Abstract
NBD-556 and the chemically and structurally similar NBD-557 are two low-molecular-weight compounds that reportedly block the interaction between the HIV-1 envelope glycoprotein gp120 and its receptor, CD4. NBD-556 binds to gp120 with a binding affinity of 2.7 × 105 M−1 (Kd = 3.7 μM) in a process characterized by a large favorable change in enthalpy partially compensated by a large unfavorable entropy change; a thermodynamic signature similar to that observed for sCD4 binding to gp120 and associated with a large structuring of the gp120 molecule as also demonstrated by CD spectroscopy. NBD-556, like CD4, activates the binding of gp120 to the HIV-1 coreceptor, CCR5, and to the 17b monoclonal antibody, which recognizes the coreceptor-binding site of gp120. NBD-556 stimulates HIV-1 infection of CD4-negative, CCR5-expressing cells. The thermodynamic signature of the binding of NBD-556 to gp120 is very different from that of another viral entry inhibitor, BMS-378806. Whereas NBD-556 binds gp120 with large favorable enthalpy and compensating unfavorable entropy changes, BMS-378806 does so with a small binding enthalpy change in a mostly entropy-driven process. NBD-556 is a competitive inhibitor of sCD4 and elicits a similar structuring of the coreceptor binding site, whereas BMS-378806 does not compete with sCD4 and does not induce coreceptor binding. These studies demonstrate that low-molecular-weight compounds can induce conformational changes in the HIV-1 gp120 glycoprotein similar to those observed upon CD4 binding revealing distinct strategies for inhibiting the function of the HIV-1 gp120 envelope glycoprotein. Furthermore, competitive and non-competitive compounds have characteristic thermodynamic signatures that can be used to guide the design of more potent and effective viral entry inhibitors.
The entry of human immunodeficiency virus (HIV-1) into target cells is mediated by the gp120 exterior envelope glycoprotein and the gp41 transmembrane envelope glycoprotein, which assemble into trimers on the virion surface (1, 2). Upon engaging the receptor, CD4, the gp120 glycoprotein undergoes extensive structural ordering; the resulting conformation of gp120 can bind the second HIV-1 receptor, CCR5 or CXCR4 (3-7). Receptor binding induces further conformational changes in the HIV-1 envelope glycoproteins that allow the gp41 glycoproteins to mediate the fusion of the viral and cell membranes.
Thermodynamically, the binding of CD4 is characterized by large favorable enthalpy and large unfavorable entropy changes that reflect the structuring of gp120, a protein with large unstructured regions in its unligated form (8). CD4 mimetic miniproteins based on scyllatoxin containing the gp120-binding epitope (9) also elicit conformational changes in gp120 resembling those triggered by CD4 binding, and consequently are characterized by similar thermodynamic signatures (10, 11).
Recently, two low-molecular weight compounds that presumably interfere with viral entry of HIV-1 into cells were reported (12) (Figure 1). The studies presented here show that these compounds are competitive inhibitors of CD4 and that they induce conformational changes in gp120 similar to those induced by CD4. These compounds activate coreceptor binding and, in our studies, enhance HIV-1 entry into CD4-negative cells expressing CCR5. This behavior is in contrast to that of the potent viral entry inhibitor BMS-378806, which binds gp120 with a small favorable enthalpy change in an entropically driven process (13). BMS-378806 does not compete with CD4 and does not induce any major structural ordering of the gp120 molecule exerting its antiviral action in an allosteric fashion. In this paper, we demonstrate experimentally that the binding mode and antiviral properties of these compounds are reflected in their binding thermodynamics and that the thermodynamic signature of a compound can be used in the design of more potent and effective viral entry inhibitors.
Figure 1.
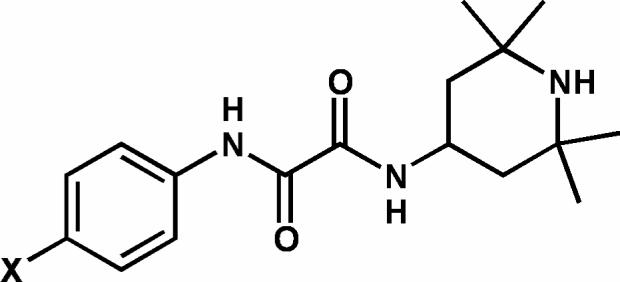
General structure of the two compounds, NBD-556 (X = Cl) and NBD-557 (X = Br).
MATERIALS AND METHODS
Synthesis of Compounds
BMS-378806 was prepared according to a published procedure (14). The synthesis of NBD-557 and NBD-556 is outlined in Figure 2. Acylation of either 4-bromo- or 4-chloroaniline provided the corresponding ethyl oxalamate 1. Hydrolysis of 1 with NaOH to the corresponding acid followed by amide formation with 4-amino-2,2,6,6-tetramethylpiperidine in the presence of EDC and HOBt furnished the desired oxalamides NBD-557 and NBD-556. Tetrahydrofuran (THF) was freshly distilled from sodium/benzophenone under argon. All reagents were purchased from Aldrich and used without further purification. Reactions were magnetically stirred and carried out under argon atmosphere. Flash chromatography was performed with silica gel 60 (particle size 0.040 − 0.062 mm) supplied by Silicycle. Infrared spectra were recorded on a Jasco Model FT/IR-480 Plus spectrometer. Proton and carbon-13 spectra were recorded on a Bruker AMX-500 spectrometer. Chemical shifts are reported relative to chloroform (δ 7.26 for 1H-NMR and δ 77.0 for 13C-NMR). High-resolution mass spectra were measured at the University of Pennsylvania Mass Spectrometry Service Center.
Figure 2.
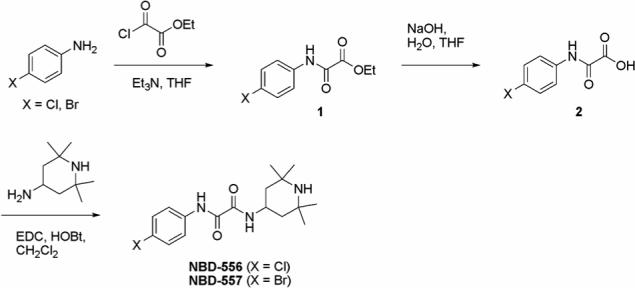
Scheme for the synthesis of NBD-556 and NBD-557.
