Abstract
Heparan sulfate (HS) is a highly sulfated polysaccharide and is present in large quantities on the cell surface and in the extracellular matrix. Herpes simplex virus type 1(HSV-1) utilizes a specialized cell surface HS, known as 3-O-sulfated HS, as an entry receptor to establish infection. Here, we exploit an approach to inhibit HSV-1 infection by using a 3-O-sulfated octasaccharide, mimicking the active domain of the entry receptor. The 3-O-sulfated octasaccharide was synthesized by incubating a heparin octasaccharide (3-OH octasaccharide) with HS 3-O-sulfotransferase isoform 3. The resultant 3-O-sulfated octasaccharide has a structure of ΔUA2S-GlcNS6S-IdoUA2S-GlcNS6S-IdoUA2S-GlcNS3S6S-IdoUA2S-GlcNS6S (where ΔUA is 4-deoxy-α-l-threo-hex-4-enopyranosyluronic acid, GlcN is d-glucosamine and IdoUA is l-iduronic acid). Results from cell based assays revealed that the 3-O-sulfated octasaccharide has stronger activity in blocking HSV-1 infection than that of the 3-OH octasaccharide, suggesting that the inhibition of HSV-1 infection requires a unique sulfation moiety. Our results suggest the feasibility of inhibiting HSV-1 infection by blocking viral entry with a specific oligosaccharide.
Keywords: Heparin, heparan sulfate, herpes simplex virus, glycoprotein D, sulfotransferase, oligosaccharides
Heparan sulfate (HS) is a highly sulfated linear polysaccharide present ubiquitously on the cell surface and in the extracellular matrix. HS plays a role in regulating embryonic development, inflammatory response, blood coagulation and assisting viral/bacterial infections (1). Heparin, a commonly used anticoagulant drug, is a special form of HS containing glucuronic (GlcUA)/iduronic acid (IdoUA) and glucosamine, each carrying sulfo groups (Fig. 1A). The uniquely distributed sulfation pattern of HS polysaccharide is believed to regulate its functional specificity (2-4). Thus, understanding the structure and function relationship of HS attracts considerable interest in improving the anticoagulant efficacy of heparin and exploiting heparin or heparin-like molecules for the development of anticancer and antiviral drugs (5-7). The major difficulty in dissecting the structure and function relationship of HS is to obtain HS oligosaccharides or polysaccharides with defined structures. Chemical synthesis has been the major route to prepare structurally defined oligosaccharides to mimic the functions of HS. However, the synthesis of those molecules larger than hexasaccharides is extremely difficult. Using HS biosynthetic enzymes to prepare biologically active polysaccharides and oligosaccharides has recently gained momentum (8-12).
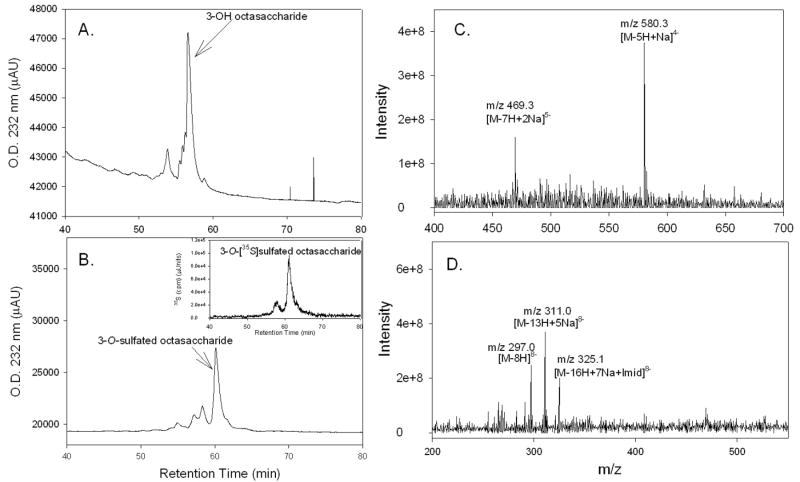
Figure 1. The HS structure and the preparation of 3-O-sulfated octasaccharide by 3-OST-3.
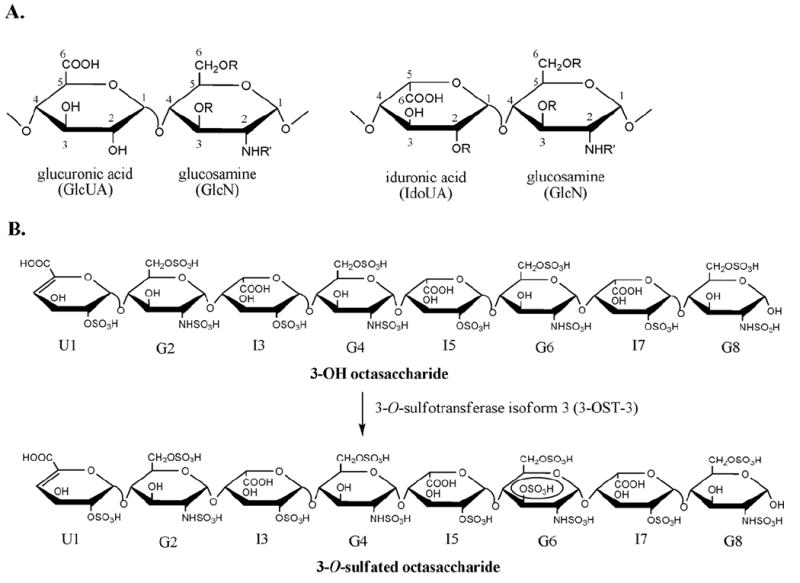
Panel A shows the structures of the disaccharide building blocks that make up heparin and heparan sulfate. Numbers represents positions. Panel B illustrates the scheme to prepare 3-O-sulfated octasaccharide using 3-OST-3 enzyme. For clarity, each sugar unit of the octasaccharide is labeled as U1, G2, etc, where U is uronic acid, G is glucosamine, and I is iduronic acid. Also, R can be either H or SO3H while R′ can be either H, Ac, or SO3H where Ac is an acetyl group.
The biosynthesis of HS occurs in the Golgi apparatus. HS is initially synthesized as a linear copolymer of GlcUA and GlcNAc, which is then followed by various modifications. These modifications are carried out by specialized sulfotransferases and a glucuronyl C5-epimerase. The glucosaminyl N-deacetylase/N-sulfotransferase converts an N-acetylated glucosamine (GlcNAc) residue to an N-sulfo glucosamine (GlcNS) residue. After N-sulfation, C5-epimerase converts some GlcUA residues to IdoUA residues. The resultant polysaccharide is further modified by 2-O-sulfotransferase, 6-O-sulfotransferase and 3-O-sulfotransferase to introduce the 2-O-sulfo group to IdoUA/GlcUA, and 6-O-sulfo/3-O-sulfo groups to the glucosamine residue, respectively. Unlike for proteins and DNA, biosynthesis of HS is not a template-driven process. The control of the sulfation pattern at the polysaccharide is largely unknown. However, enzymatic synthesis of structurally defined oligosaccharides appears to be possible by using a structurally defined oligosaccharide substrate (11).
