Abstract
Activation of C3 into C3b is a critical step in the complement immune response against pathogenic, immunogenic and apoptotic particles. Ajees et al.1 report a crystal structure of C3b, which deviates from the one reported by Janssen et al.2 and Wiesmann et al.3. Analysis of the data deposited by Ajees et al.1 reveals features that are inconsistent with known physical properties of macromolecular structures and their diffraction data. Therefore, Ajees et al. do not provide substantial evidence for the deviating crystal structure of C3b.
Three structures of the 12-domain protein C3b were reported1-3. In the structure of C3b reported by Ajees et al.1 the CUB domain adopts an unfolded conformation and the TED domain has a C3-like (instead of an activated, C3d-like) conformation and is positioned away from the main body of the molecule. The structure of the remaining 10 domains is similar among the three structures and resembles the C3c structure as determined by Janssen et al. in 20054. The conformation and locations of the CUB and TED domains are of specific interest, because they are critical to the biological functions of this central molecule of the complement system.
While analysing the structural differences among the three structures of C3b, we noticed that the deposited coordinates of Ajees et al. (PDB entry 2HR0) do not form a connected network of molecules in the crystal lattice. The crystal structure forms layers that are separated by a large void in the c-direction (an approximately 30-40 Å thick slab that spans the entire unit cell). First, we considered whether this highly unusual, and unreported, feature was due to a book-keeping error. We reproduced the reported refinement statistics using the diffraction data deposited in the PDB using two independent computer programs (Refmac5 and CNS 1.26). We conclude that the published statistics, the deposited coordinate model and diffraction data are consistent with each other. Second, we considered the possibility of a missing protein molecule. Re-determination by molecular replacement using the program Phaser7 resulted in the same overall molecular arrangement as observed by Ajees et al. We found no evidence for potentially missing protein molecules, neither by features in the electron density map nor by increased scores in the log-likelihood function when searching for additional components in Phaser. In addition to the absence of crystal contacts in the c-direction, we noticed other physically unrealistic features. The deposited diffraction data do not show the features that should arise from the presence of bulk solvent (Figure 1), while the molecular arrangement indicates large regions not occupied by protein molecules. In other words, the diffraction data are consistent with protein molecules in vacuum, but not with molecules surrounded by disordered solvent as always seen for macromolecular crystals8. The B-factors of the model (both the deposited B-factors as well as the B-factors obtained by re-refinement) do not display any significant variations throughout the molecule, even though long segments of the chain are almost completely solvent exposed (see Figure 2). B-factors describe the size of displacements available to the atoms, so they are correlated with disorder for surface-exposed residues and rigid-body like motion of domains9. The Rfree and R distributions are exceptionally low at low resolution, and the Rfree-R difference is unusually small for a structure refined at 2.3-Å resolution using an amplitude-based target function (Figure 1B). Given these physically unrealistic features we question the validity of the deposited diffraction data and the associated model presented in the paper by Ajees et al. Only with the experimental diffraction images made available can the deviating C3b model be either verified or falsified.
Figure 1.
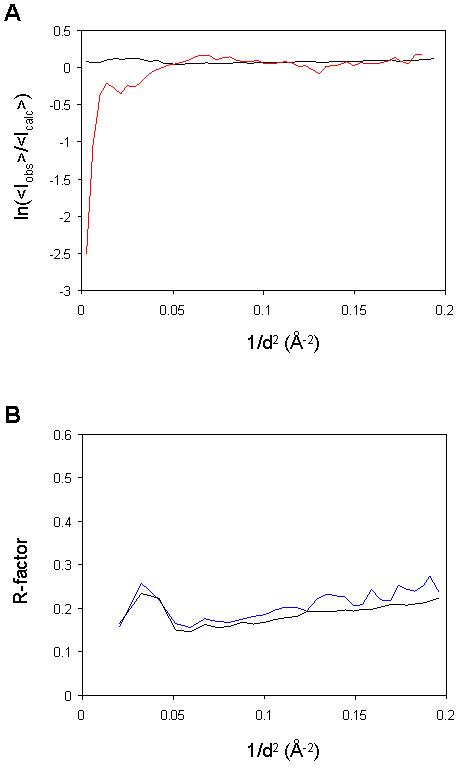
Lack of bulk solvent in the deposited diffraction data. (A) Plot, as a function of resolution, of the logarithm of the ratio between average observed intensities and average intensities calculated from a model lacking a bulk solvent contribution. The black line shows the results for the structure from Ajees et al. (2HR0), while the red line shows results for a control experiment with data from PDB entry 1H1810, which has a similar size and resolution limits. The plot is expected to fall off at low resolution, as seen for 1H18, because the presence of disordered solvent reduces the contrast and hence the average diffraction intensity. (B) R (black line) and Rfree (blue line) plotted as a function of resolution, calculated with CNS 1.2 using the data and the structure of Ajees et al. (2HR0) without a bulk solvent model. In the absence of a bulk solvent model a large discrepancy is expected at low resolution (with R-factors beyond 0.5), because the contribution of the bulk solvent in the crystal to the diffraction data is not accounted for in the model8.