(1) N-(4-Bromo-phenyl)-N′-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamide (NBD-557)
Ethyl oxalyl chloride (0.20 mL, 1.74 mmol) was added to a round bottom flask containing anhydrous THF (10 mL), and the resulting solution was cooled in an ice bath. Triethylamine (0.24 mL, 1.74 mmol) and 4-bromoaniline (300 mg, 1.74 mmol) were then added, and the resulting mixture was removed from the ice bath and allowed to come to room temperature. The reaction mixture was stirred for 6 h at room temperature, and the solid was removed by vacuum filtration. The filtrate was concentrated in vacuo, and the residue was re-dissolved in EtOAc (20 mL). The organic phase was washed with 1M HCl (2 × 10 mL) and saturated NaHCO3 (10 mL), and dried over MgSO4. The solvent was removed under reduced pressure to give the desired ester (408 mg, 88%), which was used in the subsequent reaction without further purification. M.p. 153−154°C. 1H NMR (500 MHz, CDCl3) δ 8.90 (br s, 1H), 7.54 (d, J = 8.9 Hz, 2H), 7.48 (d, J = 8.9 Hz, 2H), 4.40 (q, J = 7.1 Hz, 2H), 1.41 (t, J = 7.1 Hz, 3H); 13C NMR (125 MHz, CDCl3) δ 160.7, 153.9, 135.4, 132.2, 121.3, 118.3, 63.8, 13.9; IR (thin film): 3334, 3113, 2980, 1729, 1710, 1542, 1491, 1477, 1302, 1176 cm−1; high resolution mass spectrum (ES+) m/z 270.9851 [M+; calculated for C10H10BrNO3: 270.9844].
To a flask containing the above ester (400 mg, 1.47 mmol) was added 1M NaOH solution (7.4 mL, 7.40 mmol) and THF (8 mL), and the resulting mixture was stirred at room temperature for 20 h. EtOAc (20 mL) was added and the organic layer was washed with 1 M NaOH (2 × 10 mL). The combined aqueous layer was acidified to pH 2 with 2 M HCl, and extracted with EtOAc (3 × 20 mL). The combined organic layer was dried over MgSO4 and concentrated to give the crude acid (272 mg, 76%). The acid was re-dissolved in anhydrous THF (3 mL) and added to a flask containing 4-amino-2,2,6,6-tetramethylpiperidine (0.21 mL, 1.23 mmol). HOBt (196 mg, 1.45 mmol) and EDC (279 mg, 1.45 mmol) were added and the resulting mixture was stirred at room temperature for 20 h. The reaction mixture was diluted with CH2Cl2 (30 mL) and the organic layer was washed with water (4 × 20 mL) and dried over MgSO4. After solvent was removed in vacuo, the crude product was chromatographed (EtOAc/MeOH, 1:1) to give NBD-557 (176 mg, 41%): Rf = 0.3; m.p. 204−205°C; 1H NMR (500 MHz, CDCl3) δ 9.25 (br s, 1H), 7.53 (d, J = 9.0 Hz, 2H), 7.49 (d, J = 9.0 Hz, 2H), 7.29 (br d, J = 7.9 Hz, 1H), 4.20−4.33 (m, 1H), 1.92 (dd, J = 12.6, 3.8 Hz, 2H), 1.29 (s, 6H), 1.17 (s, 6H), 1.03−1.13 (m, 3H); 13C NMR (125 MHz, CDCl3) δ 158.8, 157.6, 135.5, 132.2, 121.3, 118.1, 51.0, 44.6, 43.8, 34.9, 28.5; IR (thin film): 3311, 3249, 2959, 2917, 1658, 1511 cm−1; high resolution mass spectrum (ES+) m/z 382.1137 [(M+H)+; calculated for C17H25BrN3O2: 382.1130].
(2) N-(4-Chloro-phenyl)-N′-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamide (NBD-556)
Ethyl oxalyl chloride (0.26 mL, 2.35 mmol) was added to a round bottom flask containing anhydrous THF (10 mL), and the resulting solution was cooled in an ice bath. Triethylamine (0.33 mL, 2.35 mmol) and 4-chloroaniline (300 mg, 2.35 mmol) were then added, and the resulting mixture was removed from the ice bath and allowed to come to room temperature. The reaction mixture was stirred for 6 h at room temperature, and the solid was removed by vacuum filtration. The filtrate was concentrated in vacuo, and the residue was re-dissolved in EtOAc (20 mL). The organic phase was washed with 1M HCl (2 × 10 mL) and saturated NaHCO3 (10 mL), and dried over MgSO4. The solvent was removed under reduced pressure to give the desired ester (505 mg, 94%), which was used in the subsequent reaction without further purification. M.p. 149−150°C. 1H NMR (500 MHz, CDCl3) δ 8.92 (br s, 1H), 7.59 (d, J = 8.9 Hz, 2H), 7.33 (d, J = 8.9 Hz, 2H), 4.40 (q, J = 7.2 Hz, 2H), 1.41 (t, J = 7.1 Hz, 3H); 13C NMR (125 MHz, CDCl3) δ 160.8, 153.9, 134.9, 130.6, 129.2, 121.0, 63.8, 13.9; IR (thin film): 3333, 3120, 2988, 1728, 1700, 1597, 1546, 1496, 1476, 1302, 1177 cm−1; high resolution mass spectrum (ES+) m/z 227.0338 [M+; calcd for C10H10ClNO3: 227.0349].
To a flask containing the above ester (580 mg, 2.55 mmol) was added 1M NaOH solution (12.8 mL, 12.8 mmol) and THF (15 mL), and the resulting mixture was stirred at room temperature for 20 h. EtOAc (20 mL) was added and the organic layer was washed with 1M NaOH (2 × 10 mL). The combined aqueous layer was acidified to pH 2 with 2M HCl, and extracted with EtOAc (3 × 20 mL). The combined organic layer was dried over MgSO4 and concentrated to give the crude acid (430 mg, 85%). The acid was re-dissolved in anhydrous THF (6 mL) and added to a flask containing 4-amino-2,2,6,6-tetramethylpiperidine (0.41 mL, 2.37 mmol). HOBt (379 mg, 2.80 mmol) and EDC (540 mg, 2.80 mmol) were added and the resulting mixture was stirred at room temperature for 20 h. The reaction mixture was diluted with CH2Cl2 (30 mL) and the organic layer was washed with water (4 × 20 mL) and dried over MgSO4. After solvent was removed in vacuo, the crude product was chromatographed (EtOAc/MeOH, 1:1) to give NBD-556 (247 mg, 34%): Rf = 0.3; m.p. 197−198°C; 1H NMR (500 MHz, CDCl3) δ 9.35 (br s, 1H), 7.59 (d, J = 8.8 Hz, 2H), 7.30−7,40 (m, 3H), 4.20−4.33 (m, 1H), 1.92 (dd, J = 12.6, 3.8 Hz, 2H), 1.27 (s, 6H), 1.17 (s, 6H), 1.03−1.13 (m, 3H); 13C NMR (125 MHz, CDCl3) δ 158.8, 157.6, 134.9, 130.4, 129.2, 121.0, 50.9, 44.5, 43.7, 34.8, 28.4; IR (thin film): 3297, 2959, 1660, 1592, 1512, 1403 cm−1; high resolution mass spectrum (ES+) m/z 322.1312 [(M-CH3)+; calculated for C16H21ClN3O2: 322.1322].