Herpes simplex virus type 1 (HSV-1) belongs to the alphaherpesviruses subfamily of the herpesvirus family. Infections with HSV-1 are highly prevalent in human and cause localized but recurrent mucocutaneous lesions and encephalitis in rare cases (13). Further, infection of HSV-1 in the eye is a leading cause of blindness in the US (14). Cell surface HS plays an important role in assisting HSV-1 attachment to host cells, as well as in inducing viral entry into target cells (15). The attachment process primarily involves the interaction between HS and the virion envelope glycoproteins gC and/or gB (16). Following attachment, HSV-1 enters the target cells by interacting with specific cell surface entry receptors to establish the infection (17). Three families of HSV-1 entry receptors are known, and all of the known receptors interact with HSV-1 envelope protein gD. HVEM (herpesvirus entry mediator) and nectin-1 represent two families of those receptors, which belong to the TNFR (tumor necrosis factor receptor) family and the immunoglobulin superfamily respectively (18, 19). The 3-O-sulfated HS, which is generated by six 3-OST isoforms, represents the third family of HSV-1 entry receptors (20-24). This receptor is unique as it is a polysaccharide and contains a specific saccharide structure. Further structural analysis of a gD-binding oligosaccharide revealed that the binding of HS to gD requires a 3-O-sulfated glucosamine unit (25).
Our research attention has been focused on identifying a specific HS structure to inhibit herpes infections. Numerous reports have been published on using sulfated compounds, especially sulfated polymers, to block herpes infections (26, 27). However, the sulfated polymers that are structurally heterogeneous could bind to a variety of physiologically important proteins, and thus the risk of toxicity to the host is increased. In this paper, we exploited the possibility of inhibiting HSV-1 infection by targeting the gD-mediated membrane fusion step that is required for virus penetration. The inhibition was achieved by using a 3-O-sulfated heparin octasaccharide, which mimics the gD-binding site of the 3-O-sulfated HS polysaccharide. This octasaccharide was synthesized by incubating purified 3-O-sulfotransferase isoform 3 (3-OST-3) and a heparin-derived octasaccharide, 3-OH octasaccharide (Figure 1B). The structure of the 3-O-sulfated octasaccharide was proven to be ΔUA2S-GlcNS6S-IdoUA2S-GlcNS6S-IdoUA2S-GlcNS3S6S-IdoUA2S-GlcNS6S (where ΔUA is 4-deoxy-α-l-threo-hex-4-enopyranosylurmic acid, S is sulfate, GlcN is d-glucosamine and IdoUA is l-iduronic acid). The synthetic octasaccharide exhibited much more effective inhibition of HSV-1 infection than that of 3-OH octasaccharide at 40 μM or higher in a cell-based assay. Our results suggest the feasibility of inhibiting HSV-1 infection with a specific oligosaccharide as a therapeutic reagent to treat herpes simplex virus infection.
Experimental Procedures
Preparation of heparin tetra-, hexa- and octa-saccharides
A solution containing 1.0 g of heparin (150 U/mg from Celsus, Cincinnati, OH) in 15 ml of 50 mM sodium phosphate buffer (pH 7.0) and heparin lyase I (92 mU) was incubated in a polyethylene vial at 37°C for 10 hrs. When the reaction was 30% completed it was heated to 100°C for 5 min to inactivate the enzyme. The denatured protein was removed by centrifugation at 12,000 g for 10 min. The low molecular weight heparin oligosaccharides (average molecular weight < 5000 Da) obtained by pressure filtration. Fractionation was carried out on a Bio Gel P-10 (fine) column (4.8 cm × 1.5 m) eluted with 200 mM sodium chloride at a flow rate of 0.5 ml/min afforded sized oligosaccharide fractions (detected at 232 nm). The tetrasaccharides, hexasaccharides and octasaccharides were collected, and desalted on a Bio Gel P-2 column (4.8 × 70 cm) using water as the mobile phase and detected at 232 nm. The resulting sized oligosaccharide mixtures were freeze-dried and charge separated using strong anion-exchange high-pressure liquid chromatography (SAX-HPLC) on a semipreparative SAX S5 Spherisorb column (Waters) with a 0.1-2 M NaCl (pH 3.5) linear gradient elution. The major peak from each oligosaccharide mixture was collected, desalted and freeze dried. The structures isolated were confirmed by ESI-MS (28) and 1H-NMR spectroscopy (29).
Preparation of 3-O-[35S]sulfated oligosaccharides
To prepare 3-O-sulfated oligosaccharides, individual reactions consisting of 1-5 μg of the oligosaccharide (either tetra-, hexa-, or octasaccharide) were mixed with approximately 140-240 ng of purified 3-OST-3 enzyme (30) and [35S]PAPS (2 × 106 cpm) in a buffer containing 50 mM MES, 1% Triton X-100, 5 mM MgCl2, 10 mM MnCl2, 100 μg/ml bovine serum albumin, pH 7, in a final volume of 65 μl. The reaction mixture was incubated for 1.5 hrs. at 37°C. The reaction was stopped by boiling at 100°C for 2 min. The resultant solution was centrifuged at 14,000 rpm for 2 min. to remove any insoluble materials. The supernatant was then subjected to a 200 μl DEAE column, and 3-O-[35S]sulfated oligosacccharides were eluted with 1000 mM NaCl.
Expression and purification of 3-OST-3 enzyme
N-terminal (His)6-tagged 3-OST-3 catalytic domain (Gly139-Gly406) was expressed in E. coli. The enzyme was purified by a Ni2+-agarose column as described previously (30).
Determination of the binding of 3-O-[35S]sulfated oligosaccharides to gD
The assay for determining the binding of various sized 3-O-[35S]sulfated oligosaccharides to gD was carried out by an immunoprecipitation procedure using purified gD and DL6, an anti-gD monoclonal antibody (20). Either the 3-O-[35S]sulfated tetra-, hexa-or octasaccharide (1-10 pmole) was incubated in 50 μl of buffer containing 50 mM Tris, 0.001% Triton X-100, pH 7.4 (binding buffer), and 20 μg gD at room temperature for 30 min. The anti-gD monoclonal antibody DL6 (5 μl) was added and the reaction mixture was incubated on ice for 1 hr, followed by the addition of protein A-agarose beads with agitation at 4°C for an additional hour. The protein A-agarose beads were then washed stepwise with 150 mM NaCl in the above binding buffer. The bound oligosaccharides were eluted with 1000 mM NaCl.
Affinity Co-Electrophoresis
The binding affinity between the 3-O-[35S]sulfated oligosaccharides and gD was determined by an affinity co-electrophoresis approach as previously described (20, 31). Briefly, purified gD was casted in 1 % low melting point agarose separation zones in a degassed gel buffer containing 125 mM sodium acetate and 50 mM 3-(N-morpholino)-2-hydroxypropane-sulfonic acid, pH 7, at six final concentrations ranging from 0 to 60 μM of gD in each zone. The 3-O-[35S]sulfated octasaccharide (≈ 28,000 cpm) or 3-O-[35S]sulfated hexasaccharide (≈ 28,000 cpm) was loaded into each separation zone. The gel electrophoresis was performed at 350 mA for 3 hrs in circulated cold water. The gel was dried, and analyzed on a PhosphorImager (Amersham Biosciences, Storm 860). The autoradiography image was obtained by exposing the gel to X-ray film at -80°C for 1 week.
Digestion with Δ4,5-Glycuronate-2-sulfatase
The 3-O-[35S]sulfated octasaccharide was digested with Δ4,5-glycuronate-2-sulfatase (Sigma) as described previously (30).
