal cascades that would inhibit their growth, as happens in other SI species such as Papaver rhoeas (McClure and Franklin-Tong, 2006). A clearer picture of the role of NaStEP in the pollen rejection response may be gained in future experiments involving RNA interference (RNAi)-mediated silencing of NaStEP.
Both NaStEP and NaSoEP contain the pentapeptide NPIVL near the N-terminus. This sequence is similar to the ssVSS required for vacuolar targeting of sweet potato sporamin (Matsuoka and Nakamura, 1999), Brazil nut 2S albumin (Saalbach et al., 1996), castor bean ricin (Frigerio et al., 2001), and barley aleurain (Holwerda et al., 1992). Although there is no strict vacuolar targeting consensus sequence, conserved physicochemical properties combined with limited sequence conservation are sufficient (Matsuoka and Nakamura, 1999). The NPIVL sequence in NaStEP and NaSoEP is a good match to the signal proposed by Matsuoka (2000): X1–X2–I/L–X3–X4, where X1 lacks a small hydrophobic side chain (N is preferred), X2 may not be acidic, X3 is any amino acid, and X4 is large and preferably hydrophobic. A similar motif is present in the NTPP of some potato Kunitz-type proteinase inhibitor precursors (Ishikawa et al., 1994), the aureusidin synthase (AmAS1) of snapdragon (Ono et al., 2006), the C-terminal propeptide of Na-PI from N. alata (Atkinson et al., 1993; Miller et al., 1999; Matsuoka, 2000), and the N. attenuata proteinase inhibitor (Zavala et al., 2004). Interestingly, if the vacuolar sorting signal is proteolytically processed in the ER, proteins can be secreted instead of sorted to the vacuole, as has been shown in N. alata (Johnson et al., 2006).
The process by which NaStEP is discharged from the vacuole is a mechanism for presenting potential pollination factors for interaction not described so far. The present results show dramatic changes in some stigmatic cells after pollination (Fig. 8). Perforation of the cell wall associated with the discharge of cytoplasmic contents into the stigmatic exudate was observed (Fig. 8d). This discharge includes organelles such as chloroplasts and vacuolar materials (the osmiophilic bodies containing NaStEP). The phenomenon seems to be a generalized process because it happens independently whether the N. alata Breakthrough stigmas are pollinated with self-pollen or with pollen from N. plumbaginifolia or N. tabacum (Fig. 8j–l and data not shown). However, the increase of NaStEP synthesis seen 24 hap with either compatible or incompatible pollen (Fig. 9) suggests that not all stigmatic cells suffer cell damage after pollination. Release of material into the stigmatic exudate has also been reported in P. hybrida and Solanum tuberosum (Herrero and Dickinson, 1979, 1980; MacKenzie et al., 1990). However, in these previous studies the dramatic degeneration of papillary cells 6 h after pollination seen here was not observed. Perhaps this was because these authors worked either with unpollinated stigmas before and after anthesis or with stigmas 2 h after pollination (Herrero and Dickinson, 1979, 1980). Whether these changes are due to a pollination-specific effect or an acceleration of stigma senescence, they represent a novel mechanism for delivery of organellar proteins such as NaStEP to a position where they are available for pollen–pistil interactions.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. DNA gel-blot analysis of NaStEP.
Figure S2. NaStEP and NaSoEP are homologous to Kunitz-type protease inhibitors.
Figure S3. Anti NaStEP antibody specificity.
Figure S4. Affinity chromatography of a protein crude extract bound to Con A-Sepharose resin and PNGase F deglycosylation analysis.
Table S1. Pollination phenotypes of the Nicotiana species studied.
Table S2. Identity percentages between NaStEP/NaSoEP and the proteinase inhibitors in clade V.
Appendix S1. Materials and methods and references.
Supplementary Material
Acknowledgments
We thank Laurel Fábila, Javier Andrés Juárez-Díaz, Yuridia Cruz González-Zamora, and Carlos Mújica for technical support. We also thank Dr K O'Grady and Dr William B Gurley for critical review of the manuscript. This material is based upon work partially supported by the Consejo Nacional de Ciencia y Tecnología (Grant 40614Q, 54586), Dirección General de Asuntos del Personal Académico-UNAM (Grant IN207406) and National Science Foundation under Grant No. 0315647 and 0614962.