Cell Lines
The codon-optimized env sequences encoding the gp120 glycoprotein from the YU2 HIV-1 strain with a CD5 signal peptide and a carboxy-terminal (His)6 tag were cloned into the pCIG plasmid. This plasmid was modified from the pIRES-GS glutamine synthetase amplification vector (15). The pCIG-YU2 gp120 plasmid was transfected into CHO-K1 cells, which were subjected to selection for glutamine synthetase, as described (15). Highly expressing clones were selected for production of the gp120 glycoprotein. CHO-K1 cells expressed YU2 gp120 in 100 mL suspension cultures of ProCHO5 (Cambrex) with 2 % GS supplement (JRH), 1% HT Supplement (Gibco), 100 U/L penicillin G/ 100 mg/L streptomycin (Gibco), 275 μM MSX (Sigma) and 1 mL/L Protease Inhibitor Cocktail Set III (Calbiochem). 293T human embryonic kidney and Cf2Th canine thymocytes (ATCC) were grown at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum (Sigma) and 100 μg/mL penicillin-streptomycin (Mediatech, Inc.). Cf2Th cells stably expressing human CCR5 were grown in medium supplemented with 0.4 mg/mL G418 (Invitrogen).
Preparation and Purification of Protein
Full-length gp120 from the YU2 strain of HIV-1 with a C-terminal His tag was produced using CHO-K1 cells described above. Cell supernatants were dialyzed against 20 mM sodium phosphate, 500 mM NaCl, pH 7.4. Approximately 50 mL dialyzed supernatant was passed through a 5 mL HisTrap HP column (Amersham Bioscience) and the protein was eluted using a linear imidazole gradient (ÄKTA FPLC system, Amersham Bioscience). Eluted fractions were immediately neutralized by dilution in 200 mM Tris, pH 7.4, then dialyzed into PBS (Roche Diagnostics). Approximately 30 ml of this solution was passed through a 5 mL HiTrap NHS-activated HP column (Amersham Bioscience) coupled to mAb F105 (Strategic Biosolutions) and the protein was eluted using a stepwise gradient of 100 mM glycine, 150 mM NaCl, pH 2.4 (ÄKTA FPLC system, Amersham Bioscience). Eluted fractions were immediately neutralized by dilution in 0.2 M Tris, pH 7.4, then dialyzed into PBS. Purified gp120 was analyzed by SDS-PAGE (Invitrogen) and shown to be a single species of approximately 90 kDa, of which 37 kDa is due to glycosylation. The purified YU2 gp120 was stored at −20 °C in 1 mL aliquots of 0.5 mg.
Soluble D1-D2 CD4 (sCD4) was kindly provided by Dr. Richard Wyatt (National Institutes of Health Vaccine Research Center, Bethesda, MD). Monoclonal antibody 17b was produced by Strategic Biosolutions (Newark, DE). sCD4 and MAb 17b were both dialyzed into PBS and stored in approximately 6 and 5 mg aliquots, respectively.
Isothermal Titration Calorimetry
Isothermal titration calorimetric experiments were performed using a high-precision VP-ITC titration calorimetric system from MicroCal Inc. (Northampton, MA). For the direct determination of the binding of the inhibitors to gp120 the calorimetric cell (about 1.4 mL), which contained gp120 dissolved in PBS (Roche Diagnostics GmbH), pH 7.4, with 2 % DMSO, was titrated with the inhibitors NBD-557, NBD-556 or BMS-378806 dissolved in the same buffer. The concentration of gp120 was about 3 μM and the concentration of inhibitor in the injection syringe was between 100 and 175 μM for NBD-557 and NBD-556 and 45 μM for BMS-378806. The binding of each inhibitor was studied at different temperatures in the range 15 − 25 °C for NBD-557 and NBD-556 and 15 − 37 °C for BMS-378806. CD4 binding to gp120 was studied in experiments where the calorimetric cell, which contained 3 μM gp120, was titrated with a solution of 30 μM sCD4. The effect of NBD-556 and BMS-378806 on CD4 binding was studied by equilibrating 3 μM gp120 with 300 μM of either NBD-556 or BMS-378806 prior to titration with 30 μM CD4. Binding of MAb 17b to gp120 was studied by stepwise additions of 15 μM (30 μM per Fab-site) to the calorimetric cell containing 3 μM gp120 by itself or equilibrated with 300 μM NBD-556. The effect of 17b on the binding of NBD-556 to gp120 was studied by titration of gp120 pre-bound to 17b with NBD-556. To that end, 2.5 μM gp120 equilibrated with 5 μM 17b was titrated with NBD-556 at a concentration of 100 μM. All titrations were performed by adding the titrant in steps of 10 μL. All solutions were properly degassed to avoid any formation of bubbles in the calorimeter during stirring. The heat evolved upon each injection of inhibitor was obtained from the integral of the calorimetric signal. The heat associated with binding to gp120 in the cell was obtained by subtracting the heat of dilution from the heat of reaction. The individual heats were plotted against the molar ratio and the enthalpy change, ΔH, and association constant, Ka = 1/Kd, were obtained by non-linear regression of the data. Because of the low affinity, the stoichiometry was fixed to a value of one for the binding of NDB-556 and NBD-557 to gp120.