Reducing end labeling with 2-AB (2-aminobenzamide)
The coupling of the 3-O-[35S]sulfated oligosaccharides with 2-aminobenzamide (2-AB) was achieved using a similar procedure as previously described (32, 33). The 3-O-[35S]sulfated oligosaccharide (1-50 nmoles) was dried. An aliquot (5-20 μl) of a freshly prepared coupling reagent containing 0.35 M [2-AB], 1 M NaBH3CN, 30% (v/v) acetic acid in DMSO was added to the dried sample and incubated for 3 hrs at 65°C. The resultant reaction mixture was spotted on paper chromatography. The paper strips were washed with 1 ml of acetonitrile 3 times while the labeled oligosaccharide was eluted with 1 ml of distilled water. The labeled oligosaccharides were dried and reconstituted in 50-100 μl distilled water for further analysis.
Preparation of larger quantities of 3-O-sulfated octasaccharide
Scale up preparation of the 3-O-sulfated octasaccharide was achieved by coupling 3-OST-3 modification with a PAPS regeneration system (10). The reaction mixture containing 50 mM MES pH 7, 40 μM PAP, 1 mM PNPS, 0.1 mg/ml arylsulfotransferase IV, and 10 μg/ml of purified 3-OST-3 was incubated at room temperature for 15 min. To the reaction, 3-OH octasaccharide (50 μg/ml) substrate was added, and the reaction was rotated at room temperature overnight. The reaction was terminated by boiling for 2 min. The resultant material was loaded onto a 1 ml DEAE column equilibrated with 150 mM NaCl. The 3-O-sulfated octasaccharide was eluted from the DEAE column with 1 M NaCl.
Expression and purification of recombinant gD
Truncated form of gD (Lys25-His332) with a (His)6 tag at the C-terminus was expressed in E. coli. The cDNA of gD was isolated from recombinant baculovirus expressing gD306t (a gift from Drs. Cohen and Eisenberg, University of Pennsylvania) amplified by PCR using two primers, 5′-ATTATTATCATATGAAATATGCCTTGGCGGATGC-3′ (NdeI site underlined), and 5′-ATAATATAAAGCTTATGGTAAGGCGTCGCGGCGT-3′ (HindIII site underlined). The PCR product was cloned into the pET21b vector (Novagen) using NdeI and Hind III sites, and the plasmid was sequenced to confirm the product (University of North Carolina DNA Sequencing Core Facility). The gD expression was carried out in Origami-B cells (Novagen) carrying the pGro7 plasmid (Takara, Japan) expressing chaperonin proteins GroEL and GroES of E. coli. Transformed cells were grown in LB media supplemented with 12.5 μg/ml tetracycline, 15 μg/ml kanamycin, 20 μg/ml chloramphenicol, and 50 μg/ml carbenicillin at 37°C. When the O.D.600nm reached 0.6-0.8, isopropyl-β-thiogalactopyranoside (IPTG) (200 μM) and arabinose (1 mg/ml) were added to induce the expression of gD and chaperon, respectively. The cells were then allowed to shake overnight at 22°C. The cells were harvested, and recombinant gD was purified by a Ni2+-Sepharose column (Amersham). The protein was further purified by a HiLoad™ 16/60 Superdex™ (Amersham Biosciences) which was eluted with a buffer containing 20 mM Tris, 1 M NaCl, pH 7. The purity of the resulting gD was typically greater than 85% as determined by precasted 12% SDS-PAGE (Bio-Rad). This two-step purification allowed for the recovery of approximately 5 mg of purified gD from a liter of bacterial culture.
PAMN-HPLC chromatography
The 3-O-[35S]sulfated oligosaccharides were analyzed by monitoring their elution profiles when applied to a silica-based polyamine (PAMN) HPLC column (0.46 × 25 cm, Waters) (30). The column was eluted using a linear gradient of KH2PO4 from 300 mM to 1000 mM in 60 min at a flow rate of 0.5 ml/min.
Determination of the purity and concentration of 3-O-sulfated octasaccharide
The purity of the 3-O-sulfated HP octasaccharide was determined using a nonporous DEAE-NPR column (0.46 × 7.5 cm, Tosohass). The DEAE-NPR column was eluted using a linear gradient NaCl from 100 mM to 1,000 mM in 20 mM Tris (pH 7.0) in 100 min at a flow rate of 0.5 ml/min. The eluted materials were monitored by both an online radioactive and UV detector. The concentration of 3-O-sulfated octasaccharide was determined by comparing the area of the UV (232 nm) peak with that of the unmodified octasaccharide with known amount.
Preparation of 3-O-[35S]sulfated disaccharide and 3-O-[35S]sulfated tetrasaccharide as well as the corresponding 2-AB labeled standards
The 3-O-[35S]sulfated tetrasaccharide standard that has a structure of ΔUA2S-[3-O-35S]GlcNS3S6S-IdoUA2S-GlcNS6S was prepared by incubating a heparin tetrasaccharide substrate (ΔUA2S-GlcNS6S-IdoUA2S-GlcNS6S) and purified 3-OST-3 enzyme. The 3-O-sulfated tetrasaccharide was purified by a DEAE column and the structural characterization was described in a previous publication (30). The 3-O-[35S]sulfated disaccharide standard (ΔUA2S-[3-O-35S]GlcNS3S6S) was prepared by digesting the 3-O-[35S]sulfated tetrasaccharide using a mixture of heparin lyase I, II, and III followed by the purification by PAMN-HPLC as described previously (30). The 2-AB labeled tetra- and disaccharide standards were prepared by incubating the 3-O-[35S]sulfo tetrasaccharide or the 3-O-[35S]sulfated disaccharide with 2-aminobenzamide as described above. The labeled products were analyzed by PAMN-HPLC to confirm the completion of the labeling reaction. The 2-AB labeled di- or tetrasaccharides carried both 35S-radioactivity and absorbance at 310 nm, which is from the 2-AB tag.
Electrospray ionization mass spectrometry
The sample was dialyzed against 25 mM ammonium acetate using MWCO 1000 membrane (Spectrum). The sample was dissolved in a solution contained in 70% acetonitrile and 10 μM imidazole, and was introduced by direct infusion (10 μl/min) into the electrospray ionization mass spectrometer (Agilent 11090 MSD-Trap). Experiments were preformed in negative ionization mode (2000 V at 200°C, dry gas at 15 psi, nebulizing gas at 5 L/min).
Viral entry assays
Viral entry was quantified for the level of β-galactosidase in the target cells infected by a recombinant HSV-1 virus (KOS gL86), in which β-galactosidase expression is inducible by early viral protein synthesis upon HSV infection. Cells (HeLa and cornea fibroblast (CF) cells) were plated at 2 × 10 4 per well in 96-well plates at least 16 hrs prior to infection. HSV virions were preincubated with octasaccharides before infection. The activity of β-galactosidase was determined by using o-nitrophenyl-β-d-galactopyranoside (ONPG) or by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as a substrate as previously described (20). Microscopy was performed with the 20x objective of an inverted microscope (Axiovert 100 M; Zeiss). All experiments were repeated a minimum of three times unless otherwise noted.
