Abstract
A sensitive method for the analysis of single nucleotide polymorphisms (SNPs) in genomic DNA that utilizes nanoparticle-enhanced surface plasmon resonance imaging (SPRI) measurements of surface enzymatic ligation reactions on DNA microarrays is demonstrated. SNP identification was achieved by using sequence-specific surface reactions of the enzyme Taq DNA ligase, and the presence of ligation products on the DNA microarray elements was detected with SPRI through the hybridization adsorption of complementary oligonucleotides attached to gold nanoparticles. The use of gold nanoparticles increases the sensitivity of the SPRI so that single bases in oligonucleotides can be successfully identified at a concentration of 1 pM. This sensitivity is amply sufficient for performing multiplexed SNP genotyping by using multiple PCR amplicons, and should also allow for the direct detection and identification of SNP sequences from 1 pM unamplified genomic DNA samples with this array-based and label-free SPRI methodology. As a first example of SNP genotyping, three different human genomic DNA samples were screened for a possible point mutation in the BRCA1 gene that is associated with breast cancer.
I. Introduction
There is currently a major effort to develop rapid, multiplexed methods for the identification and analysis of single nucleotide polymorphisms (SNPs) in human genomic DNA samples both for the detection of specific disease-related genetic mutations and for the large scale analysis of human genetic variations.1, 2 Analysis of a SNP requires the detection and identification of single base pairs in a DNA sequence either through hybridization-based methods or enzymatic reactions.3 Many of the current multiplexed SNP analysis methods employ DNA microarrays on a variety of substrates in conjunction with fluorescence-based detection.4-7 A multiplexed SNP analysis is sometimes performed directly on unamplified human genomic DNA samples,8, 9 but more frequently a multiplexed polymerase chain reaction is first employed to create a mixture of PCR amplicons from the various SNPs of interest.4-7
Surface plasmon resonance imaging (SPRI) is a rapid, multiplexed method for the label-free detection of surface bioaffinity interactions,10-14 and has been used extensively with DNA microarrays.15-17 Can SPRI of DNA microarrays be used for SNP genotyping? SPRI and single channel SPR methods have been used previously to detect single base mismatches in DNA by hybridization adsorption,18-24 but multiplexed SNP genotyping with SPRI has not yet been demonstrated. A successful multiplexed SNP genotyping measurement with SPRI requires the detection and identification of single base pairs and mismatches in genomic DNA at very low concentrations. Mismatch identification with enzymatic reactions such as ligation or polymerase extension work well in solution, but DNA enzyme reaction conditions typically need to be modified to work well on the gold surfaces required by SPRI.25, 26 In addition, the detection limit of SPRI for DNA hybridization adsorption of 1 nM16, 17 is sufficient for SNP analysis of mixtures of PCR amplicons, but a lower detection limit (1 pM or better8, 27) is required to detect un-amplified genomic DNA. Amplification of the SPRI response to hybridization adsorption by DNA modified gold nanoparticles,28 coupled bioconjugates19, 20 or surface enzyme reactions16, 27 can in principle be used to achieve sufficient sensitivity for direct multiplexed SNP analysis of genomic DNA samples.
In this paper we demonstrate that SPRI can be used for SNP genotyping in a microarray-based format amenable to multiplexed analysis. To achieve single base pair selectivity in the SPRI measurements, we employ the sequence specific surface enzyme reaction of Taq DNA ligase with a DNA microarray. The sequence-specific ligation of DNA has been used previously both in solution29, 30 and in thin polymer films31 for SNP analysis; Barany et al. recently described a solution-based Taq DNA ligase SNP genotyping methodology that used zip code DNA microarrays to detect the solution ligation products.32, 33 To achieve sufficient sensitivity in our SPRI measurements, the surface ligation reaction is detected by the subsequent hybridization adsorption of DNA-modified gold nanoparticles to the surface. This method of nanoparticle enhancement for SPRI was originally employed for DNA detection by He et al. in a sandwich assay format.28 Using this nanoparticle-enhanced surface ligation strategy, we are able to detect and identify single base mismatches (SBMs) in single stranded DNA (ssDNA) down to a concentration of 1 pM. As an initial demonstration, SNP genotyping was performed on PCR amplicons from three human genomic samples for a specific point mutation in the BRCA1 gene associated with breast cancer.34
II. Experimental Considerations
Materials
11-amino-1-undecanethiol hydrochloride (MUAM; Dojindo), sulfosuccinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (SSMCC; Pierce), 9-fluorenylmethoxycarbonyl-N-hydroxysuccinimide (Fmoc-NHS; Novabiochem) and N-hydroxysuccinimidyl ester of methoxypoly (ethylene glycol) propionic acid MW 2000 (PEG-NHS; Nektar Therapeutics) were all used as received. All rinsing steps were performed with either absolute ethanol or Millipore filtered water. Taq DNA ligase (40,000 units/mL) and its reaction buffer (20 mM Tris-HCl, 25 mM potassium acetate, 10 mM magnesium acetate, 1 mM NAD, 10 mM dithiothreitol, 0.1% Triton X-100, pH 7.6) were purchased from New England Biolabs and used for all ligation reactions. All experiments were performed at room temperature unless stated otherwise.
