Abstract
Although geneticists have extensively debated the mode by which agriculture diffused from the Near East to Europe, they have not directly examined similar agropastoral diffusions in Africa. It is unclear, for example, whether early instances of sheep, cows, pottery, and other traits of the pastoralist package were transmitted to southern Africa by demic or cultural diffusion. Here, we report a newly discovered Y-chromosome-specific polymorphism that defines haplogroup E3b1f-M293. This polymorphism reveals the monophyletic relationship of the majority of haplotypes of a previously paraphyletic clade, E3b1-M35*, that is widespread in Africa and southern Europe. To elucidate the history of the E3b1f haplogroup, we analyzed this haplogroup in 13 populations from southern and eastern Africa. The geographic distribution of the E3b1f haplogroup, in association with the microsatellite diversity estimates for populations, is consistent with an expansion through Tanzania to southern-central Africa. The data suggest this dispersal was independent of the migration of Bantu-speaking peoples along a similar route. Instead, the phylogeography and microsatellite diversity of the E3b1f lineage correlate with the arrival of the pastoralist economy in southern Africa. Our Y-chromosomal evidence supports a demic diffusion model of pastoralism from eastern to southern Africa ≈2,000 years ago.
Keywords: pastoralism, phylogeography, population genetics, Y-chromosome, Khoisan
A major focus of early research on the autosomal and mitochondrial population genetics of Europeans was to assess the genetic impact of the Neolithic agricultural expansion throughout Europe, ≈8,000 years ago (1–4). Contrasting models of demic and cultural diffusion for the spread of agriculture from the Near East to Europe predict different genetic signatures. Namely, demic diffusion predicts the replacement of old indigenous European lineages with Near Eastern ones or substantial admixture with Near Eastern lineages across the continent. The cultural diffusion model predicts low levels of gene flow from the Near East to Europe, allowing the majority of European lineages to reflect earlier Upper Paleolithic migration patterns. Although detailed genetic investigations of the European Neolithic are ongoing (5, 6), similar investigations of agropastoral expansions within Africa have been limited by patchy genetic sampling and a sparser archaeological record. The migration of agricultural Bantu-speaking cultures from western Africa appears to have had a major impact on patterns of genetic variation in Africa, but specific models have rarely been tested. In this article, we have investigated Y-chromosomal data from pastoral, agricultural, and hunter-gatherer populations in both eastern and southern Africa, to explore the possibility of demic diffusion of agropastoral practices during the Holocene.
Previously, Arredi et al. (7) suggested that a subset of E3b1-related Y chromosomes were associated with a demic diffusion of the Neolithic culture within northern and eastern Africa. Although representative subclades of E3b1-M35 are broadly distributed across Africa and Europe, reaching informative frequencies in many instances, a paraphyletic subset of chromosomes, E3b1-M35*, has a more restricted distribution. Frequencies of E3b1-M35* >25% are primarily found in samples from eastern African populations, including: the Hema in northeastern Democratic Republic of Congo; the Maasai in Kenya; and the Wairak, Datog, Sandawe, and Burunge of Tanzania (8–11). Paragroup E3b1-M35* is also found at moderate frequencies in Ethiopia and Somalia (8). Within southern Africa, the Khwe (Kxoe) and !Kung of Namibia and Angola, respectively, show high to moderate frequencies of M35*. Additionally, E3b1-M35* Y-chromosomes are scattered throughout Europe and northern Africa at very low frequencies (8, 12).
The relatively high frequency of M35* in both eastern (e.g., Sandawe, Datog, Maasai) and southern (e.g., Kxoe) African populations is intriguing given previous studies showing substantial isolation between the two regions (10, 11, 13, 14). Jointly, these studies suggest two temporally distinct migration events between the regions. The earlier event has been dated at >30,000 years ago and may have involved people with Khoisan linguistic affiliations moving into southern Africa, as evidenced by the Y-chromosome A and B clades [supporting information (SI) Fig. S1] (10, 13, 14) and mtDNA L0d clade (10). The second event involves the migration of Bantu-speaking agropastoralists from eastern Africa into southern Africa ≈1,500 years ago (15).
Here, we report a Y-chromosome-specific polymorphism that defines Y-chromosome haplogroup E3b1f-M293, accounting for many of the previous E3b1-M35*s. We generated Y-chromosome M293 SNP and microsatellite data for 13 populations of eastern and southern Africa to test the possibility of a pastoralist migration between these regions. Given its estimated age (below); the M293 locus is informative regarding migrations and demographic events occurring during the Holocene in Africa.