Virus-free cell-to-cell fusion assay
The CHO-K1 cells designated effector cells were cotransfected with plasmids expressing four HSV-1(KOS) glycoproteins, pPEP98 (gB), pPEP99 (gD), pPEP100 (gH), and pPEP101 (gL), along with plasmid pT7EMCLuc, which expresses the firefly luciferase gene under the control of the T7 promoter (22). Wild-type CHO-K1 cells lack functional entry receptors, therefore, the cells are resistant to both HSV entry and virus-induced cell fusion (20, 22-24). The effector cells after a 30 min incubation with octasaccharides were co-cultured with CHO-K1 target cells cotransfected with the plasmid expressing 3-OST-3 to synthesize an entry receptor and pCAGT7, which expresses T7 RNA polymerase with the chicken actin promoter and the CMV enhancer (20, 23). Effector cells expressing pT7EMCLuc and pcDNA3 (devoid of any viral glycoproteins) and target CHO-K1 transfected with T7 RNA polymerase alone (no expression of 3-OST-3) were used as negative controls. Activation of the reporter luciferase gene, as a measurement of cell fusion, was examined by reporter lysis assay (Promega) at 24 hrs post mixing as previously described.
Results
Enzymatic synthesis of 3-O-sulfated octasaccharide
The starting material, 3-OH octasaccharide, was isolated from partially depolymerized heparin (29). The octasaccharide was eluted as a major component with absorbance at 232 nm when analyzed by DEAE-NPR HPLC (Fig. 2A), suggesting that the purity of the starting octasaccharide is adequate. The 3-O-sulfated octasaccharide was prepared by incubating 3-OST-3 enzyme, 3-OH octasaccharide and 35S-labeled PAPS as illustrated in Fig. 1B. The resultant octasaccharide product, carrying a 3-O-[35S]sulfo group, afforded a prominent peak at 62 min (Fig. 2B) that carried 35S-radioactivity (Fig. 2B, inset). We estimated that the purity of the 3-O-sulfated octasaccharide was between 70 and 80% based on the elution profile. We noted that about 20 to 30% of the 35S-labeled oligosaccharides were eluted at 58 min, raising the question whether this fraction of oligosaccharides binds to gD. To address this question, we affinity purified some 3-O-sulfated octasaccharides by an immunoprecipitation approach using gD and anti-gD antibody as described under “Experimental Procedures”. We then compared the elution profile of gD-affinity purified octasaccharides to that of the unpurified ones. Quite noticeably, we did not find any difference between the relative ratio in the intensity of the 35S-peak eluted at 58 min vs the one at 62 min (chromatograms are not shown), suggesting both components bind to gD. Our efforts for the structure characterization of the octasaccharide were then focused on the component that was eluted at 62 min.
Figure 2. HPLC Chromatograms and MS spectra of the 3-OH octasaccharide and 3-O-sulfated octasaccharide.
The 3-OH octasaccharide (Panel A) and the 3-O-sulfated octasaccharide (Panel B) were subjected to DEAE-NPR-HPLC. Panel B insert shows the [35S] elution profile of 3-O-[35S]sulfated octasaccharide. Panel C shows the mass spectrum 3-OH octasaccharide. Panel D shows the mass spectrum of 3-O-sulfated octasaccharide. The expected ions are indicated where “Na” and “Imid” represents sodium and imidazole adducts respectively.
Electrospray ionization mass spectrometric analyses were performed for the 3-OH and 3-O-sulfated octasaccharides. The MS spectrum of 3-OH octasaccharide is shown in Fig 2C. The sample showed two prominent molecular ion peaks. These were [M-7H+2Na]5- at 469.3 m/z (Mr = 2307.5 Da) and [M-5H+Na]4- at 580.3 m/z (Mr = 2303.2 Da). From this data the molecular mass of the 3-OH octasaccharide was determined to be 2305.4 ± 2.2 Da which is close to the anticipated molecular mass (Mr = 2306.9 Da). The 3-O-sulfated octasaccharide was purified by DEAE-NPR-HPLC, and it gave three molecular ions, which were [M-8H]8- at 297 m/z (Mr = 2384.0 Da), [M-13H+5Na]8- at 311 m/z (Mr = 2386.1 Da), and [M-16H+7Na+Imid]8- at 325.1 m/z (Mr = 2387.9 Da) (Fig 2D). From these data the molecular weight of the 3-O-sulfated octasaccharide was determined to be 2386.0 ± 1.6 Da. This determination is consistent with the molecular mass of 2386.8 Da calculated for an octasaccharide carrying one 3-O-sulfo group. These results confirm that the product is the octasaccharide carrying one extra sulfo group.
Determination of the binding affinity of the 3-O-sulfated octasaccharide to gD
The binding constant (Kd) of gD and 3-O-[35S]sulfated octasaccharide was determined by using an affinity coelectrophoresis method (31). The 3-O-[35S]sulfated octasaccharide was separated under electrophoresis conditions through agarose gel zones containing gD concentrations ranging from 0 to 60 μM. The electrophoretic migration profile of the 3-O-[35S]sulfated octasaccharide was visualized using a Phosphor-Imager and by autoradiography (Fig. 3A). The migration of the 3-O-[35S]sulfated octasaccharide was clearly retarded by gD in a concentration dependent manner (Fig. 3A), suggesting that this octasaccharide interacts with gD. The Kd for the 3-O-[35S]sulfated octasaccharide to gD was determined to be 19 μM (Fig. 3B), which is very similar to the Kd of a previously characterized gD binding 3-O-sulfated octasaccharide isolated from HS (18 μM) (25).
Figure 3. Determining the Binding Constant (Kd) between gD and 3-O-[35S]sulfated HP octasaccharide.
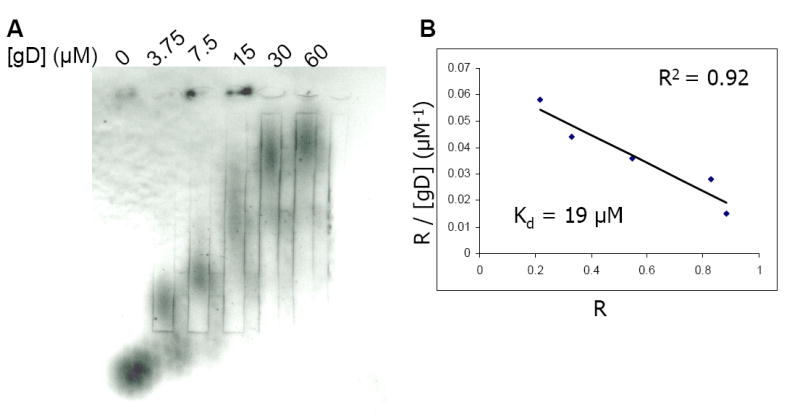
Panel A presents the autoradiography of the agarose gel in which purified 3-O-[35S]sulfated octasaccharide was subjected to electrophoresis through separation zones containing gD at concentrations indicated. Approximately 28,000 cpm (4 × 10-12 mol)/lane of the 3-O-[35S]sulfated octasaccharide was loaded into each zone. Panel B represents the plot of R/[gD]total versus R, where R = (M0-M)/M0. M0 is the migration of free 3-O-[35S]sulfated octasaccharide and M is the observed migration of 3-O-[35S]sulfated octasaccharide in the presence of gD at various concentrations. The plot yields a straight line with a slope of -1/Kd. The linear coefficient is presented as an R2 value and is shown in panel B.
We also determined the bindings of 3-O-sulfated tetrasaccharide and 3-O-sulfated hexasaccharide to gD. In these experiments, the 3-O-sulfated tetrasaccharide and 3-O-sulfated hexasaccharide were prepared by incubating a tetrasaccharide (ΔUA2S-GlcNS6S-IdoUA2S-GlcNS6S) or a hexsaccharide substrate (ΔUA2S-GlcNS-IdoUA2S-GlcNS6S-IdoUA2S-GlcNS6S) with 3-OST-3 enzyme and [35S]PAPS. The resultant 3-O-sulfated hexasaccharide and tetrasaccharide did not show any significant binding to gD (data not shown), suggesting that octasaccharide represents the minimum size of a 3-O-sulfated HS to exhibit a detectable binding affinity to gD. We did not determine the binding affinity between gD and 3-OH octasaccharide because it is known that full length heparin does not bind to gD (20).