DNA Array Fabrication
A multistep chemical protection/deprotection process described previously was used for the creation of DNA microarrays.35 Thin gold films (45 nm) with a 1 nm underlayer of chromium were first deposited onto SF-10 glass (Schott Glass) using a Denton DV-502A metal evaporator. The gold substrates were then modified with a self-assembled monolayer of an amine-terminated alkanethiol MUAM. This MUAM surface was next reacted with the hydrophobic protecting group Fmoc-NHS. UV photopatterning of the Fmoc surface with a quartz mask containing 500 μm square features was used to create bare gold patches within the hydrophobic Fmoc background. The bare gold spots were then exposed to a MUAM solution for 2 h before manually spotting SSMCC onto the MUAM array elements to form a thiol-reactive maleimide-terminated surface. Thiol-modified sequences of DNA were then manually spotted onto the individual SSMCC array elements. After the DNA attachment reaction, the Fmoc background was removed with a mildly basic solution and replaced by a poly(ethylene glycol) (PEG) surface that is resistant to non-specific adsorption of oligonucleotides and Taq DNA ligase.
Preparation of Synthetic Oligonucleotides
All the DNA sequences used in this paper are listed in Table 1. Synthetic oligonucleotides were purchased from Integrated DNA Technologies (IDT). All thiol-modified oligonucleotides were deprotected and purified using binary reversed-phase HPLC prior to use. Thiol-free oligonucleotides were HPLC purified by IDT and used as received. Array probe oligonucleotides were attached to the DNA microarray via a 5′-thiol modification. The array probes (PA, PC and PG) differ only at the last nucleotide at the 3′-termini with PN used as a negative control. All array probes are 20mers with an additional (T)20 spacer at the 5′-end to improve the accessibility of the surface-bound DNA to DNA hybridization and Taq DNA ligase. The ligation probe (L) has a 5′-phosphate group modification. The oligonucleotide (LC), which is complementary to L, has a 5′-thiol modification for attachment onto gold nanoparticles.
Table 1.
Summary of all DNA sequences used for the ligation reaction, nanoparticle-enhanced SPRI detection and PCR amplification
| Array probes (Immobilized on the DNA microarray): |
| PG = 5′-S-S-(CH2)6-(T)20 GTG CTT TGT TCT GGA TTT CG -3′ |
| PC = 5′-S-S-(CH2)6-(T)20 GTG CTT TGT TCT GGA TTT CC -3′ |
| PA = 5′-S-S-(CH2)6-(T)20 GTG CTT TGT TCT GGA TTT CA -3′ |
| PN = 5′-S-S-(CH2)6-(T)40 -3′ |
| Oligonucleotide Target (Location of the targeted single base pair is underlined): |
| T = 5′-TGC CCT TGA GGA CCT GCG AAA TCC AGA ACA AAG CAC -3′ |
| Ligation Probe: |
| L = 5′-PO4 CAG GTC CTC AAG GGC A -3′ |
| Ligation Probe Complement (Attached to gold nanoparticles): |
| LC = 5′-S-S-(CH2)6-(T)20 TGC CCT TGA GGA CCT G -3′ |
| PCR primers for exon 13 of BRCA1 gene: |
| Forward: 5′-AAT GGA AAG CTT CTC AAA GTA -3′ |
| Reverse: 5′-ATG TTG GAG CTA GGT CCT TAC -3′ |
PCR Amplification of Genomic DNA
Three different genomic DNA samples were individually PCR amplified: a wild-type genomic DNA sample (Promega), and two different genomic DNA samples with a point mutation in exon 13 of the BRCA1 gene (NA13710 and NA14637 from the Coriell Institute). HPLC purified PCR primers from IDT were used as received. Primer sequences (Table 1) for the amplification of exon 13 were obtained from the literature.36 PCR was performed in multiple 50-μL reaction sets each containing 250 ng of genomic DNA, 0.2 mM dNTP (Promega), 50 pmol each of BRCA1 gene primer, one Hot Start Polymerase TaqBead (Promega), 5 mM MgCl2, and 1× thermophilic DNA polymerase reaction buffer (Promega). The amplification reaction was performed on a thermal cycler (MJ Research, Model PTC-100) with 5 min at 95 °C followed by 30 cycles of 1 min at 95 °C, 1 min annealing at 58 °C, 1 min of primer extension at 72 °C, and a final extension at 72 °C for 5 min. PCR products were purified using the Wizard PCR Preps DNA Purification System (Promega), lyophilized and resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5). The presence of each expected 260 bp amplicon was confirmed by gel electrophoresis using a 1% agarose gel. The concentration of each amplicon was determined by measuring UV absorption at 260 nm. PCR products were stored at -20 °C until required for the surface ligation reaction.
Surface Ligation Reaction Conditions
Ligation with synthetic oligonucleotides
Probe DNA microarrays were reacted with a mixture of Taq DNA ligase (800 units/mL), 1 μM ligation probe DNA and target oligonucleotide in Taq DNA ligase reaction buffer for 2 h. Ligation with PCR products: PCR amplicon was first diluted to 1 nM with Taq DNA ligase reaction buffer before heating at 95 °C for 5 min. Both ligation probe and Taq DNA ligase were then added to the heated solution with final concentrations of 1 μM and 800 units/mL, respectively. Next, the 95 °C solution mixture was immediately introduced to the DNA microarray and allowed to react at room temperature for 2 h.