Results
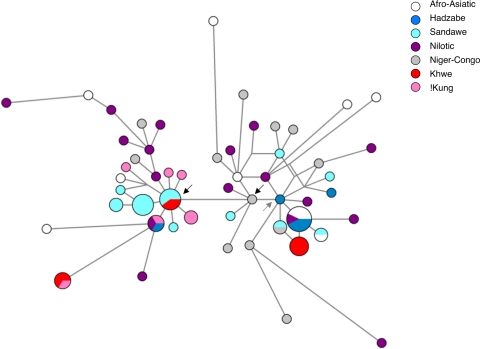
To date the time of origin of E3b1f-M293, we generated and analyzed microsatellite data for 10 associated Y-chromosome loci. Because the DYS389I locus did not appear to follow the single-step mutation model, we initially excluded this locus in our temporal estimates of haplogroup E3b1f based upon network analysis (data not shown) and the ρ estimator (16, 17). Analysis of the nine locus Y-chromosome short tandem repeat (STR) dataset yielded an estimate of 9,300–11,100 years for the age of the M293 mutation, assuming the mutation rate (μ) calculated in Zhivotovsky et al. (18). With DYS389I included, we also dated the two components of the network separately [i.e., just DYS389I (repeat size 10) representatives vs. DYS389I (repeat size 13 or higher) individuals] (Fig. 1). The inclusion of the DYS389I locus but exclusion of DYS389I-10+ haplotypes yielded a date of 12,200–10,100 years ago (ya) (ρ = 3.024, min ρ = 2.512). The M293 (DYS389I-10) associated Y-STR network yielded a date of 9,400 ya (ρ = 2.081, SE = 1,070 y, μ adjusted for nine loci given the invariant status of DYS389I). The difference in the ages of the two network hemispheres indicates that the DYS389I-10 allele is most likely derived.
Fig. 1.
Median-joining network of 10-locus Y-STR haplotypes for E3b1f-M293 positive individuals. Network was generated using Network 4.1.1.1 (Fluxus Engineering). Black arrows indicate roots used for ρ estimates; gray arrow indicates the node giving a minimum ρ estimate for M293*. Networks were processed first by the reduced-median method, and then by the median-joining method (16, 42). DYSA7.2 was removed from the analyses due to an apparently high level of homoplasy. Networks were generated initially without weighting any of the STR loci; then the three most homoplastic mutations were down-weighted (by half) for the final network. Such weighting did not significantly change the structure of the network (data not shown). ε was set to zero. ρ estimates were converted to years using the average Y-STR mutation rate estimated in ref. 18.
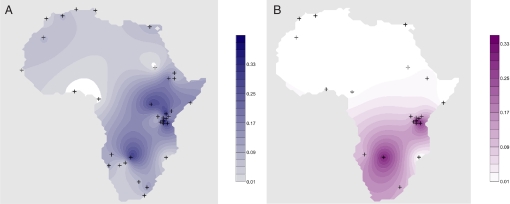
While the spatial frequency distribution of paragroup E3b1-M35*(former, uniformed by M293) is pan-African, these haplotypes are rarely identified in western and central African populations (8, 11, 12, 19) or in North Africa (7) (Fig. 2a). Rather, its regionalization is greatest in both eastern and southern Africa. The geographic distribution of the E3b1f-M293 clade, while more confined, has corresponding frequency maxima in Tanzania and southern Africa (Fig. 2b).
Fig. 2.
Contour maps of the frequencies of E3b1-M35*(former) (A) and E3b1f-M293 (B) in Africa. Populations without M293 in B are based on unpublished data. The geographic distributions of M35*(former) and M293 frequencies across Africa were created using the Kringing method in Surfer 8 (Golden Software). Locations of populations in Table 1 are indicated by cross hatches. Because M35* is a paraphyletic haplogroup, the sharing of M35* does not indicate a close genetic relationship. Areas of high frequency are similar in the two maps as 90% of the M35*(former) samples are M293+. M293 is only found in sub-Saharan Africa, indicating a separate phylogenetic history for M35*(former) samples further north.