Determination of the site of 3-O-sulfation in the octasaccharide
The 3-OH octasaccharide has four glucosamine residues, G2, G4, G6, and G8 (Fig. 1B) that might accept a 3-O-sulfo group (also see Supplementary Materials). We then determined which saccharide unit carried the 3-O-[35S]sulfo group using a combination of chemical and enzymatic degradations from both non-reducing and reducing ends.
The strategy from the reducing end analysis of 3-O-sulfated octasaccharide to identify the position of the 3-O-[35S]sulfo group is illustrated in Fig. 4. First, 2-aminobenzamide (2-AB) was coupled to the reducing end of the octasaccharide by reductive amination. Then, the 2-AB labeled 3-O-sulfated octasaccharide was subjected to partial digestion with heparin lyases to yield a 35S-labeled tetrasaccharide and a 35S-labeled disaccharide that could be resolved by PAMN-HPLC. As expected, heparin lyases treated 2-AB labeled 3-O-sulfated octasaccharide exhibited two 35S-labeled components as analyzed by PAMN-HPLC (Fig 5A). The 35S-labeled component that was eluted at 55 min was confirmed to be ΔUA2S-[3-O-35S]GlcNS3S6S as it coeluted with the standard (Supplementary Fig 2). The other 35S-labeled component that was eluted at 94 min coeluted with a tetrasaccharide standard with a structure of ΔUA2S-[3-35S]GlcNS3S6S-IdoUA2S-GlcNS6S-(2-AB)(Fig. 5C), but not with ΔUA2S-[3-35S]GlcNS3S6S-IdoUA2S-GlcNS6S (Fig. 5B), clearly indicating that the 3-O-[35S]sulfo group is located on the G6 glucosamine residue. Additional experiments were carried out to further confirm that the 3-O-[35S]sulfo group was located at G6 but not at the G2 residue (Supplementary Materials).
Figure 4. Schematic diagram for the structural characterization of the 3-O-sulfo octasaccharide.
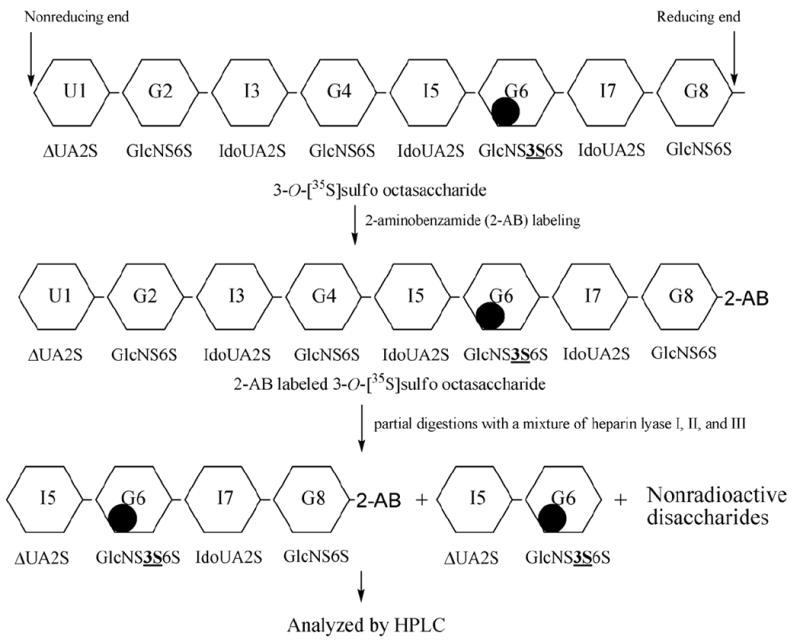
The 3-O-[3-35S]sulfo octasaccharide is subjected to reducing end labeling with 2-AB followed by partial digestion with a mixture of heparin lyases (I,II,III). The resultant [35S] labeled species can be resolved by HPLC. Black circles represents location of the 3-O-[3-35S]sulfo group.
Figure 5. Reducing end sequence analysis of 3-O-sulfated octasaccharide.
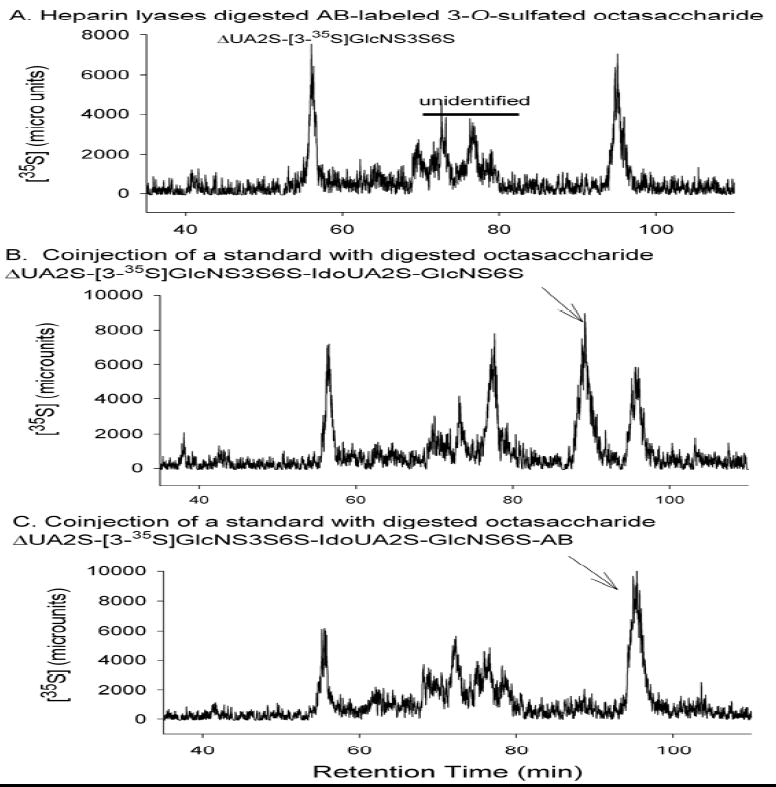
Purified 3-O-sulfated octasaccharide was digested with heparin lyases, including lyase I, II and III. The products were analyzed by PAMN-HPLC. The identities of the digested products were confirmed by coinjecting with appropriate tetrasaccharide standards. Panel A shows the digestion of the 2-AB labeled 3-O-sulfated octasaccharide alone, where both di- and tetrasaccharides are eluted. Panel Bshows the profile of the digested octasaccharide coinjected with ΔUA2S-[3-35S]GlcNS6S3S-IdoUA2S-GlcNS6S. Panel C shows the profile of the digested octasaccharide coinjected with ΔUA2S-[3-35S]GlcNS6S3S-IdoUA2S-GlcNS6S-AB. Arrow in Panel B indicates the elution position of ΔUA2S-[3-35S]GlcNS3S6S-IdoUA2S-GlcNS6S. Arrow in Panel C indicates the elution position of ΔUA2S-[3-35S]GlcNS3S6S-IdoUA2S-GlcNS6S-AB.