Circular Dichroism Spectroscopy
CD experiments were conducted using a Jasco J-710 spectropolarimeter. Wavelength scans were performed with 3.7 μM gp120 in presence of 0, 5, 10, 20 and 300 μM NBD-556 in a 0.1 cm cuvette in a water-jacketed cell. NBD-556 was dissolved in DMSO and dispensed in test tubes. The test tubes were placed in a lyophilizer to remove any DMSO prior to the addition of gp120 prepared in PBS. The solution were mixed thoroughly and allowed to equilibrate for 1 hour. Each complete spectrum was the average of 5 consecutive scans collected from 195 to 240 nm. The individual scans were recorded using a scan rate of 20 nm/min, a bandwidth of 1 nm and a response time of 2 s per point. Buffer scans were accumulated and subtracted from the sample scans and the mean residue ellipticity was computed. The temperature was kept constant at 25 °C.
Recombinant Luciferase Viruses
293T human embryonic kidney cells were co-transfected with plasmids expressing the pCMVΔP1Δenv HIV Gag-Pol packaging construct (16), the R5 YU2 envelope glycoproteins, or the envelope of control Amphotropic Murine Leukemia virus (A-MLV), and the firefly luciferase-expressing vector at a DNA ratio of 1:1:3 μg using the Effectene transfection reagent (Qiagen). Co-transfection produced single-round, replication-defective viruses. The virus-containing supernatants were harvested 24−30 h after transfection, filtered (0.45 μm), aliquoted, and frozen at −80 °C until further use. The reverse transcriptase (RT) activities of all viruses were measured as described previously (17).
Infection by Single-round Luciferase Viruses
Cf2Th-CCR5 target cells were seeded at a density of 6 × 103 cells/well in 96-well luminometer-compatible tissue culture plates (Dynex) 24 h before infection. On the day of infection, NBD-556 (0−100 μM) was incubated with recombinant viruses (10,000 RT units) at 37 °C for 30 min. The mixtures were then added to the target cells and incubated for 48 h at 37 °C; after this time, the medium was removed from each well, and the cells were lysed by the addition of 30 μL passive lysis buffer (Promega) and three freeze-thaw cycles. An EG&G Berthold Microplate Luminometer LB 96V was used to measure the luciferase activity of each well after the addition of 100 μL of luciferin buffer (15 mM MgSO4, 15 mM KPO4, pH 7.8, 1 mM ATP, and 1 mM dithiothreitol) and 50 μL of 1 mM D-luciferin potassium salt (BD Pharmingen).
Radiolabeling of Env gGlycoprotein
293T cells were seeded at 3.5 × 106 cells in a T75 tissue culture flask one day before transfection. Cells were co-transfected with 9 μg of pSVIIIEnv (YU2) and 1 μg of pLTR-Tat using the Polyfect transfection reagent (Qiagen). One day after transfection the cells were labeled for 48 hrs with [35S]Express protein labeling mix (100 μCi/mL) (Perkin-Elmer). The supernatant were harvested 48 h later, cleared by centrifugation at 2000 rpm for 5 min, and stored at 4 °C. The amount of labeled gp120 was quantitated by immunopercipitation with AIDS patient sera and protein A-Sepharose bead (Amersham Bio-Sciences), followed by SDS-PAGE gels and autoradiography.
Env-CCR5 Binding Assay
Cf2Th cells expressing high levels of CCR5 were lifted using 5 mM EDTA pH 7.5. The cells were washed with serum free DMEM, added to micorcentrifuge tubes (2−3 × 106 cells/tube), and incubated with 500 μL of labeled YU2 gp120 in the presence and the absence of varying concentrations of CD4-Ig or NBD-556 at 37 °C for 1.5 h with gentle agitation. The supernatants were removed following incubation and the cells were washed 2 times with cold DMEM before lysis in 0.5 mL of IP buffer containing 0.5 M NaCl, 10 mM Tris, pH 7.5 and 0.5% [vol/vol] NP40 and a cocktail of protease inhibitors. The cells were incubated in IP buffer for 30 min at 4 °C with gentle agitation. The lysates were cleared by centrifugation at 14,000 rpm for 30 min at 4 °C and immunoprecipitated with AIDS patient sera and protein A-sepharose beads and visualized by autoradiography of 3−8% SDS-PAGE gel.
RESULTS
Isothermal Titration Calorimetric Studies of Inhibitor Binding to HIV-1 gp120
Figure 3 shows a microcalorimetric titration of gp120 from the YU2 strain of HIV-1 with NBD-556. The binding of NBD-556 to gp120 at 25°C is characterized by a binding affinity of 3.7 μM, which corresponds to a change in Gibbs energy of −7.4 kcal/mol. The large favorable binding enthalpy change of −24.5 kcal/mol is partially compensated by a large unfavorable entropy change of −57.4 cal/(K × mol). Under similar conditions, sCD4 binds to gp120 with a binding enthalpy of −34.5 kcal/mol and an entropy change of −79 cal/(K × mol). The binding of NBD-557 to gp120 was also characterized by an enthalpy change and affinity similar to those measured for NBD-556 (data not shown). The change in heat capacity upon binding of NBD-556 to gp120, calculated from the temperature dependence of the binding enthalpy, is −1000 ± 50 cal/(K × mol) (Insert, Figure 3). Under similar conditions, the heat capacity change associated with the binding of sCD4 to gp120 is −1800 cal/(K × mol). As presented in an earlier report (13), the binding of BMS-378806 to gp120 is characterized by an affinity of 43 nM and an enthalpy change that is close to zero at 25 °C. The change in heat capacity for the binding of BMS-378806 to gp120 is only −292 cal/(K × mol). Thus, NBD-556 exhibits a thermodynamic signature similar to that of sCD4 and much different from that of BMS-378806. Figure 4 shows the thermodynamic profiles for sCD4, NBD-556 and BMS-378806. The large favorable enthalpy change coupled to a large unfavorable entropy and large negative heat capacity is characteristic of a binding process that induces a significant structuring in the target protein (8).
Figure 3.