The haplogroup E3b1f distribution spans different language phyla and subsistence economies. When the Wafiome are excluded because of low sample size (n = 2), the Tanzanian Datog population has both the highest haplogroup E3b1f-M293 frequency (43%) and Y-STR diversity (Table 1) of any group surveyed. The Datog are pastoralists who speak a Southern Nilotic language. The Burunge are Afro-Asiatic agropastoralists, who display the M293 clade at a frequency of 28%. M293 associated Y-STR diversity is also quite high in the Burunge, second only to the Datog (Table 1). Network analysis indicates that the Burunge have several haplotypes that fall on fairly long branches distinct from other populations, but they also share Y-STR haplotypes with individuals representing the Hadza, Datog, and Sandawe (Fig. 1). The frequency of M293 chromosomes in the Sandawe, a Tanzanian click-speaking (Khoisan) population, is 24%. Sandawe haplotypes are broadly scattered across the M293 network with the majority concentrated within the M293(DYS389I-10) clade (Fig. 1). The sample from the Hadza, a small click-speaking population that until recently subsisted via hunting and gathering, has considerably less M293-specific Y-STR diversity than do the other Tanzanian samples (Table 1). In two cases, Y-chromosomes of Hadza and Datog individuals share identical haplotypes. This pattern reflects the current geographic proximity of these two populations in northern Tanzania. The Sandawe, Hadza, and Datog populations all have frequencies of the residual E3b1-M35* at ≈5%.
Table 1.
Population descriptions and corresponding M35* and M293 frequencies
| Population | Language† | Location‡ | N§ | M35*¶ | M293‖ | DYS389I-10†† | Gene diversity (H)‡‡, variance | |
|---|---|---|---|---|---|---|---|---|
| Hadza | Kh | Tz | 54 | 0.04 | 0.11 | 0.02 | 0.122 | 0.698 |
| Sandawe | Kh | Tz | 70 | 0.04 | 0.24 | 0.17 | 0.240 | 1.545 |
| Datog | NS | Tz | 40 | 0.05 | 0.43 | 0.23 | 0.403 | 2.073 |
| Burunge | AA | Tz | 25 | 0.28 | 0.08 | 0.367 | 1.811 | |
| Wafiome | AA | Tz | 2 | 1.00 | 0.50 | 0.250 | 1.250 | |
| Mbugwe | NK | Tz | 14 | 0.21 | 0.333 | 1.704 | ||
| Turu | NK | Tz | 22 | 0.14 | 0.178 | 0.691 | ||
| Sukuma | NK | Tz | 30 | 0.03 | 0.03 | 0.000 | 0.000 | |
| Kenyan Bantu | NK | Ky | 11 | 0.18 | 0.213 | 1.203 | ||
| Ethiopian | NS/AA | Et | 88 | 0.05 | 0.02 | 0.050 | 0.250 | |
| Khwe (Kxoe) | Kh | SA§§ | 26 | 0.31 | 0.15 | 0.300 | 1.688 | |
| !Kung/!Xun | Kh | SA§§ | 64 | 0.11 | 0.11 | 0.282 | 1.599 | |
| SA Bantu | NK | SA | 8 | 0.13 | 0.13 | 0.000 | 0.000 | |
†Language families have been abbreviated as follows: Kh, Khoisan; NS, Nilo-Saharan; AA, Afro-Asiatic; and NK, Niger-Kordofanian.
‡Locations of population samples have been abbreviated as follows: Tz, Tanzania; Ky, Kenya; Et, Ethiopia; and SA, South Africa.
§N indicates the sample size for each population.
¶The frequency of E3b1-M35* individuals for each population, where M35* is defined as M35+, M78-, M81-, M123-, M281-, V6- and M293- (data from present study and refs. 8 and 10).
‖The frequency of E3b1f-M293 positive individuals in each population.
††The frequency of E3b1f-M293 individuals displaying the DYS389I-10 allele, for each population.
‡‡We calculated gene diversity (H) using GenAlEx6 (42).
§§Although the Khwe (Kxoe) and !Kung samples were obtained in South Africa, these individuals are recent immigrants to the area. The Kxoe were relocated from the Caprivi strip in Namibia and the !Kung are from southern Angola.
The presence of M293 at low to moderate frequencies in Bantu-speaking populations of eastern and southern Africa (Table 1) likely reflects recent admixture with local populations after the Bantu-speakers migrated out of western Africa (8). Paragroup E3b1-M35*(former) is also rare in northern African populations, with a maximum frequency of 9% for a sample from an Egyptian population (7, 11, 12, 20). No E3b1-M35*(former) individual has yet been reported in central Africa (8, 11, 19). We focus therefore on populations of eastern and southern Africa as potential sources of M293 diversification.