3-O-sulfated octasaccharide inhibited herpes simplex virus 1 infection and cell-cell fusion
Because the binding of cellular entry receptor and gD is needed for the entry of HSV-1 to the target cells (20, 21, 34), we examined whether the 3-O-sulfated octasaccharide could serve as an inhibitor for this interaction by binding to gD in a cell based assay. First, we monitored the entry of a recombinant HSV-1 expressing β-galactosidase (18) to primary cultures of human corneal fibroblasts (CF) (Fig. 6A). We have previously demonstrated that HSV-1 entry into CF is mediated by 3-O-sulfated HS (35). As shown in Fig. 6A, the 3-OH octasaccharide showed no obvious inhibition of the viral infection as demonstrated by the fact that the number of blue cells observed with 3-OH octasaccharide treatment were comparable to those untreated. However, under the concentration used, 3-O-sulfated octasaccharide completely blocked the viral entry, suggesting that the 3-O-sulfated octasaccharide has stronger ability to inhibit HSV-1 infection.
Figure 6. 3-O-sulfated octasaccharide inhibits HSV-1 entry.
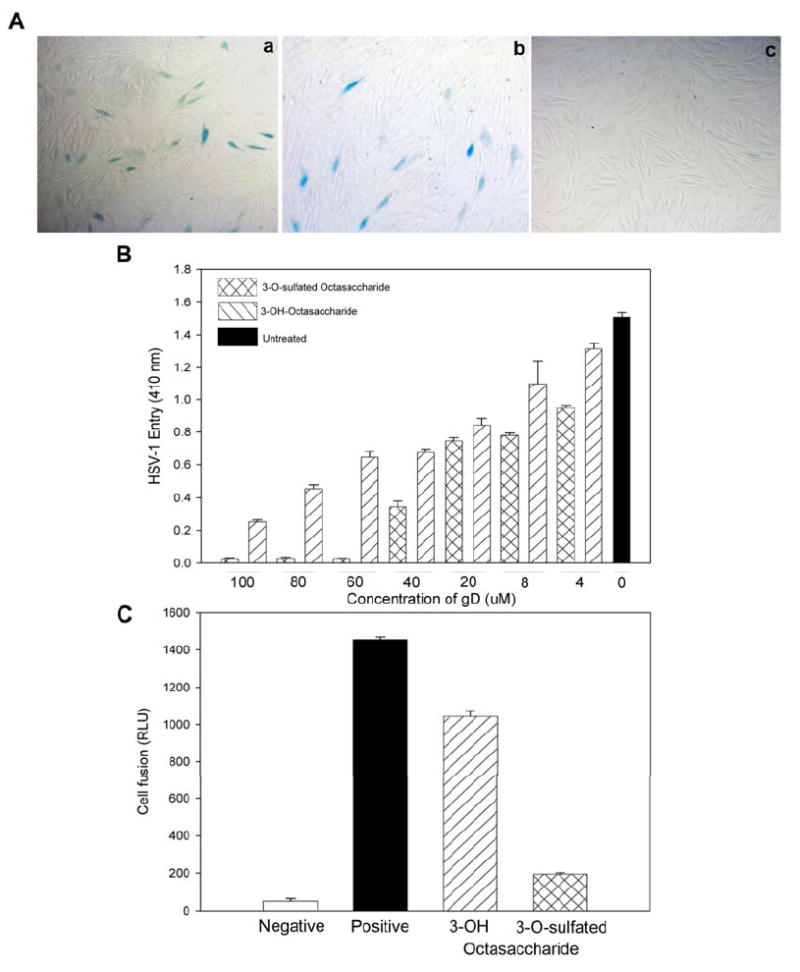
Panel A shows the nhibition of HSV-1 entry into cultured corneal fibroblasts (CFs). CFs were infected with 1PFU (plaque-forming units)/well of HSV-1(KOS-gL86) preincubated with buffer alone (a), 100 μM of 3-OH octasaccharide (b) or 100 μM of 3-O-sulfated octasaccharide (c). At 6 hrs later, the cells were washed, fixed and incubated with X-gal to identify infected cells (dark cells). Panel B shows the dose dependent inhibition of HSV-1 entry into HeLa cells. HeLa cells were infected with 1PFU (plaque-forming units)/well of HSV-1(KOS-gL86) preincubated with indicated concentration of 3-OH octasaccharide or 3-O-sulfated octasaccharide as shown. Untreated cells were used as a control. About 6 hrs post infection the cells were lysed for the quantification of β-galactosidase activity as a measure of viral entry. Absorbance at 410 nm (OD 410 nm) of ONPG reaction products were plotted against the concentration of the octasaccharides used. Panel C shows the specific inhibition of HSV-1 glycoproteins-induced membrane fusion. CHO-K1 cells were used as effector and target cells. Effector cells were transfected with plasmid expressing HSV-1 glycoproteins and luciferase reporter plasmids. Target cells were transfected with T7 RNA polymerase and the plasmid expressing 3-OST-3. Luciferase activity was measured 24 hrs after mixing and co-cultivating the effector and target cells. The luciferase activity is from one experiment performed in triplicate. The concentrations of 3-OH or 3-O-sulfated octasaccharides used in this experiment were 100 μM.
Further, we discovered that 3-O-sulfated octasaccharide also exhibited stronger efficacy in inhibiting the infection of HSV-1 to HeLa cells in a dose-response manner (Fig. 6B). In this experiment, the recombinant HSV-1 was incubated with different concentrations of octasaccharides before it was exposed to the cells. The inhibition activity of 3-O-sulfated octasaccharide becomes significantly greater at concentrations above 40 μM. At 60 μM, 3-O-sulfated octasaccharide nearly completely blocked the entry of HSV-1 to HeLa cells, while 3-OH octasaccharide only inhibited about 50% of the viral entry.
We next examined whether 3-O-sulfated octasaccharide could inhibit the fusion of cells mediated by HSV-1 glycoproteins. Fusion of cells mimics the fusion of virion and target cells that occurs during viral entry (36). In this experiment, Chinese hamster ovary (CHO-K1) cells, designated “effector” cells, were generated. The effector cells express all viral glycoproteins essential for the entry via transfection of plasmids expressing gB, gD, gH, gL, along with the plasmid pT7EMCLuc. Another group of CHO-K1 cells, designated “target” cells, were generated that expressed viral entry receptor 3-O-sulfated HS and T7 RNA polymerase, which induces the expression of luciferase in effector cells when the target cells fuse with the effector cells (36). Both effector and target cells were mixed for 18 hrs before the luciferase activity was measured to determine the extent of cell-cell fusion and the inhibitory effects of the octasaccharides. The 3-O-sulfated octasaccharide significantly decreased luciferase expression compared to the 3-OH octasaccharide (Fig. 6C), suggesting that 3-O-sulfated octasaccharide but not the 3-OH octasaccharide has the ability to block the fusion process. Since fusion is not only important for entry of the exogenous virions but also for the spread of the endogenous viruses, our data also suggest that 3-O-sulfated octasaccharide might be capable of stopping viral spread among neighboring cells.