Figure 2.
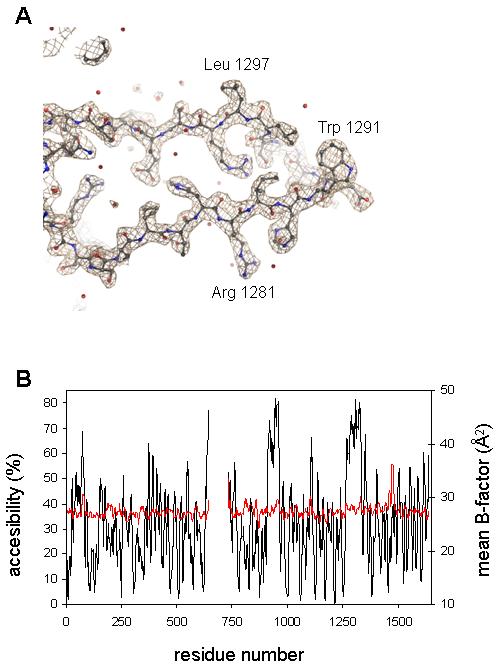
Lack of correlation between surface exposure and disorder. (A) The electron density (calculated with coefficients 2mFobs-DFcalc, φcalc) of a region in the unfolded CUB domain is contoured at 2.5 σ (0.55 e/Å3). (B) Plot of percent surface accessibility (black, computed with NACCESS11) and atomic B-factors (Å2) after re-refinement (red), both averaged over a window of 9 residues. Normally the two measures are highly correlated.
References
- 1.Abdul Ajees A, et al. The structure of complement C3b provides insights into complement activation and regulation. Nature. 2006;444:221–5. doi: 10.1038/nature05258. [DOI] [PubMed] [Google Scholar]
- 2.Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–6. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 3.Wiesmann C, et al. Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature. 2006;444:217–20. doi: 10.1038/nature05263. [DOI] [PubMed] [Google Scholar]
- 4.Janssen BJ, et al. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437:505–11. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 5.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 6.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 7.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–64. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 8.Jiang JS, Brunger AT. Protein hydration observed by X-ray diffraction. Solvation properties of penicillopepsin and neuraminidase crystal structures. J Mol Biol. 1994;243:100–15. doi: 10.1006/jmbi.1994.1633. [DOI] [PubMed] [Google Scholar]
- 9.Kuriyan J, Weis WI. Rigid protein motion as a model for crystallographic temperature factors. Proc Natl Acad Sci U S A. 1991;88:2773–7. doi: 10.1073/pnas.88.7.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker A, Kabsch W. X-Ray structure of pyruvate formate-lyase in complex with pyruvate and CoA: how the enzyme uses the Cys-418 thiyl radical for pyruvate cleavage. J Biol Chem. 2002;277:40036–42. doi: 10.1074/jbc.M205821200. [DOI] [PubMed] [Google Scholar]
- 11.Hubbard SJ, Thornton JM. ‘NACCESS’, Computer Program. Department of Biochemistry and Molecular Biology, University College London; 1993. [Google Scholar]