The M293 mutation is common in the Datog (43%), Kxoe (31%), and Burunge (28%) samples (in descending order of frequency). The high level of Y-STR diversity on the M293 background in the Datog population, coupled with highest frequency, suggests that the Datog have carried M293 longer than any other population. Datog haplotypes occur within both hemispheres of the E3b1f-M293 network (Fig. 1). The Burunge also carry a significant percentage of M293 haplotypes, although the frequency and genetic diversity is somewhat reduced compared with that of the Datog. The Kxoe (known in earlier studies as the Khwe), from southern Africa, have only one Y-STR haplotype on the M293* ancestral background which is shared by four individuals. They are thus unlikely to be the source population for M293. Two of the 11 Bantu-speaking Kenyan males in the Human Genome Diversity Panel (HGDP) panel reported as E-M35*(former) (8) are derived at M293. Without information about M293 in the Maasai, Hema, and other populations in Kenya, Sudan, and Ethiopia, we cannot pinpoint the precise geographic source of M293 with greater confidence. However, the available evidence points to present-day Tanzania as an early and important geographic locus of M293 evolution.
Population genetic studies of migration from eastern Africa to southern Africa have confirmed the impact of Bantu-speaking populations arriving in southern Africa ≈1,500 ya (21–24). Before this contact, separation of eastern and southern African populations is likely to have occurred at least 15 ka, and may have occurred as early as 40–50 ka, with little subsequent gene flow (10, 13, 14, 25). The apparent molecular age and distribution of the M293 haplogroup underscores that this locus is exceptionally informative with regards to the context of this important intermediate time period in the history of eastern and southern Africa.
E3b1-M35*(former) was found at relatively high frequencies in southern African click-speaking populations (31% and 11% in the Kxoe and !Kung, respectively) (8). All of these individuals now exhibit the M293 mutation (present study). With the exception of one haplotype found in the Kxoe sample, the southern African M293+ Y-chromosomes all carry the derived M293(DYS389I-10) allele (Fig. 1). Three Sandawe and two Kxoe share an identical M293(DYS389I-10) Y-STR haplotype. Most !Kung individuals have Y-chromosomes that appear to derive from this central haplotype (see Fig. 1). One !Kung individual shares a Y-STR haplotype with Hadza and Datog individuals. These Tanzanian and Khoe-San individuals with identical Y-STR haplotypes across 10 loci are likely to have very recent common ancestry. Using analytical methods developed in Walsh (26), we can estimate the time to the most recent common ancestor (TMRCA) of two individuals who share identical haplotypes across 10 loci. Assuming an infinite alleles model and an average Y-STR mutation rate from Zhivotovsky et al. (18), the mean TMRCA is 1,800y (95% credible region 40–6,680 y). Assuming a binary stepwise STR model, the mean TMRCA of two identical 10 locus haplotypes is 2,065 y. The median estimate under a binary stepwise model is 1,200 y (95% credible region 40–5,070 y). The mean and median estimates are somewhat different due to the exponential shape of the TMRCA curve.
The recent common ancestry between southern African Khoe-San- and northern Tanzanian M293-derived individuals seems to be independent of the Bantu-speaking populations. Out of our sample of 94 individuals from eastern African Bantu-speaking populations, only one individual carried the M293(DYS389I-10) allele. The only other Bantu-speaking individual with this allele was sampled from South Africa. Furthermore, only the M293* haplotype from one set of Kxoe individuals falls within three steps of the haplotype of an eastern African Bantu-speaking individual (Fig. 1). The distance between M293 Y-STR haplotypes of Bantu speakers and southern African Khoe-San speakers strongly suggests a migration of non-Bantu-speakers to southern Africa distinct from the Bantu migration 1,500 ya. The direct haplotype sharing between Sandawe/Kxoe and !Kung/Hadza/Datog leads us to argue for a migration between Tanzania and southern-central Africa (specifically, northern Namibia and southern Angola).