Discussion
HS assists HSV infection at both viral attachment and viral entry steps by interacting with different viral envelope proteins. The critical role of HS has attracted considerable interest in developing HS-based compounds as anti-herpes agents. Soluble HS and heparin inhibit HSV infections effectively (16, 37). Furthermore, synthetic polymers carrying sulfate or sulfonate groups also display a strong activity for blocking HSV infections (26, 27). However, because the HS polysaccharide and synthetic polymers carry a large number of sulfate or sulfonate groups, these molecules interact with a variety of proteins, raising concerns about potential toxicity issues. Although the toxic effects can generally be reduced by narrowing the structural complexity of the polymers, it is still extremely difficult to prepare homogeneous sulfated or sulfonated polymers with existing techniques. Thus, it has been quite challenging to develop any structurally homogeneous preparations of sulfated polymers for use as antiviral drugs. Further, the structural selectivity of HS in HSV attachment has not been clearly established, questioning the viability of polymers with specific sulfation patterns in inhibiting HSV attachment. Because a specific 3-O-sulfated HS is involved in serving as an entry receptor for HSV-1 a structurally defined mimetic such as 3-O-sulfated oligosaccharide should provide the selectivity in inhibiting the infection. However, the feasibility of this approach has not been tested previously because such an oligosaccharide has not been available in sufficient amount to carry out the test.
In this article, we synthesized a structurally defined HS octasaccharide using an enzymatic approach. The structure of the 3-O-sulfated octasaccharide is ΔUA2S-GlcNS6S-IdoUA2S-GlcNS6S-IdoUA2S-GlcNS3S6S-IdoUA2S-GlcNS6S as determined by the mass spectrometry and sequencing analysis. This octassccharide is structurally distinct from a previous HS-derived octasaccharide, ΔUA-GlcNS-IdoUA2S-GlcNAc-UA2S-GlcNS-IdoUA2S-GlcNH23S6S (where Ac is acetyl and UA is either glucuronic or iduronic acid), which was synthesized through a completely different route (25). The previous structure was prepared in extremely small quantities from 3-OST-3 enzyme modified HS hexasaccharide library. This method did not afford sufficient quantities for antiviral testing. The 3-O-sulfated octasaccharide used in the current study was prepared from one step modification of a heparin octasaccharide substrate, permitting us to prepare submilligram quantities and thus, allowing us to test its antiviral efficacy. This 3-O-sulfated octasaccharide binds to HSV-1 gD with an affinity of 19 μM, which is very similar to that of the previously reported HS-derived octasaccharide, suggesting that gD can bind to octasaccharides with somewhat different structures.
HSV-1 utilizes three types of cell surface molecules as entry receptors for infection, HVEM (herpesvirus entry mediator), nectin-1, and 3-O-sulfated HS (15). All three receptors bind to gD at submicromolar affinities. HeLa cells express HVEM and nectin-1 and probably 3-OST-3 (18, 38), while CF represents a unique cell type in which HSV-1 entry is primarily mediated by 3-O-sulfated HS (35). We observed that the 3-O-sulfated octasaccharide inhibited the entry of HSV-1 into both HeLa and CF, suggesting that saturating gD with the octasaccharide can serve as a generic viral entry inhibitor independent of which entry receptor is present on the cell surface. Our finding is also important given the origin of CF. These primary cells were cultured from the stroma of the human cornea (35). Infection of the stroma is the leading cause of infectious blindness in the developed countries (14).
Our data support the possibility that the 3-O-sulfated octasaccharide inhibits the infection of HSV-1 at both the attachment and entry steps. We note that the unmodified octasaccharide shows modest inhibition of HSV-1 infection, while the 3-O-sulfated octasaccharide shows a significantly stronger inhibitory effect. Perhaps, the difference between the mechanisms of action of the unmodified and 3-O-sulfated octasaccharide appears to lie in the ability of 3-O-sulfated octasaccharide to inhibit the membrane fusion. It is noteworthy that while 3-OH octasaccharide could only block about 50% of entry to HeLa cells at 60 μM, an identical concentration of the 3-O-sulfated octasaccharide nearly completely inhibited the entry (Fig 6B). This effect raises the possibility that the 3-O-sulfated octasaccharide has acquired an additional ability to specifically block membrane fusion event by saturating gD. Since both are expected to block the attachment process equally, it is likely that the increased effectiveness of the 3-O-sulfated octasaccharide is derived from its ability to effectively block membrane fusion. In any case, ours is the first report describing an octasaccharide that can specifically block a membrane fusion event. Previous reports using full-length heparin or HS have only found inhibition of viral attachment to cell surfaces (39).
In summary, we describe a novel approach to inhibit HSV-1 infection by targeting the entry mechanism. This inhibition is achieved by using a unique 3-O-sulfated octasaccharide, which was prepared from heparin using an enzymatic approach. Our results demonstrate, for the first time, the infection of HSV-1 can be blocked by saturating the viral envelop glycoprotein gD using a small molecule of defined structure. Although the potency of this inhibitory effect is not very high, the compound exhibits structural selectivity, suggesting that we could design oligosaccharides with specific sulfation patterns to inhibit the infection. The binding affinity of these oligosaccharides to gD will be improved by undertaking extensive structure and activity relationship studies. For example, the binding affinity of oligosaccharides for gD might be increased by increasing the oligosaccharide size to 10 or 12 saccharide units. Continued efforts to synthesize oligosaccharides having different sulfation patterns could also lead to additional novel molecules that inhibit HSV infections with high potency.
Supplementary Material
Supplementary materials are available free of charge via the internet at http://pubs.acs.org.
Acknowledgments
Authors thank Drs. Gary Cohen and Roselyn Eisenberg (University of Pennsylvania) for providing recombinant baculovirus expressing gD306t and DL6 anti-gD antibody.
This work is supported in part by National Institutes of Health Grants AI50050 (to J.L.) AI057860 (to D.S.) and HL62244 and GM38060 (to RJL). RC is a recipient of the predoctoral fellowship from the David and Lucille Packard Foundation.