Preparation of Oligonucleotide-modified Au Nanoparticles
Gold nanoparticles preparation
Gold colloid was prepared following procedures described previously by Natan et al.37 Briefly, 20 mL of 38.8 mM sodium citrate solution was added quickly to a 200 mL boiling solution of 1 mM HAuCl4 while stirring vigorously under reflux conditions. Boiling and stirring was continued for 15 min after no further change in solution color was observed. This colloidal solution, which has a λmax at 518 nm, was then stored at 4 °C after cooling. Gold nanoparticles prepared using this method are known to have average diameters of 12-13 nm with a standard deviation of less than 10%.37 DNA:Au conjugate preparation: DNA:Au nanoparticle conjugates were prepared using procedures described by Keating et al.38 Briefly, 50 μL of freshly purified 100 μM thiol-modified oligonucleotide was added to 800 μL of Au nanoparticle solution in an eppendorf tube. The tube was then placed in a heat block at 37 °C for a minimum of 4 h before the addition of 230 μL of H2O and 120 μL of 1 M NaCl/100 mM phosphate (pH 7.4). After aging overnight at 37 °C, excess oligonucleotide was extracted by centrifugation at 9500 rpm (7500 g) for 50 min with the removal of supernatant and resuspension of the Au pellet in 0.1 M NaCl/10 mM phosphate buffer (pH 7.4) repeated three times. After the final centrifugation, the Au pellet was resuspended in 0.3 M NaCl/10 mM phosphate buffer (pH 7.4) with a final concentration of ∼10 nM. This concentration was obtained from UV-vis spectroscopy using an extinction coefficient of ∼2 × 108 M-1 cm-1 as described previously.38 All surface hybridization adsorption experiments using DNA-modified nanoparticles were performed with 0.3 M NaCl/10 mM phosphate buffer (pH 7.4) at room temperature.
SPR Imaging Measurements
An SPR imager (GWC Technologies)17 using near-infrared excitation from an incoherent white light source was used for SNP genotyping. Briefly, a polychromatic beam of p-polarized light is used to illuminate a sample assembly at a fixed incident angle near the SPR angle. The reflected light is passed through a narrow band-pass filter centered at 830 nm and collected with a CCD camera. Changes in the measured reflected light intensity are a direct result of changes in the thickness and/or refractive index of the material adsorbed at the gold surface. Data are collected using the software package V++ 4.0 (Digital Optics, NZ) and further analyzed using the software package NIH Image V.1.63.
III. Results and Discussion
A. SPRI SNP Analysis Methodology
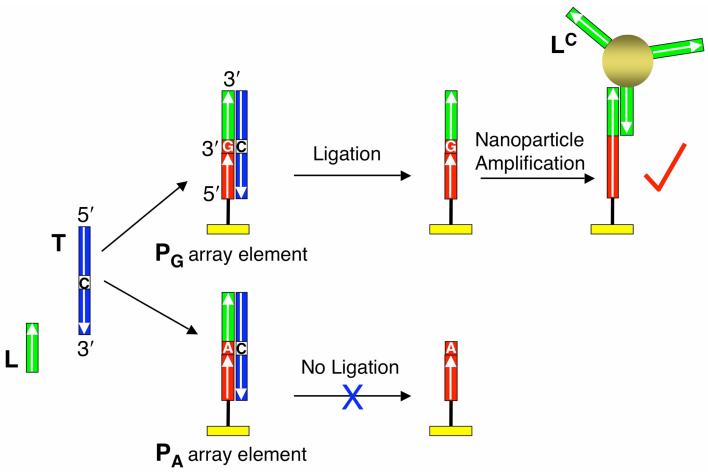
A combination of surface hybridization, surface ligation and nanoparticle adsorption on DNA microarrays is used to detect and identify SNPs with SPRI. The general scheme of this SNP genotyping method is illustrated in Figure 1, which depicts how to identify a single base pair or mismatch in the middle of a 36mer single stranded DNA (ssDNA) target T. A solution containing the target T, a 5′-phosphorylated ssDNA 16mer ligation probe L, and the enzyme Taq DNA ligase is introduced to a two-component ssDNA microarray. The two array elements in Figure 1 contain the ssDNA 20mer array probes PG and PA. The two array probe DNA sequences are covalently attached at the 5′-ends to a chemically modified gold surface, and only differ by the last nucleotide at their 3′-termini (G for PG and A for PA). The target DNA molecules hybridize simultaneously to the ligation probe and the array probes, resulting in the formation of two different three-component surface complexes shown in the figure, L-T-PG and L-T-PA. In this example, PG is perfectly complementary to the target molecule, whereas PA forms a single A-C mismatch. Since Taq DNA ligase will only catalyze the formation of a phosphodiester bond between the juxtaposed 5′-phosphate and 3′-hydroxyl groups of two ssDNA when they are perfectly complementary to a hybridized target, the ligation probe will be specifically ligated to PG but not to PA. On completion of the ligation reaction, an 8 M urea wash is used to denature and remove any target, non-ligated L and Taq DNA ligase from the surface, resulting in an array with ligated L-PG array elements and unreacted PA array elements. The creation of only ligated L-PG array elements indicates that the target sequence contained a C at that specific base location, and not a T.