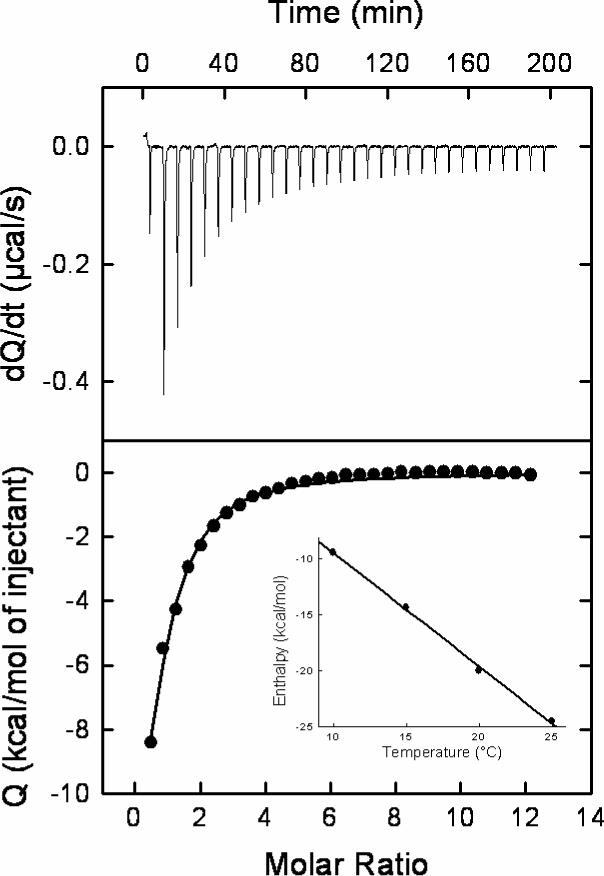
Calorimetric titration of gp120 with NBD-556 at 25 °C in PBS, pH 7.4, with 2% DMSO. The concentration of gp120 was 3.3 μM and the syringe contained NBD-556 at a concentration of 175 μM. The change in heat capacity was calculated from the temperature dependence of the binding enthalpy (insert).
Figure 4.
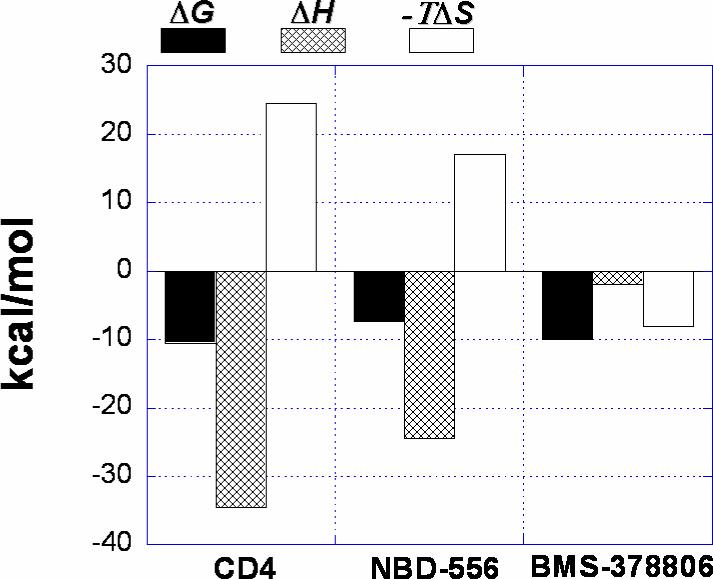
Dissection of the binding thermodynamics of sCD4, NBD-556 and BMS-378806 to gp120 at 25 °C.
CD of gp120 Bound to NBD-556
The gp120 structuring is also reflected in the CD spectrum of gp120 bound to NBD-556 (Figure 6). The specific increase in secondary structure induced by NBD-556 is most pronounced between the isodichroic points at 221 nm and 238 nm suggesting a larger helical content. The concomitant reduction in random coil is reflected in the positive increase in ellipticity around 200 nm (18). The effect of NBD-556 is similar to the change in CD spectrum observed for gp120 upon binding to CD4 (19).
Figure 6.
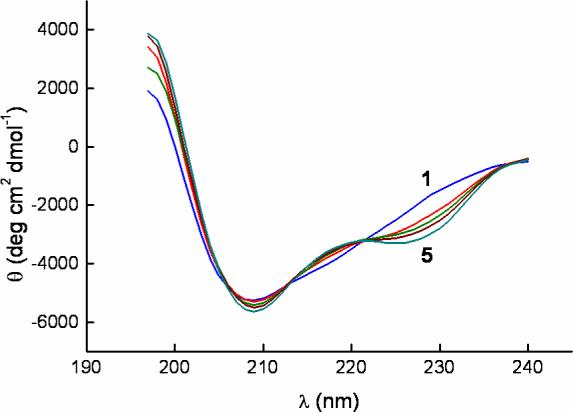
CD spectra of gp120 alone and in the presence of NBD-556. The concentrations of NBD-556 were 0, 5, 10, 20 and 300 μM from curve 1 to 5 as indicated in the figure. The concentration of gp120 was kept constant at 3.7 μM in all experiments.
The effect of NBD-556 and BMS-378806 on CD4 Binding to gp120
The effect of the binding of NBD-556 or BMS-378806 on CD4 binding to gp120 was assessed by performing sCD4 binding experiments on gp120 pre-bound to either compound. The expected apparent affinity in the presence of a competitive inhibitor can easily be calculated using the following equation:
| (1) |
where Ka is the association constant for sCD4 in the absence of inhibitor, Ka, app is the expected apparent association constant for sCD4 in the presence of inhibitor, Ka,I is the association constant of the inhibitor and [I] is the concentration of inhibitor. Similarly, the expected enthalpy change in the presence of a competitive inhibitor, ΔHapp, is calculated according to the equation:
| (2) |
where ΔH and ΔHI are the enthalpy changes for the binding of sCD4 and the inhibitor, respectively. Fb is the degree of saturation of gp120 with the pre-bound inhibitor.
According to Eqs. 1, 2, in the presence of 300 μM NBD-556, sCD4 is expected to bind gp120 with an association constant of about 1 × 106 M−1 (Kd = 1 μM) and a change in enthalpy of −10 kcal/mol if NBD-556 is a competitive inhibitor. Figure 5 shows the microcalorimetric titrations of gp120 (3 μM) with sCD4 in the absence of inhibitor and in the presence of 300 μM NBD-556, corresponding to more than 99 % saturation of gp120. The experimentally measured binding enthalpy of sCD4 to gp120 in the presence of a saturating concentration of NBD-556 is reduced to −11 ± 1 kcal/mol, which agrees well with the expected enthalpy change calculated with Eq. 2. The experimentally determined affinity under these conditions is 0.3 ± 0.1 μM, which is close to the expected value for a purely competitive inhibitor; in terms of the Gibbs energy of binding, the difference is 0.65 kcal/mol, which is well within experimental error. Taken together, the binding enthalpy change and affinity of sCD4 for gp120 in the presence of NBD-556 indicate that NBD-556 and sCD4 compete for the same binding site of gp120. The results are completely different for BMS-378806; in this case, the titrations in which 3 μM gp120 is incubated with 300 μM BMS-378806 show only a 10-fold reduction in the binding affinity of sCD4. Under those conditions, if BMS-378806 were a competitive inhibitor, the apparent affinity of CD4 for gp120 would have been reduced 7000-fold instead of 10-fold. The results clearly indicate that BMS-378806 is not a competitive inhibitor of sCD4. Albeit 700 times weaker than the affinity reduction expected from a competitive inhibitor, the 10-fold reduction in the affinity of CD4 for gp120 in the presence of excess BMS-378806 is real and most likely originates from some non-specific conformational effect.