Glossary
Abbreviations
- BSA
bovine serum albmin
- ECM
extracellular matrix
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- GST
glutathione S-transferase
- hap
hours after pollination
- IEF
isoelectric fosusing
- 120K
120 kDa glycoprotein
- Na-PI
N. alata serine proteinase inhibitor
- NaSoEP
N. alata sexual organs expressed protein
- NaStEP
N. alata stigma expressed protein
- NTPP
N-terminal propeptide
- PBS
phosphate-buffered saline
- PPAL
pistil pollen allergen-like
- SC
self-compatible
- SI
self-incompatible/self-incompatibility
- SsVSS
sequence-specific vacuolar sorting signal
- STIG1
stigma-specific protein
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson AH, Heath RL, Simpson RJ, Clarke AE, Anderson MA. Proteinase inhibitors in Nicotiana alata stigmas are derived from a precursor protein which is processed into five homologous inhibitors. The Plant Cell. 1993;5:203–213. doi: 10.1105/tpc.5.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecher B, McClure BA. Effects of RNases on rejection of pollen from Nicotiana tabacum and N. plumbaginifolia. Sexual Plant Reproduction. 2001;14:69–76. [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wang H, Wu H-M. A floral transmitting tissue specific glycoprotein attracts pollen tubes and stimulates their growth. Cell. 1995;82:383–393. doi: 10.1016/0092-8674(95)90427-1. [DOI] [PubMed] [Google Scholar]
- Clarke AE, Gleeson P, Harrison S, Knox RB. Pollen–stigma interactions: identification and characterization of surface components with recognition potencial. Proceedings of the National Academy of Sciences, USA. 1979;76:3358–3362. doi: 10.1073/pnas.76.7.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-García F, Hancock N, Kim D, McClure B. Stylar glycoproteins bind to S-RNase in vitro. The Plant Journal. 2005;42:295–304. doi: 10.1111/j.1365-313X.2005.02375.x. [DOI] [PubMed] [Google Scholar]
- de Nettancourt D. Incompatibility and incongruity in wild and cultivated plants. 2nd edn. Berlin: Springer; 2001. [Google Scholar]
- Frigerio L, Jolliffe NA, Di Cola A, Felipe DH, Paris N, Neuhaus JM, Lord JM, Ceriotti A, Roberts LM. The internal propeptide of the ricin precursor carries a sequence-specific determinant for vacuolar sorting. Plant Physiology. 2001;126:167–175. doi: 10.1104/pp.126.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell A, Bacic A, Clarke AE. Arabinogalactan-proteins of the female sexual tissue of Nicotiana alata. Changes during floral development and pollination. Plant Physiology. 1986;82:885–889. doi: 10.1104/pp.82.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson PA, Clarke AE. Structural studies on the major component of Gladiolus style mucilage, an arabinogalactan-protein. Biochemical Journal. 1979;181:607–621. doi: 10.1042/bj1810607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman MHS, Golberg RBJ, Mariani C. Female sterile tobacco plants are produced by stigma-specific cell ablation. EMBO Journal. 1994;13:2976–2984. doi: 10.1002/j.1460-2075.1994.tb06596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman MHS, Pezzotti M, Seurinck J, Mariani C. Developmental expression of tobacco pistil-specific genes encoding novel extensin-like protein. The Plant Cell. 1992;4:1041–1051. doi: 10.1105/tpc.4.9.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldraij A, Kondo K, Lee CB, Hancock CN, Sivaguru M, Vazquez-Santana S, Kim S, Phillips TE, Cruz-Garcia F, McClure B. Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature. 2006;439:805–810. doi: 10.1038/nature04491. [DOI] [PubMed] [Google Scholar]
- Graaf BHJ, Knuiman BA, Weerden GM, Feron R, Derksen J, Mariani C. The PELPIII glycoproteins in Solanaceae: stylar expression and transfer into pollen tube walls. Sexual Plant Reproduction. 2004;16:245–252. [Google Scholar]
- Hancock CN, Kent L, McClure B. The 120 kDa glycoprotein is required for S-specific pollen rejection in Nicotiana. The Plant Journal. 2005;43:716–723. doi: 10.1111/j.1365-313X.2005.02490.x. [DOI] [PubMed] [Google Scholar]
- Heath RL, Barton PA, Reid GEM, Lim G, Anderson MA. Characterization of the protease processing sites in a multidomain proteinase inhibitor precursor from Nicotiana alata. European Journal of Biochemistry. 1995;230:250–257. doi: 10.1111/j.1432-1033.1995.tb20558.x. [DOI] [PubMed] [Google Scholar]