Figure 1.
Schematic representation of the SNP genotyping method based on a combination of surface ligation chemistry and nanoparticle enhanced SPRI. Two array elements with different DNA probes are shown. The array probes (PA and PG) differ only by the last nucleotide at their 3′ ends. When target DNA (T), ligation probe DNA (L) and Taq DNA ligase are simultaneously introduced to the array, surface duplexes form at both array elements. However, ligation only occurs if the duplex is perfectly complementary. After denaturation with 8 M urea, the perfectly matched PG is extended with the L sequence while PA returns to its original state. The presence of L is then detected by the hybridization adsorption of gold nanoparticles modified with oligonucleotides (LC) complementary to L.
The detection of the ligated L-PG array elements is achieved through the hybridization adsorption of gold nanoparticles modified with ssDNA 16mers LC which are complementary to the ligation probe sequence. The SPRI signal due to this nanoparticle adsorption will be substantially larger than the SPRI signal change due solely to the surface ligation reaction. DNA-nanoparticle conjugates have been employed previously in a sandwich assay format for the amplified SPR detection of ssDNA, and can lower the ssDNA detection limit of SPRI measurements from nanomolar down to picomolar concentrations.28
B. Single Base Pair Identification in Oligonucleotides
To verify the single base specificity of this SNP analysis method and to determine the sensitivity of this novel SPRI detection methodology, we first performed an experiment to identify a single base pair in a 36mer ssDNA target T. The sequence of this 36mer is given in Table 1. A four-component array consisting of probes PA, PC, PG and PN was used to determine the identity of the base at position 20 from the 3′-end of the target molecule. The array probes PA, PC and PG were terminated at their 3′-ends with 20mers that were all complementary to the 19 bases at the 3′-end of T but differed only by the last nucleotide at their 3′-termini (see Table 1 for complete DNA sequences). Probe PN was a poly T negative control sequence. To create the surface ligation reaction, a solution containing T, a 5′-phosphorylated 16mer ssDNA ligation probe L that is complementary to the 16 bases at the 5′-end of T, and Taq DNA ligase were introduced simultaneously to the DNA microarray. Various target DNA concentrations were investigated while the concentrations of L and enzyme remained fixed at 1 μM and 800 units/mL respectively. All probes PA, PC and PG hybridized with T and L to form surface duplexes. However, the surface ligation of L should only occur with the probe PG since probes PA and PC both form SBMs with T. After 2 h, the DNA microarray was rinsed with 8 M urea.
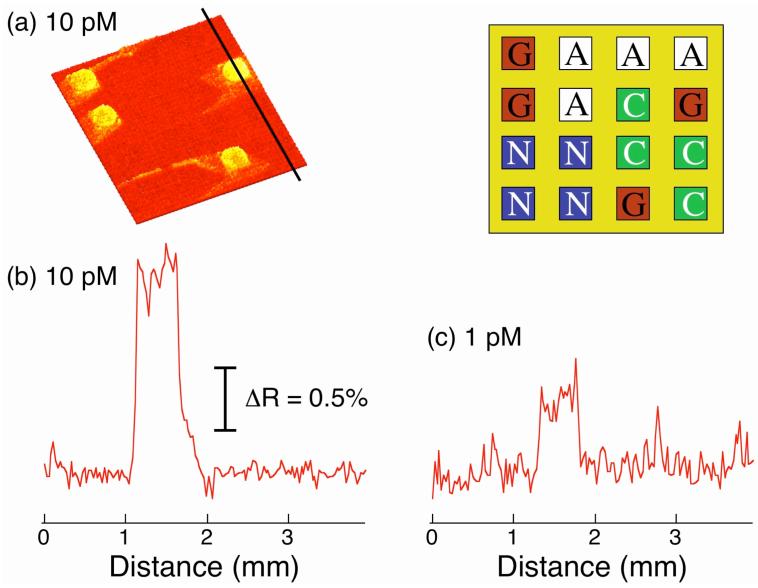
To detect any surface ligation reactions, the DNA microarray was then exposed to a ∼10 nM solution of gold nanoparticles coated with the 16mer ssDNA LC that is complementary to the ligation probe L. Figure 2a shows the SPR difference image obtained from a T concentration of 10 pM. Specific hybridization adsorption was only observed on the PG array elements, and no non-specific adsorption was observed either on the other array elements or on the PEG array background. The signal increase on the PG array elements is about 1.5% ΔR as shown in the line profile in Figure 2b. This experiment clearly demonstrates that this SPRI detection methodology can be used to identify single base pairs and mismatches at picomolar concentrations.
Figure 2.
Detection and identification of single base pairs and mismatches in short synthetic DNA sequences. A four-component DNA microarray consisting of DNA probes (PA, PC, PG and PN) was used. The upper right inset depicts the array pattern. The ligation reaction was first performed in the presence of 10 pM target DNA (T) for 2 h at room temperature. The surface was then denatured with 8 M urea and rinsed with hybridization buffer. (a) An SPR difference image was obtained by subtracting images taken before and after the exposure of the ligated array to LC-modified gold nanoparticles. SPR signal increase due to hybridization adsorption of the nanoparticles was observed only at the PG array elements, where the probe sequence is perfectly matched to T. Non-specific adsorption was not observed either on the other probe array elements or on the PEG background. (b) A line profile taken from the SPR difference image in (a) for 10 pM target DNA. (c) A line profile taken from an SPR difference image obtained from a separate experiment using 1 pM T. The percent reflectivity scale is the same for both (b) and (c). The ligation reaction with 1 pM DNA was performed at a temperature of 45 °C.