Figure 5.
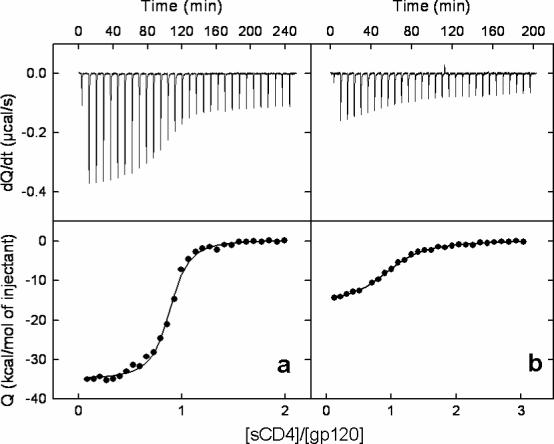
Microcalorimetric titrations of gp120 (3 μM) with sCD4 a) in absence of inhibitor and b) in presence of 300 μM NBD-556. The concentration of sCD4 in the syringe was 30 μM in both experiments.
NBD-556 Enhances gp120 Binding to CCR5 and to the 17b Antibody
CD4 binding stabilizes a gp120 conformation that efficiently binds CCR5 and CD4-induced antibodies, such as 17b, that recognize the gp120 coreceptor-binding site (20, 21). To determine whether NBD-556 would induce similar changes in gp120, the binding of radiolabeled gp120 glycoprotein to CCR5-expressing cells was examined in the absence or presence of NBD-556. As expected, the binding of gp120 to the Cf2Th-CCR5 cells was increased by incubation with CD4-Ig (Figure 7). Likewise, both NBD-556 and NBD-557 enhanced gp120 binding to the Cf2Th-CCR5 cells. The observed binding was inhibited by several different CCR5 ligands, including TAK-779, the 2D7 anti-CCR5 antibody, and Compound A. In contrast to NBD-556, BMS-378806 exerted no effect on the interaction of gp120 with the CCR5-expressing cells. Neither NBD-557/NBD-556 nor BMS-378806 inhibited gp120 binding to CCR5 in the presence of sCD4. We conclude that NBD-556 and NBD-557, but not BMS-378806, activate the CCR5-binding ability of HIV-1 gp120.
Figure 7.
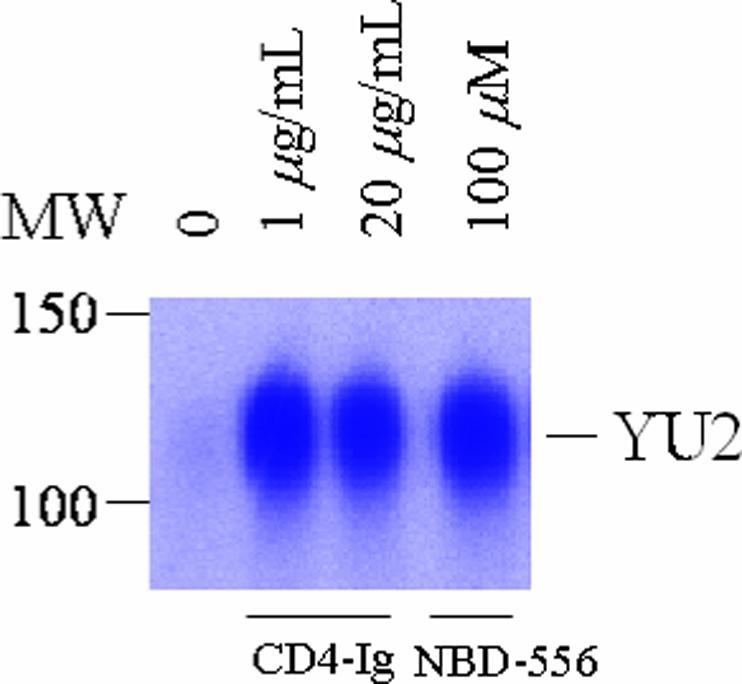
Effect of NBD-556 on gp120 binding to CCR5. Radiolabeled gp120 from the YU2 strain of HIV-1 was incubated with Cf2Th-CCR5 cells in the presence of the indicated ligands. The gp120 bound to the cells is shown.
Isothermal titration calorimetry was used to test whether the presence of saturating concentrations of NBD-556 altered the affinity of 17b for gp120. Direct binding of 17b to gp120 (22) is characterized by an affinity of 5 nM and an enthalpy change of −28.9 kcal/mol at 25 °C. The presence of NBD-556 increases the affinity of 17b for gp120 more than 7-fold and reduces the enthalpy change to −10.9 kcal/mol. As expected, the presence of 17b also enhances the affinity of NBD-556 for gp120. The binding of NBD-556 to gp120 pre-bound to 17b is characterized by an affinity of 0.5 μM and an enthalpy change of −6.5 kcal/mol. The binding of NBD-556 and 17b to gp120 is presented as a thermodynamic cycle in Figure 8.
Figure 8.
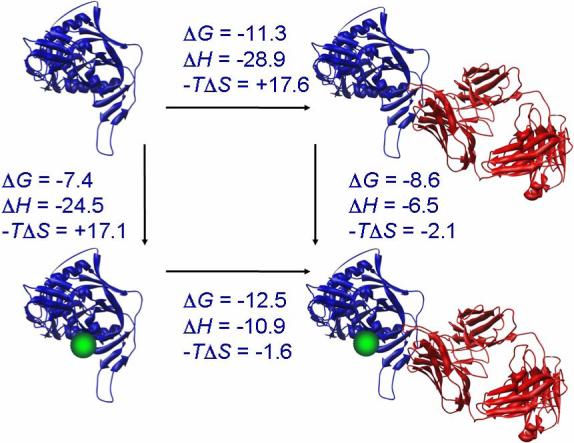
Binding thermodynamics for NBD-556 and 17b to gp120. All values were determined by isothermal titration calorimetry at 25 °C, except values for ΔG and -TΔS for the binding of 17b to gp120 with pre-bound NBD-556 that were calculated by completing the cycle. The structures of gp120, sCD4 and 17b correspond to those in pdb file 1GC1 (27). The compound NBD-556 is represented by the green-colored sphere.