- Heath RL, McDonald G, Christeller JT, Lee M, Bateman K, West J, van Heeswijk R, Anderson MA. Proteinase inhibitors from Nicotiana alata enhance resistance to insect pests. Journal of Insect Physiology. 1997;43:833–842. doi: 10.1016/s0022-1910(97)00026-7. [DOI] [PubMed] [Google Scholar]
- Herrero M, Dickinson HG. Pollen–pistil incompatibility in Petunia hybrida: changes in the pistil following compatible and incompatible intraspecific crosses. Journal of Insect Physiology. 1979;36:1–18. doi: 10.1242/jcs.36.1.1. [DOI] [PubMed] [Google Scholar]
- Herrero M, Dickinson HG. Ultrastructural and physiological differences betwwen buds and mature flowers of Petunia hybrida prior to the following pollinations. Planta. 1980;148:138–145. doi: 10.1007/BF00386414. [DOI] [PubMed] [Google Scholar]
- Holwerda BC, Padgett HS, Rogers JC. Proaleurain vacuolar targeting is mediated by short contiguous peptide interactions. The Plant Cell. 1992;4:307–318. doi: 10.1105/tpc.4.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou W-C, Lin Y-H. Activity staining on polyacrylamide gels of trypsin inhibitors from leaves of sweet potato (Ipomoea batatas L. Lam) varieties. Electrophoresis. 1998;19:212–214. doi: 10.1002/elps.1150190212. [DOI] [PubMed] [Google Scholar]
- Johnson DJ, Miller EA, Anderson MA. Dual location of a family of proteinase inhibitors within the stigmas of Nicotiana alata. Planta. 2006;225:1265–1276. doi: 10.1007/s00425-006-0418-6. [DOI] [PubMed] [Google Scholar]
- Johnson MA, Preuss D. On your mark, get set, grow! LePRK2–LAT52 interactions regulate pollen tube growth. Trends in Plant Science. 2003;8:97–99. doi: 10.1016/S1360-1385(03)00009-8. [DOI] [PubMed] [Google Scholar]
- Juárez-Díaz JA, McClure B, Vázquez-Santana S, Guevara-García A, León-Mejía P, Márquez-Guzmán J, Cruz-García F. A novel thioredoxin h is secreted in Nicotiana alata and reduces S-RNases in vitro. Journal of Biological Chemistry. 2006;281:3418–3424. doi: 10.1074/jbc.M511687200. [DOI] [PubMed] [Google Scholar]
- Kao TH, Tsukamoto T. The molecular and genetic bases of S-RNase-based self-incompatibility. The Plant Cell. 2004;16:S72–S83. doi: 10.1105/tpc.016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Mollet J-C, Dong J, Zhang K, Park S-Y, Lord EM. Chemocyanin, a small, basic protein from the lily stigma, induces pollen tube chemotropism. Proceedings of the National Academy of Sciences, USA. 2003;100:16125–16130. doi: 10.1073/pnas.2533800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboyama T. A novel thaumatin-like protein gene of tobacco is specifically expressed in the transmitting tissue of stigma and style. Sexual Plant Reproduction. 1998;11:251–256. [Google Scholar]
- Kuboyama T, Yoshida KT, Takeda G. An acidic 39-kDa protein secreted from stigmas of tobacco has an amino-terminal motif that is conserved among thaumatin-like proteins. Plant and Cell Physiology. 1997;38:91–95. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee J-Y, Lee D-H. Use of serial analysis of gene expression technology to reveal changes in gene expression in Arabidopsis pollen undergoing cold stress. Plant Physiology. 2003;132:517–529. doi: 10.1104/pp.103.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MCS, Scanlon MJ, Craik DJ, Anderson MA. A novel two-chain proteinase inhibitor generated by circularization of a multidomain precursor protein. Nature Structural Biology. 1999;6:526–530. doi: 10.1038/9293. [DOI] [PubMed] [Google Scholar]
- Leung DWM. Involvement of plant chitinase in sexual reproduction of higher plants. Phytochemistry. 1992;31:1899–1900. [Google Scholar]
- Lord EM, Sanders LC. Roles of the extracellular matrix in plant development and pollination: a special case of cell movement in plants. Developmental Biology. 1992;153:16–28. doi: 10.1016/0012-1606(92)90088-x. [DOI] [PubMed] [Google Scholar]
- Lu Z, Szafron D, Greiner R, Lu P, Wishart DS, Poulin B, Anvik J, Macdonell C, Eisner R. Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics. 2004;20:547–556. doi: 10.1093/bioinformatics/btg447. [DOI] [PubMed] [Google Scholar]