An essential component of this technique is the Taq ligase surface ligation; a high surface ligation efficiency will lead to high SNP detection sensitivity. In a separate set of experiments (not shown), the surface ligation efficiency was roughly estimated by comparing the SPR signals obtained from the hybridization adsorption of the same complementary ssDNA to i) a microarray fabricated by ligating the probe DNA onto the surface and ii) a microarray where the same probe DNA was chemically attached via thiol modification. Assuming a similar hybridization efficiency for the two DNA microarrays, a surface ligation efficiency of approximately 75 ± 15% was obtained. This surface reaction efficiency is comparable to what we have observed previously with T4 DNA ligase.39, 40
To determine the detection limit of these nanoparticle enhanced SPRI single base pair identification and SBM detection measurements, the same experiment was repeated at a target concentration of 1 pM. This experiment was performed at 45 °C instead of room temperature as in the case of the 10 pM target solution, resulting in a slight improvement in ligation efficiency. As can be seen from the line profile in Figure 2c, the 0.4% increase in SPRI reflectivity associated with the detection of L at PG array elements is clearly distinguishable from the background. This detection limit for SBMs is better than the previously reported 10 pM for the direct detection of oligonucleotides via hybridization adsorption using nanoparticle-enhanced SPRI measurements.28 In addition, a detection limit of 1 pM is comparable to that obtained from the fluorescence imaging measurements that are typically used in multiplexed DNA microarray analyses.41, 42
C. SNP Genotyping in the BRCA1 gene
Having demonstrated the concept of single base pair identification and SBM detection with synthetic oligonucleotides, we then applied this method to the SNP genotyping of PCR products from human genomic DNA samples. A SNP located in the BRCA1 gene linked to breast cancer was chosen. Mutations in the BRCA1 gene are thought to be responsible for more than 80% of hereditary breast and ovarian cancers.43 A variety of methods such as single-strand conformation polymorphisms assay (SSCP) and DNA sequencing has been utilized previously for the screening of BRCA1 mutations.34, 36 DNA microarray based SNP genotyping methods are attractive because they are more amenable to automation and multiplexed analysis.
The SNP that we chose to determine is a common point mutation in the BRCA1 gene located in codon 1443 of exon 13.34 The genomic DNA samples used in this study include one wild-type sample (Promega) and two mutant DNA samples (Coriell Institute). The wild-type DNA from breast cancer free population is homozygous with a base C at nucleotide 4446. The mutant DNA samples are both heterozygous containing a wild type and a different mutant allele at this position. DNA sample NA13710 has a C to G missense mutation, and NA14637 has a C to T nonsense mutation. To identify the genotype of each of these genomic DNA samples, exon 13 of the BRCA1 gene was PCR amplified (see Table 1 for primer sequences) to create a 260 base pair PCR amplicon that contained nucleotide 4446.
SNP genotyping was then performed using a four-component DNA microarray containing array probes PA, PC, PG and PN (see Table 1 for sequences). These DNA probes were specifically designed to bind to the alleles containing the SNP of interest. Probe PG is the perfect complement to the wild-type allele; probe PC is the perfect complement to the mutant allele in NA13710; and probe PA is the perfect complement to the mutant allele in NA14637. The PCR product was first denatured at 95 °C for 5 min and quickly mixed with the ligation probe (L) and Taq DNA ligase. This solution was then immediately introduced to the four-component microarray and allowed to react for 2 h. The array probe and ligation probe both bound to the allele of interest and formed a ligase substrate similar to that depicted in Figure 1. After surface ligation and urea denaturation, the SNP genotype was identified by the detection of nanoparticle hybridization adsorption to any ligated array elements.
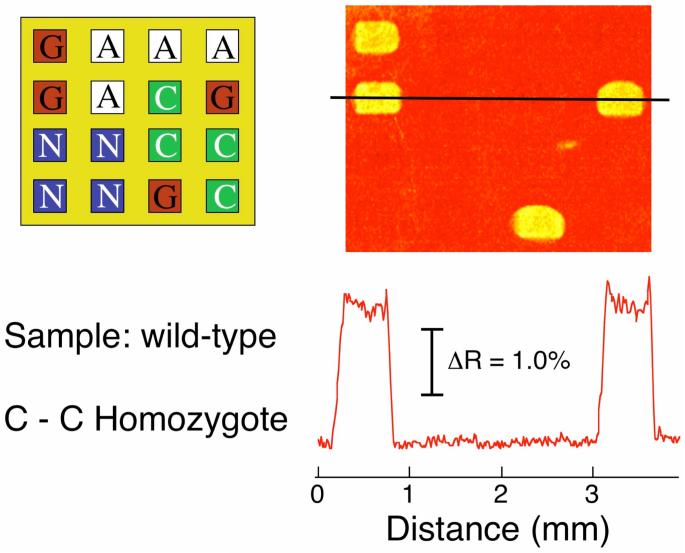
The SNP genotyping results for the three different genomic DNA samples are shown in Figures 3-5. For the wild-type genomic DNA, ligation was only observed on the PG array elements to give the SPRI difference image for the hybridization adsorption of DNA-nanoparticle conjugates in Figure 3. This result clearly verified that the genotype of the wild-type DNA sample is a C-C homozygote. Note that in all of the SPRI images in Figures 3-5 there is negligible non-specific adsorption of DNA-nanoparticle conjugates on any of the other array elements or the PEG background.