NBD-556 and NBD-557 Enhance Viral Entry in the Absence of CD4
The results described above suggested that NBD-556 might promote conformational changes in gp120 related to virus entry. To test this hypothesis, we examined the effect of NBD-556 on the infection of CD4-negative, CCR5-expressing cells by recombinant HIV-1 bearing the envelope glycoproteins from the ADA strain. Both NBD-556 and NBD-557 exhibited a dramatic enhancing effect on CD4-independent infection (Figure 9 and data not shown). The infection of Cf2Th-CCR5 cells by the HIV-1 (ADA) virus was increased by a factor of more than two orders of magnitude in the concentration range 0 − 40 μM of NBD-556. The effect of NBD-556 on HIV-1 entry is specific, as no enhancement was observed for an HIV-1 bearing the envelope glycoproteins of the unrelated amphotropic murine leukemia virus (A-MLV), under the same conditions. Thus, NBD-556 and NBD-557 both induce entry-related conformational changes in the HIV-1 envelope glycoproteins similar to those induced by CD4.
Figure 9.

The effect of NBD-556 on viral entry. The effect of NBD-556 on the infection of Cf2Th-CCR5 cells by recombinant luciferase-expressing HIV-1 bearing the envelope glycoproteins of the YU2 strain of HIV-1 or the amphotropic murine leukemia virus (A-MLV) is shown. Virus infection is expressed as the percentage of infection (measured by luciferase activity in the target cells) observed in the presence of NBD-556 relative to the level of infection observed in the absence of drug.
DISCUSSION
Previous studies (8) have shown that unliganded gp120 is characterized by the presence of a significant number (up to 50 %) of unstructured residues. Furthermore, the relevant binding events associated with its biological function are coupled to large structuring events. For example, the binding of gp120 to CD4, the first event in HIV-1 entry into cells, is coupled to the structuring of approximately 130 gp120 residues, many of which define the coreceptor-binding epitope. The so-called activation of the coreceptor-binding site induced by CD4 binding is, at the thermodynamic level, a reduction in the conformational energy penalty associated with structuring the binding site. In the absence of the CD4 or a surrogate molecule, the coreceptor-interactive region on gp120 is not in a binding-competent conformation, and consequently the binding to coreceptor or a surrogate (e.g. 17b) is much weaker because part of the binding energy is used in structuring the site.
The thermodynamic signature characteristic of binding events that are coupled to large structuring processes includes large favorable enthalpies, large unfavorable entropies and large negative heat capacity changes. For example, at 25°C the binding of sCD4 to gp120 is characterized by an enthalpy change of −34.5 kcal/mol, an entropy change of −79 cal/(K × mol) and a heat capacity change of −1.8 kcal/(K × mol). A negative change in heat capacity is associated with the burial of hydrophobic surface, a process also characterized by a positive entropy change. The experimental observation of an overall negative entropy indicates that the positive entropy associated with the burial of hydrophobic surface is more than compensated by the negative conformational entropy associated with structuring.
The two compounds, NBD-557 and NBD-556, bind gp120 with large favorable enthalpy and unfavorable entropy changes and with a large negative heat capacity change, resembling CD4 itself. This signature contrasts with that of the low-molecular-weight inhibitor of viral entry, BMS-378806, (14, 23, 24), that binds gp120 with a small enthalpy change in a process that is mostly entropy driven (13). The effect of NBD-556 on gp120 binding to CCR5 and to the CD4-induced antibody, 17b, is qualitatively similar to the effect observed for CD4. Binding of NBD-556 to gp120 induces the structuring of the coreceptor-binding site, thus enhancing the affinity of CCR5 and 17b due to a much reduced conformational energy penalty. An estimate of the change in conformational entropy can be made from the measured entropy and heat capacity changes, as described in detail elsewhere (25). Analysis of the data presented here shows that the binding of NBD-556 to gp120 induces the structuring of approximately 67 residues. This number is not as large as the number observed for sCD4 but sufficient to enhance co-receptor binding and remarkable for a compound with a molecular weight of less than 400 g/mol (NBD-556 338 g/mol; NBD-557 382 g/mol). Conformational changes to a more structured state are further confirmed by CD spectroscopy, which show an increase in secondary structure in gp120 after binding to NBD-556.
The results from the cell-based assays of viral entry show that the sCD4-induced structuring of the coreceptor-binding site in gp120 measured microcalorimetrically is similar to that induced by CD4 on the cell surface, resulting in a gp120 conformation with higher affinity for the coreceptor CCR5 or CXCR4. NBD-556 acts like a CD4 surrogate, enhancing viral entry in the absence of CD4. This also supports the relevance of the calorimetry measurements, made on monomeric HIV-1 gp120, to the gp120 glycoprotein in the context of the functional envelope glycoprotein trimer on the virion. The implied conformational flexibility of the HIV-1 gp120 glycoprotein in the virion trimer is consistent with the relationship between the binding thermodynamics and neutralizing activity of anti-gp120 antibodies (22), and with recent cross-linking studies of HIV-1 envelope glycoproteins on the surface of infected cells (26). An understanding of this conformational flexibility will assist the rational design of drugs and vaccines targeting the HIV-1 envelope glycoproteins.
ACKNOWLEDGMENTS
We thank Ms. Yvette McLaughlin for manuscript preparation. J.S. also acknowledges support from the International AIDS Vaccine Initiative, the Bristol-Myers Squibb Foundation, the William A. Haseltine Foundation for the Arts and Sciences, and the late William F. McCarty-Cooper.
Supported by the National Institutes of Health grant GM56550 (E.F., J.S. and A.B.S.) AI24755, AI41851(J.S.) and the National Science Foundation MCB0131241 (E.F.). N.M. was supported by NRSA postdoctoral fellowship (F32 NS43260 M) and D.N was supported by NRSA postdoctoral fellowship (F32 GM072111) from the National Institutes of Health.