- McCormick S, Twell D, Vancanneyt G, Yamaguchi J. Molecular analysis of gene regulation and function during male gametophyte development. In: Jenkins GI, Schuch W, editors. Molecular biology of plant development. London: Company of Biologists; 1991. pp. 229–244. [PubMed] [Google Scholar]
- MacKenzie CJ, Bong YY, Seabrook JEA. Stigma of Solanum tuberosum cv Shepody: morphology, ultrastructure and secretion. American Journal of Botany. 1990;77:1111–1124. [Google Scholar]
- Matsuoka K. C-terminal propeptides and vacuolar sorting by BP-80-type proteins: not all C-terminal propeptides are equal. The Plant Cell. 2000;12:181–182. doi: 10.1105/tpc.12.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Nakamura K. Large alkyl side chains of isoleucine and leucine in the NPIRL region constitute the core of the vacuolar sorting determinant of sporamin precursor. Plant Molecular Biology. 1999;41:825–835. doi: 10.1023/a:1006357413084. [DOI] [PubMed] [Google Scholar]
- McClure B, Franklin-Tong V. Gametophytic self-incompatibility: understanding the cellular mechanism involved in ‘self’ pollen tube inhibition. Planta. 2006;224:233–245. doi: 10.1007/s00425-006-0284-2. [DOI] [PubMed] [Google Scholar]
- McClure BA, Gray JE, Anderson MA, Clarke AE. Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature. 1990;347:757–760. [Google Scholar]
- McClure BA, Mou B, Canevascini S, Bernatzky R. A small asparagine-rich protein required for S-allele-specific pollen rejection in Nicotiana. Proceedings of the National Academy of Sciences, USA. 1999;96:13548–13553. doi: 10.1073/pnas.96.23.13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EA, Lee MCS, Atkinson AHO, Anderson MA. Identification of a novel four-domain member of the proteinase inhibitor II family from the stigmas of Nicotiana alata. Plant Molecular Biology. 2000;42:329–333. doi: 10.1023/a:1006305429013. [DOI] [PubMed] [Google Scholar]
- Murfett J, Atherton TL, Mou B, Gasser CS, McClure BA. S-RNase expressed in transgenic Nicotiana causes S allele-specific pollen rejection. Nature. 1994;367:563–566. doi: 10.1038/367563a0. [DOI] [PubMed] [Google Scholar]
- Murfett J, Strabala TJ, Zurek DM, Mou B, Beecher B, McClure BA. S-RNase and interspecific pollen rejection in the genus Nicotiana: multiple pollen-rejection pathways contribute to unilateral incompatibility between self-incompatible and self-compatible species. The Plant Cell. 1996;8:943–958. doi: 10.1105/tpc.8.6.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschietti J, Dircks L, Vancanneyt G, McCormick S. LAT52 protein is essential for tomato pollen development: pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. The Plant Journal. 1994;6:321–338. doi: 10.1046/j.1365-313x.1994.06030321.x. [DOI] [PubMed] [Google Scholar]
- Mutschler MA, Liedle BE. Interspecific crossing barriers in Lycopersicon and their relationship to selfincompatibility. In: Williams EG, Clarke AE, Knox RB, editors. Genetic control of self-incompatibility and reproductive development in flowering plants. Dordrecht: Kluwer Academic Publishers; 1994. pp. 164–188. [Google Scholar]
- Myers E, Miller W. Optimal alignments in linear space. Computater Applications in the Biosciences. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Krogh A. In Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology (ISMB 6) Menlo Park, CA: AAAI Press; 1998. Prediction of signal peptides and signal anchors by a hidden Markov model; pp. 122–130. [PubMed] [Google Scholar]
- Nieuwland J, Feron R, Huisman BAH, Fasolino A, Hilbers CW, Derksen J, Mariani C. Lipid transfer proteins enhance cell wall extension in tobacco W. The Plant Cell. 2005;17:2009–2019. doi: 10.1105/tpc.105.032094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien M, Kapfer C, Major G, Laurin M, Bertrand C, Kondo K, Kowyama Y, Matton DP. Molecular analysis of the stylar-expressed Solanum chacoense small asparagine-rich protein family related to the HT modifier of gametophytic self-incompatibility in Nicotiana. The Plant Journal. 2002;32:985–996. doi: 10.1046/j.1365-313x.2002.01486.x. [DOI] [PubMed] [Google Scholar]
- Ono E, Hatayama M, Isono Y, et al. Localization of a flavonoid biosynthetic polyphenol oxidase in vacuoles. The Plant Journal. 2006;45:133–143. doi: 10.1111/j.1365-313X.2005.02625.x. [DOI] [PubMed] [Google Scholar]