Figure 3.
SNP genotyping of a 1 nM PCR amplicon of wild-type genomic DNA. A four-component probe DNA microarray (PA, PC, PG and PN) was first exposed to a solution containing target PCR amplicon, ligation probe DNA and Taq DNA ligase for 2 h followed by a surface denaturation step. The SPR difference image after specific hybridization adsorption of LC-modified nanoparticles onto the ligated microarray is shown along with the corresponding array pattern and line profile. An SPRI signal increase observed at only the PG array elements identifies the wild-type sample as being a C-C homozygote.
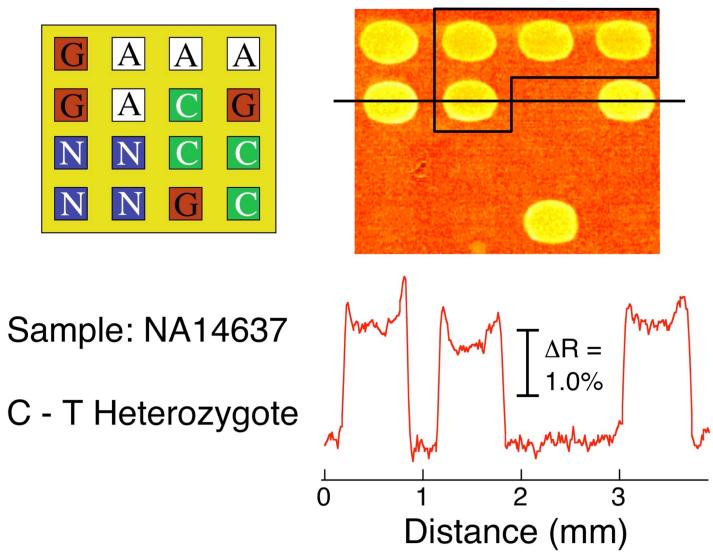
Figure 5.
SNP genotyping of a 1 nM PCR amplicon of genomic DNA sample NA14637. An SPR difference image was obtained by subtracting images taken before and after the exposure of LC-modified nanoparticles to a ligated microarray prepared in an identical manner to that in Figures 3 and 4. The array pattern and corresponding line profile are also shown. The SPRI signal increase observed at both the PG and PA (boxed) elements identifies the NA14637 sample as a C-T heterozygote.
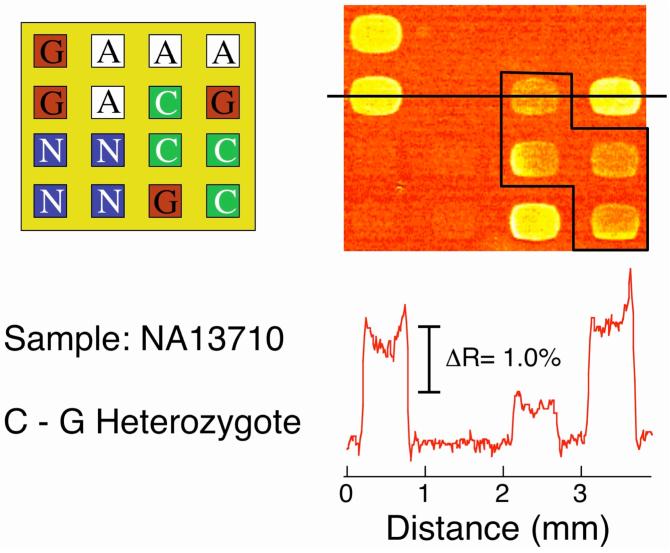
SPRI difference images from the two mutant samples are shown in Figure 4 and 5. For the NA13710 Coriell sample, the SPRI difference image in Figure 4 showed a signal increase at both the PG and PC array elements, which indicates the presence of both the wild-type C allele and the G mutant allele. This unequivocally identifies NA13710 as a C-G heterozygote. Similarly, the SPRI difference image for the NA14637 sample in Figure 5 shows a signal increase at the PG and PA array elements and identifies this sample as a C-T heterozygote.
Figure 4.
SNP genotyping of a 1 nM PCR amplicon of genomic DNA sample NA13710. The SPR difference image shows the hybridization adsorption of LC-modified nanoparticles onto a four-component DNA microarray (PA, PC, PG and PN) following surface ligation and denaturation. The array pattern and corresponding line profile are also shown. In contrast to the wild-type DNA sample in Figure 3, an increase in SPRI signal was observed at both the PG and PC (boxed) array elements, identifying NA13710 as being a C-G heterozygote.