ABBREVIATIONS
- ITC
Isothermal Titration Calorimetry
- CD
Circular Dichroism Spectroscopy
- HIV-1
Human Immunodeficiency Virus, type 1
- sCD4
The soluble form of the human CD4 receptor
- Env
Envelope glycoprotein
- GS
Glutamine synthetase
- HT
Hypoxanthine/Thymidine
- MSX
Methionine sulfoximine
- SDS-PAGE
Sodium Dodecyl Sulfate Polyacrylamide Electrophoresis
- DMSO
Dimethyl sulfoxide
- EDC
N-(3-Dimethylaminopropyl)-N'-ethylcarbodiimide hydrochloride
- HOBt
1-Hydroxybenzotriazole
- THF
Tetrahydrofuran
- EtOAc
Ethyl acetate
- MeOH
Methanol
- ES+
Electron ionization
- m/z
mass-to-charge ratio
- A-MLV
Amphotropic Murine Leukemia Virus
REFERENCES
- 1.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–73. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 2.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–8. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 3.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–48. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 4.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–6. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 5.Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–58. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 6.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CCCKR-5. Nature. 1996;381:667–73. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 7.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–7. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 8.Leavitt SA, Schon A, Klein JC, Manjappara U, Chaiken IM, Freire E. Interactions of HIV-1 proteins gp120 and Nef with cellular partners define a novel allosteric paradigm. Curr Protein Pept Sci. 2004;5:1–8. doi: 10.2174/1389203043486955. [DOI] [PubMed] [Google Scholar]
- 9.Vita C, Drakopoulou E, Vizzavona J, Rochette S, Martin L, Menez A, Roumestand C, Yang YS, Ylisastigui L, Benjouad A, Gluckman JC. Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein. Proc Natl Acad Sci U S A. 1999;96:13091–6. doi: 10.1073/pnas.96.23.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Canziani G, Plugariu C, Wyatt R, Sodroski J, Sweet R, Kwong P, Hendrickson W, Chaiken I. Conformational changes of gp120 in epitopes near the CCR5 binding site are induced by CD4 and a CD4 miniprotein mimetic. Biochemistry. 1999;38:9405–16. doi: 10.1021/bi990654o. [DOI] [PubMed] [Google Scholar]
- 11.Dowd CS, Leavitt S, Babcock G, Godillot AP, Van Ryk D, Canziani GA, Sodroski J, Freire E, Chaiken IM. Beta-turn Phe in HIV-1 Env binding site of CD4 and CD4 mimetic miniprotein enhances Env binding affinity but is not required for activation of co-receptor/17b site. Biochemistry. 2002;41:7038–46. doi: 10.1021/bi012168i. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Q, Ma L, Jiang S, Lu H, Liu S, He Y, Strick N, Neamati N, Debnath AK. Identification of N-phenyl-N'-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology. 2005;339:213–25. doi: 10.1016/j.virol.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Si Z, Madani N, Cox JM, Chruma JJ, Klein JC, Schon A, Phan N, Wang L, Biorn AC, Cocklin S, Chaiken I, Freire E, Smith AB, 3rd, Sodroski JG. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc Natl Acad Sci U S A. 2004;101:5036–41. doi: 10.1073/pnas.0307953101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang T, Zhang Z, Wallace OB, Deshpande M, Fang H, Yang Z, Zadjura LM, Tweedie DL, Huang S, Zhao F, Ranadive S, Robinson BS, Gong YF, Ricarrdi K, Spicer TP, Deminie C, Rose R, Wang HG, Blair WS, Shi PY, Lin PF, Colonno RJ, Meanwell NA. Discovery of 4-benzoyl-1-[(4-methoxy-1H- pyrrolo[2,3-b]pyridin-3-yl)oxoacetyl]-2- (R)-methylpiperazine (BMS-378806): a novel HIV-1 attachment inhibitor that interferes with CD4-gp120 interactions. J Med Chem. 2003;46:4236–9. doi: 10.1021/jm034082o. [DOI] [PubMed] [Google Scholar]
- 15.Pu H, Cashion LM, Kretschmer PJ, Liu Z. Rapid establishment of high-producing cell lines using dicistronic vectors with glutamine synthetase as the selection marker. Mol Biotechnol. 1998;10:17–25. doi: 10.1007/BF02745860. [DOI] [PubMed] [Google Scholar]
- 16.Parolin C, Taddeo B, Palu G, Sodroski J. Use of cis- and transacting viral regulatory sequences to improve expression of human immunodeficiency virus vectors in human lymphocytes. Virology. 1996;222:415–22. doi: 10.1006/viro.1996.0438. [DOI] [PubMed] [Google Scholar]
- 17.Rho HM, Gallo RC. Characterization of reverse transcriptase from filine leukemia virus by radioimmunoassay. Virology. 1979;99:192–6. doi: 10.1016/0042-6822(79)90054-0. [DOI] [PubMed] [Google Scholar]
- 18.Woody RW. Circular dichroism. Methods Enzymol. 1995;246:34–71. doi: 10.1016/0076-6879(95)46006-3. [DOI] [PubMed] [Google Scholar]
- 19.Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci U S A. 2000;97:9026–31. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sattentau QJ, Moore JP, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–93. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–88. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–82. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 23.Lin PF, Blair W, Wang T, Spicer T, Guo Q, Zhou N, Gong YF, Wang HG, Rose R, Yamanaka G, Robinson B, Li CB, Fridell R, Deminie C, Demers G, Yang Z, Zadjura L, Meanwell N, Colonno R. A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc Natl Acad Sci U S A. 2003;100:11013–8. doi: 10.1073/pnas.1832214100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Q, Ho HT, Dicker I, Fan L, Zhou N, Friborg J, Wang T, McAuliffe BV, Wang HG, Rose RE, Fang H, Scarnati HT, Langley DR, Meanwell NA, Abraham R, Colonno RJ, Lin PF. Biochemical and genetic characterizations of a novel human immunodeficiency virus type 1 inhibitor that blocks gp120-CD4 interactions. J Virol. 2003;77:10528–36. doi: 10.1128/JVI.77.19.10528-10536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luque I, Freire E. Structure-based prediction of binding affinities and molecular design of peptide ligands. Methods Enzymol. 1998;295:100–27. doi: 10.1016/s0076-6879(98)95037-6. [DOI] [PubMed] [Google Scholar]
- 26.Yuan W, Bazick J, Sodroski J. Characterization of the multiple conformational states of free monomeric and trimeric human immunodeficiency virus (HIV-1) envelope glycoproteins after fixation by crosslinker. J Virol In press. 2006 doi: 10.1128/JVI.00118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–59. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]