- Park KS, Cheong JJ, Lee SJ, Suh MC, Choi D. A novel proteinase inhibitor gene transiently induced by tobacco mosaic virus infection. Biochimica et Biophysica Acta. 2000;1492:509–512. doi: 10.1016/s0167-4781(00)00126-3. [DOI] [PubMed] [Google Scholar]
- Park SY, Jauh G-Y, Mollet J-C, Eckard KJ, Nothnagel EA, Walling LL, Lord EM. A lipid transfer-like protein is necessary for lily pollen tube adhesion to an in vitro stylar matrix. The Plant Cell. 2000;12:151–163. doi: 10.1105/tpc.12.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-Y, Lord EM. Expression studies of SCA in lily and confirmation of its role in pollen tube adhesion. Plant Molecular Biology. 2003;51:183–189. doi: 10.1023/a:1021139502947. [DOI] [PubMed] [Google Scholar]
- Pezzotti M, Feron R, Mariani C. Pollination modulates expression of the PPAL gene, a pistil-specific β-expansin. Plant Molecular Biology. 2002;49:187–197. doi: 10.1023/a:1014962923278. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Tolle DP, Barrett AJ. Evolutionary families of peptidase inhibitors. Biochemical Journal. 2004;378:705–716. doi: 10.1042/BJ20031825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saalbach C, Rosso M, Schumann U. The vacuolar targeting signal of the 2s albumin from Brazil nut resides at the C terminus and involves the C-terminal propeptide as an essential element. Plant Physiology. 1996;112:975–985. doi: 10.1104/pp.112.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Hauser K, Lind JL, Atkinson AH, Pu ZY, Anderson MA, Clarke A. Molecular characterization of a cDNA sequence encoding the back-bone of a style-specific 120 kDa glycoprotein which has features of both extensins and arabinogalactan proteins. Plant Molecular Biology. 1997;35:833–845. doi: 10.1023/a:1005816520060. [DOI] [PubMed] [Google Scholar]
- Takayama S, Isogai A. Self-incompatibility in plants. Annual Review of Plant Biology. 2005;56:467–489. doi: 10.1146/annurev.arplant.56.032604.144249. [DOI] [PubMed] [Google Scholar]
- Tang W, Ezcurra I, Muschietti J, McCormick S. A cysteine-rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. The Plant Cell. 2002;14:2277–2287. doi: 10.1105/tpc.003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kelley D, Ezcurra I, Cotter R, McCormick S. LeSTIG1, an extracellular binding partner for the pollen receptor kinases LePRK1 and LePRK2, promotes pollen tube growth in vitro. The Plant Journal. 2004;39:343–353. doi: 10.1111/j.1365-313X.2004.02139.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D, Wing R, Yamaguchi J, McCormick S. Isolation and expression of an anther-specific gene from tomato. Molecular and General Genetics. 1989;217:240–245. doi: 10.1007/BF02464887. [DOI] [PubMed] [Google Scholar]
- Verhoeven T, Feron F, Wolters-Arts M, Edqvist J, Gerats T, Derksen J, Mariani C. STIG1 controls exudate secretion in the pistil of Petunia and tobacco. Plant Physiology. 2005;138:153–160. doi: 10.1104/pp.104.054809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Heijne G. The signal peptide. Journal of Membrane Biology. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- Wu H-m, Wang H, Cheung AY. A pollen tube growth stimulatory glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower. Cell. 1995;83:395–403. doi: 10.1016/0092-8674(95)90428-x. [DOI] [PubMed] [Google Scholar]
- Wu H-m, Wong E, Ogdahl J, Chueung AY. A pollen tube growth-promoting arabinogalactan protein from Nicotiana alata is similar to the tobacco TTS protein. The Plant Journal. 2000;22:165–176. doi: 10.1046/j.1365-313x.2000.00731.x. [DOI] [PubMed] [Google Scholar]
- Xie D, Li A, Wang M, Fan Z, Feng H. LOCSVMPSI: a web server for subcellular localization of eukayotic proteins using SVM and profile of PSI-BLAST. Nucleic Acids Research. 2005;33:W105–W110. doi: 10.1093/nar/gki359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala JA, Patankar AG, Gase K, Hui D, Baldwin IT. Manipulation of endogenous trypsin proteinase inhibitor production in Nicotiana attenuata demonstrates their function as antiherbivore defenses. Plant Physiology. 2004;134:1181–1190. doi: 10.1104/pp.103.035634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.