Examination of the line profiles in Figures 3-5 shows comparable SPRI reflectivity changes were observed for all of the ligation reactions except for the PC array elements in Figure 4. One possible explanation for the decreased signal intensity at PC array elements is the formation of a hairpin structure, which lowers the hybridization efficiency of the PCR amplicon resulting in a reduction in surface ligation efficiency. DNA folding calculations (http://www.bioinfo.rpi.edu/applications/mfold/) indicated the presence of a stable hairpin structure in PC (ΔG° = -2.9 kJ mol-1; Tm = 33.3 °C) but not in PA or PG.
IV. Conclusions
In this paper we have presented a new SPRI methodology for SNP genotyping that uses a unique combination of a surface enzymatic ligation reaction in conjunction with gold nanoparticle-enhanced hybridization adsorption on DNA microarrays. The SNP is identified by the direct ligation of an oligonucleotide onto a DNA microarray element; using a surface enzyme reaction allows for the facile separation of the ligation products from any reactants or enzyme in solution. The use of DNA modified gold nanoparticles to detect the surface ligation reaction greatly enhances the sensitivity of this SNP analysis. The combination of SPRI and DNA microarrays for SNP genotyping is very attractive because it can be multiplexed; the simultaneous analysis of multiple SNPs should be possible with larger DNA microarrays and multiple PCR amplicons. Moreover, this SPRI SNP analysis methodology is label-free and can in principle be used directly on unamplified genomic DNA samples. The current detection limit of 1 pM is sufficient for the analysis of concentrated genomic DNA samples,8 and by exploring different amplification methods we expect that the detection limit can be further lowered into the femtomolar range for multiplexed SNP genotyping of dilute unamplified genomic DNA samples.
Acknowledgment
This research was funded by the National Institute of Health (2RO1 GM059622-04) and the National Science Foundation (CHE-0133151).
References
- (1).Syvanen A-C. Nat. Rev. Genet. 2001;2:930–942. doi: 10.1038/35103535. [DOI] [PubMed] [Google Scholar]
- (2).Kirk BW, Feinsod M, Favis R, Kliman RM, Barany F. Nucleic Acid Res. 2002;30:3295–3311. doi: 10.1093/nar/gkf466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Kwok P-Y. Annu. Rev. Genomics Hum. Genet. 2001;2:235–258. doi: 10.1146/annurev.genom.2.1.235. [DOI] [PubMed] [Google Scholar]
- (4).De La Vega FM, Lazaruk KD, Rhodes MD, Wenz MH. Mut. Res. 2005;573:111–135. doi: 10.1016/j.mrfmmm.2005.01.008. [DOI] [PubMed] [Google Scholar]
- (5).Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. Nat. Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- (6).Mei R, Galipeau PC, Prass C, Berno A, Ghandour G, Patil N, Wolff RK, Chee MS, Reid BJ, Lockhard DJ. Genome Res. 2000;10:1126–1137. doi: 10.1101/gr.10.8.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Fan J-B, Chen X, Halushka MK, Berno A, Huang X, Ryder T, Lipshutz R, Lockhard DJ, Chakravarti A. Genome Res. 2000;10:853–860. doi: 10.1101/gr.10.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chen Y, Shortreed MR, Olivier M, Smith LM. Anal. Chem. 2005;77:2400–2405. doi: 10.1021/ac0483825. [DOI] [PubMed] [Google Scholar]
- (9).Bao YP, Huber M, Wei T-F, Marla SS, Storhoff JJ, Muller UR. Nucleic Acid Res. 2005;33:e15. doi: 10.1093/nar/gni017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Brockman JM, Nelson BP, Corn RM. Annu. Rev. Phys. Chem. 2000;51:41–63. doi: 10.1146/annurev.physchem.51.1.41. [DOI] [PubMed] [Google Scholar]
- (11).Smith EA, Corn RM. Appl. Spectrosc. 2003;57:320A–332A. doi: 10.1366/000370203322554446. [DOI] [PubMed] [Google Scholar]
- (12).Shumaker-Parry JS, Campbell CT. Anal. Chem. 2004;76:907–917. doi: 10.1021/ac034962a. [DOI] [PubMed] [Google Scholar]
- (13).Kanda V, Kariuki JK, Harrison DJ, McDermott MT. Anal. Chem. 2004;76:7257–7262. doi: 10.1021/ac049318q. [DOI] [PubMed] [Google Scholar]
- (14).Wilkop T, Wang Z, Cheng Q. Langmuir. 2004;20:11141–11148. doi: 10.1021/la048177k. [DOI] [PubMed] [Google Scholar]