Abbreviations
- HS
Heparan sulfate
- HSV-1
Herpes simplex virus type 1
- ΔUA
4-deoxy-α-l-threo-hex-4-enopyranosyluronic acid
- GlcN
d-glucosamine
- GlcNAc
N-acetylated glucosamine
- GlcNS
N-sulfoglucosamine
- IdoUA
l-iduronic acid
- GlcUA
d-glucuronic acid
- 3-OST-3
heparan sulfate 3-O-sulfotransferase isoform 3
- gD
glycoprotein D
- PAMN-HPLC
silica-based polyamine-HPLC
- 2-AB
2-aminobenzamide
- ESI-MS
electrospray ionization mass spectrometry
- CF
cornea fibroblast cells
References
- 1.Esko JD, Selleck SB. Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Ann Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Pedersen LC. Anticoagulant heparan sulfate: Structural specificity and biosynthesis. Appl Microbiol Biotechnol. 2007;74:263–272. doi: 10.1007/s00253-006-0722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kreuger J, Spillmann D, Li J-p, Lindahl U. Interactions between heparan sulfate and proteins: the concept of specificity. J Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gama C, Tully SE, Sotogaku N, Clark PM, Rawat M, Vaidehi N, Goddard WA, Nishi A, Hsieh-Wilson LC. Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat Chem Biol. 2006;2:467–473. doi: 10.1038/nchembio810. [DOI] [PubMed] [Google Scholar]
- 5.Fuster MM, Esko JD. The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 6.Shriver Z, Raguram S, Sasisekharan R. Glycomics: A pathway to a class of new and improved therapeutics. Nat Rev Drug Discov. 2004:863–873. doi: 10.1038/nrd1521. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Thorp SC. Heparan sulfate and the roles in assisting viral infections. Med Res Rev. 2002;22:1–25. doi: 10.1002/med.1026. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Jones CL, Liu J. Using an enzymatic combinatorial approach to identify anticoagulant heparan sulfate structures. Chemistry & Biology. 2007;14:986–993. doi: 10.1016/j.chembiol.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muñoz E, Xu D, Avci F, Kemp M, Liu J, Linhardt RJ. Enzymatic synthesis of heparin related polysaccharides on sensor chips: Rapids screening of heparin-protein interactions. Biochem Biophys Res Commun. 2006;339:597–602. doi: 10.1016/j.bbrc.2005.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Avci FY, Muñoz EM, McDowell LM, Chen M, Pedersen LC, Zhang L, Linhardt RJ, Liu J. Enzymatically redesigning of biologically active heparan sulfate. J Biol Chem. 2005;280:42817–42825. doi: 10.1074/jbc.M504338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balagurunathan K, Lech MZ, Beeler DL, Wu ZL, Rosenberg RD. Enzymatic synthesis of antithrombin III-binding heparan sulfate pentasaccharide. Nat Biotechnol. 2003;21:1343–1346. doi: 10.1038/nbt885. [DOI] [PubMed] [Google Scholar]
- 12.Xu D, Moon A, Song D, Pedersen LC, Liu J. Engineering sulfotransferases to modify heparan sulfate. Nat Chem Biol. 2008;4:200–202. doi: 10.1038/nchembio.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corey L, Spear PG. Infections with herpes simplex viruses. N Engl J Med. 1986;314:686–691. 749–757. doi: 10.1056/NEJM198603133141105. [DOI] [PubMed] [Google Scholar]
- 14.Brandt CR. The role of viral and host genes in corneal infection with herpes simplex virus type 1. Exp Eye Res. 2005;80:607–621. doi: 10.1016/j.exer.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 15.Shukla D, Spear PG. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J Clin Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shieh M-T, WuDunn D, Montgomery RI, Esko JD, Spear PG. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J Cell Biol. 1992;116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spear PG, Eisenberg RJ, Cohen GH. Three classes of cell surface receptors for alphaherpesvirus entry. Virology. 2000;275:1–8. doi: 10.1006/viro.2000.0529. [DOI] [PubMed] [Google Scholar]
- 18.Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 19.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 20.Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 21.Xia G, Chen J, Tiwari V, Ju W, Li J-P, Malmström A, Shukla D, Liu J. Heparan sulfate 3-O-sulfotransferase isoform 5 generates both an antithrombin-binding site and an entry receptor for herpes simplex virus, type 1. J Biol Chem. 2002;277:37912–37919. doi: 10.1074/jbc.M204209200. [DOI] [PubMed] [Google Scholar]
- 22.Xu D, Tiwari V, Xia G, Clement C, Shukla D, Liu J. Characterization of Heparan Sulfate 3-O-Sulfotransferase Isoform 6 and Its Role in Assisting the Entry of Herpes Simplex Virus, Type 1. Biochem J. 2005;385:451–459. doi: 10.1042/BJ20040908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Donnell CD, Tiwari V, Oh MJ, Shukla D. A role for heparan sulfate 3-O-sulfotransferase isoform 2 in herpes simplex virus type 1 entry and spread. Virology. 2006;346:452–459. doi: 10.1016/j.virol.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Tiwari V, O’Donnell CD, Oh MJ, Valyi-Nagy T, Shukla D. A role for 3-O-sulfotransferase isoform-4 in assisting HSV-1 entry and spread. Biochem Biophys Res Commun. 2006;338:930–937. doi: 10.1016/j.bbrc.2005.10.056. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Shriver Z, Pope RM, Thorp SC, Duncan MB, Copeland RJ, Raska CS, Yoshida K, Eisenberg RJ, Cohen G, Linhardt RJ, Sasisekharan R. Characterization of a heparan sulfate octasaccharide that binds to herpes simplex viral type 1 glycoprotein D. J Biol Chem. 2002;277:33456–33467. doi: 10.1074/jbc.M202034200. [DOI] [PubMed] [Google Scholar]
- 26.Anderson RA, Feathergill K, Diao X, Cooper M, Kirkpatrick R, Spear P, Waller DP, Chany C, Doncel GF, Herold B, Zaneveld LJ. Evaluation of Poly(styrene-4-sulfonate) as a preventive agent for conceptiona dn sextually transmitted diseases. J Androl. 2000;21:862–875. [PubMed] [Google Scholar]
- 27.Raghuraman A, Tiwari V, Zhao Q, Shukla D, Debnath AK, Desai UR. Viral inhibition studies on sulfated lignin, a chemically modified biopolymer and a potential mimic of heparan sulfate. Biomacromolecules. 2007;8:1759–1763. doi: 10.1021/bm0701651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thanawiroon C, Rice KG, Toida T, Linhardt RJ. LC/MS sequencing approach for highly sulfated heparin-derived oligosaccharides. J Biol Chem. 2004;279:2608–2615. doi: 10.1074/jbc.M304772200. [DOI] [PubMed] [Google Scholar]
- 29.Pervin A, Gallo C, Jandik KA, Han X-J, Linhardt RJ. Preparation and Structural Characterization of Large Heparin-Derived Oligosaccharides. Glycobiology. 1995;5:83–95. doi: 10.1093/glycob/5.1.83. [DOI] [PubMed] [Google Scholar]
- 30.Moon A, Edavettal SC, Krahn JX, Munoz EM, Negishi M, Linhardt RJ, Liu J, Pedersen LC. Structural analysis of the sulfotransferase (3-OST-3) involved in the biosynthesis of an entry receptor of herpes simplex virus 1. J Biol Chem. 2004;279:45185–45193. doi: 10.1074/jbc.M405013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee MK, Lander AD. Analysis affinity and strutural selectivity in the binding of proteins to glycosamineglycans: Development of a sensitive electrophoretc approach. Proc Natl Acad Sci USA. 1991;88:2768–2772. doi: 10.1073/pnas.88.7.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bigge JC, Patel TP, Bruce JA, Goulding PN, Charles SM, Parekh RB. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal Biochem. 1995;230:229–238. doi: 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
- 33.Kinoshita A, Sugahara K. Microanalysis of glycosaminoglycan-derived oligosaccharides labeled with a fluorophore 2-aminobenzamide by high-performance liquid chromatography: application to disaccharide composition analysis and exosequencing of oligosaccharides. Anal Biochem. 1999;269:367–378. doi: 10.1006/abio.1999.4027. [DOI] [PubMed] [Google Scholar]
- 34.Spear PG. Herpes simplex virus: receptors and ligands for cell entry. Cellular Microbiol. 2004;6:401–410. doi: 10.1111/j.1462-5822.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- 35.Tiwari V, Clement C, Xu D, Scanlan PM, Seth V, Chung G, Kowlessur D, Valyi-Nagy T, Yue BYJT, Liu J, Shukla D. Entry of HSV-1 into primary cultures of corneal fibroblasts is mediated by HVEM and 3-O-S HS but not nectin-1. J Virol. 2006;80:8970–8980. doi: 10.1128/JVI.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pertel PE, Fridberg A, Parish ML, Spear PG. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology. 2001;279:313–324. doi: 10.1006/viro.2000.0713. [DOI] [PubMed] [Google Scholar]
- 37.WuDunn D, Spear PG. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J Virol. 1998;72:9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herold B, Gerber SI, Belval BJ, Siston AM, Shulman N. Differences in the susceptibility of herpes simplex virus types 1 and 2 to modified heparin compounds suggest serotype differences in viral entry. J Virol. 1996;70:3461–3469. doi: 10.1128/jvi.70.6.3461-3469.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials are available free of charge via the internet at http://pubs.acs.org.