- (15).Wolf LK, Fullenkamp DE, Georgiadis RM. J. Am. Chem. Soc. 2005;127:17453–17459. doi: 10.1021/ja056422w. [DOI] [PubMed] [Google Scholar]
- (16).Lee HJ, Li Y, Wark AW, Corn RM. Anal. Chem. 2005;77:5096–5100. doi: 10.1021/ac050815w. [DOI] [PubMed] [Google Scholar]
- (17).Nelson BP, Grimsrud TE, Liles MR, Goodman RM, Corn RM. Anal. Chem. 2001;73:1–7. doi: 10.1021/ac0010431. [DOI] [PubMed] [Google Scholar]
- (18).Endo T, Kerman K, Nagatani N, Takamura Y, Tamiya E. Anal. Chem. 2005;77:6976–6984. doi: 10.1021/ac0513459. [DOI] [PubMed] [Google Scholar]
- (19).Liu J, Tian S, Tiefenauer L, Nielsen PE, Knoll W. Anal. Chem. 2005;77:2756–2761. doi: 10.1021/ac048088c. [DOI] [PubMed] [Google Scholar]
- (20).Okumura A, Sato Y, Kyo M, Kawaguchi H. Anal. Biochem. 2005;339:328–337. doi: 10.1016/j.ab.2005.01.017. [DOI] [PubMed] [Google Scholar]
- (21).Peterson AW, Wolf LK, Georgiadis RM. J. Am. Chem. Soc. 2002;124:14601–14607. doi: 10.1021/ja0279996. [DOI] [PubMed] [Google Scholar]
- (22).Song F, Zhou F, Wang J, Tao N, Lin J, Vellanoweth RL, Morquecho Y, Wheeler-Laidman J. Nucleic Acid Res. 2002;30:e72. doi: 10.1093/nar/gnf072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Nakatani K, Sando S, Saito I. Nat. Biotech. 2001;19:51–55. doi: 10.1038/83505. [DOI] [PubMed] [Google Scholar]
- (24).Heaton RJ, Peterson AW, Georgiadis RM. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3701–3704. doi: 10.1073/pnas.071623998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Lee HJ, Wark AW, Goodrich TT, Fang S, Corn RM. Langmuir. 2005;21:4050–4057. doi: 10.1021/la046822h. [DOI] [PubMed] [Google Scholar]
- (26).Fang S, Lee HJ, Wark AW, Kim HM, Corn RM. Anal. Chem. 2005;77:6528–6534. doi: 10.1021/ac051283m. [DOI] [PubMed] [Google Scholar]
- (27).Goodrich TT, Lee HJ, Corn RM. J. Am. Chem. Soc. 2004;126:4086–4087. doi: 10.1021/ja039823p. [DOI] [PubMed] [Google Scholar]
- (28).He L, Musick MD, Nicewarner SR, Salinas FG, Benkovic SJ, Natan MJ, Keating CD. J. Am. Chem. Soc. 2000;122:9071–9077. [Google Scholar]
- (29).Landegren U, Kaiser R, Sanders J, Hood L. Science. 1988;241:1070–1077. [Google Scholar]
- (30).Barany F. Proc. Natl. Acad. Sci. U.S.A. 1991;88:189–193. doi: 10.1073/pnas.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zhong X.-b., Reynolds R, Kidd JR, Kidd KK, Jenison R, Marla RA, Ward DC. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11559–11564. doi: 10.1073/pnas.1934783100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Hashimoto M, Hupert ML, Murphy MC, Soper SA, Cheng Y-W, Barany F. Anal. Chem. 2005;77:3243–3255. doi: 10.1021/ac048184d. [DOI] [PubMed] [Google Scholar]
- (33).Gerry NP, Witowski NE, Day J, Hammer RP, Barany G, Barany F. J. Mol. Biol. 1999;292:251–262. doi: 10.1006/jmbi.1999.3063. [DOI] [PubMed] [Google Scholar]
- (34).Castilla LH, Couch FJ, Erdos MR, Hoskins KF, Calzone K, Garber JE, Boyd J, Lubin MB, Deshano ML, Brody LC, Collins FS, Weber BL. Nat. Genet. 1994;8:387–391. doi: 10.1038/ng1294-387. [DOI] [PubMed] [Google Scholar]
- (35).Brockman JM, Frutos AG, Corn RM. J. Am. Chem. Soc. 1999;121:8044–8051. [Google Scholar]
- (36).Friedman LS, Ostermeyer EA, Szabo CI, Dowd P, Lynch ED, Rowell SE, King M-C. Nat. Genet. 1994;8:399–404. doi: 10.1038/ng1294-399. [DOI] [PubMed] [Google Scholar]
- (37).Grabar KC, Freeman RG, Hommer MB, Natan MJ. Anal. Chem. 1995;67:735–743. [Google Scholar]
- (38).Goodrich GP, Helfrich MR, Overberg JJ, Keating CD. Langmuir. 2004;20:10246–10251. doi: 10.1021/la048434l. [DOI] [PubMed] [Google Scholar]
- (39).Lee HJ, Wark AW, Corn RM. Anal. Chem. 2005;77:7832–7837. doi: 10.1021/ac0516180. [DOI] [PubMed] [Google Scholar]
- (40).Frutos AG, Smith LM, Corn RM. J. Am. Chem. Soc. 1998;120:10277–10282. [Google Scholar]
- (41).Lehr H-P, Reimann M, Brandenburg A, Sulz G, Klapproth H. Anal. Chem. 2003;75:2414–2420. doi: 10.1021/ac0206519. [DOI] [PubMed] [Google Scholar]
- (42).Livache T, Maillart E, Lassalle N, Mailley P, Corso B, Guedon P, Roget A, Levy Y. J. Pharm. Biomed. Anal. 2003;32:687–696. doi: 10.1016/s0731-7085(03)00176-6. [DOI] [PubMed] [Google Scholar]
- (43).Easton DF, Bishop DT, Ford D, Crockford GP. Am. J. Hum. Genet. 1993;52:678–701. [PMC free article] [PubMed] [Google Scholar]