The polarity of this migration is inferred from the restricted M293 Y-STR haplotype diversity in southern Africa relative to that of eastern Africa (Table 1, Fig. 1). Southern African individuals are also almost exclusively M293(DYS389I-10), whereas the Datog and Sandawe have a range of haplotypes both with and without M293(DYS389I-10). Several scenarios are plausible, including the hypothesis that a group carrying the M293(DYS389I-10) allele may have moved from northern Tanzania, southward through Tanzania into eastern Namibia/southern Angola. Alternatively, this group may have originated outside of Tanzania and split into two populations with one population moving into northern Tanzania and the other circumventing Tanzania via the Democratic Republic of Congo and rapidly moving southward into central-southern Africa. The almost complete absence of E3b1-M35*(former) in Rwanda and Mozambique (Paula Sánchez, personal communication) lends some support to a migration through Tanzania, but study of populations of the southern Democratic Republic of Congo would prove more conclusive.
To infer a date for this migration event, we estimated ρ by taking the M293(DYS389I-10) haplotype from which most southern African haplotypes are derived as the root (17) (see Fig. 1). Southern African individuals that were clearly derived from another root haplotype (e.g., shared haplotype with an eastern African individual) were excluded from this estimate. Using the same mutation rate described in Fig. 1, we estimated a maximum age of 2,700 ya (ρ = 0.6, n = 10) with a standard error (SE) of 1,100 y. Although a rough estimate, subject to several caveats, the restricted M293 diversity among the !Kung and Kxoe suggests a relatively recent migration of individuals with M293 into southern Africa.
Discussion
The inferred date and geographic route of this migration is particularly relevant in the context of archaeological evidence for the spread of pastoralism to southern Africa, presumably along a tsetse fly-free corridor (27). Archaeologists continue to debate the mode of transmission of pastoralism to this region (28, 29). A major unresolved question is: were early instances of sheep, cows, pottery, and other pastoralist markers transmitted to southern-central Africa by demic or cultural diffusion? According to one model, pastoralism arrived as the result of population movement from eastern Africa. This pastoralist population migrated into southern-central Africa along with livestock. Subsequently, these pastoralists either mixed with local populations, or expanded in number without substantial genetic exchange with local groups. According to a second model, pastoralism was transmitted with little to no population movement as people passed stock and pastoralist practices from eastern Africa to southern-central Africa.
Our Y-chromosome data support the former demic diffusion model. Direct sharing of two haplotypes (Sandawe/Kxoe and Hadza/Datog/!Kung) combined with the similarity of other southern African M293 haplotypes to those of Tanzanian individuals is evidence of human migration between the two regions. Given the directionality and dating discussed previously, we conclude that eastern African individuals contributed M293 to southern African populations within the last few thousand years. The scale of this migration may have been small, minimally four male individuals (based on the two shared eastern/southern haplotypes; the Kxoe haplotype, which is one step from an eastern haplotype; and the shared Kxoe/!Kung type, which is several steps removed from any neighbors). It is possible that other male individuals who did not carry M293 were also involved in this migration, but the A clade and B2b-M112 haplotypes in eastern and southern Africa are geographically structured with no evidence of recent contact (10, 14). E3a-M2 may have been involved in this earlier pastoralist migration, but we cannot differentiate between early E3a-M2 chromosomes and E3a-M2 chromosomes that are known to have been introduced later during the expansion of Bantu-speaking peoples. Furthermore, without samples from the other regions of Namibia, Botswana and South Africa we cannot address the question of how pastoralism spread after it reached south-central Africa (28, 29).
A second set of questions in the pastoralism literature focus on the linguistic/ethnic affiliation of the population that introduced pastoralism to southern Africa. Two different linguistic groups have been proposed as likely vectors. Bantu-speaking agropastoralists could have introduced sheep, and later cattle, either by direct admixture with local Khoe-San populations or in a cultural contact zone preceding the actual arrival of Bantu-speakers into southern Africa (30). Archaeological remains of Iron Age ceramics (associated with early Bantu-speaking populations) occur in southern Zambia at 2,300 ya, although the ceramics are not clearly associated with caprine or cattle remains (31, 32). A second model is based primarily on linguistic evidence, with some archaeological correlates. Ehret (33) proposed that elements of the Khwe language, specifically words associated with pastoralism, had been borrowed from an East Sahelian language. Intriguingly, the Bambata-ware pottery found at early pastoralist sites in northern Namibia, northern Botswana and Zambia has stylistic similarities to spouted pottery found at Ngamuriak on the border of Kenya/Tanzania (29, 34). Ngamuriak is a pastoralist site considered part of the Elmenteitan culture. Southern Nilotic languages (a subset of East Sahelian) correlate with the Elmenteitan archaeological culture from 2,500 ya (33, 35).
As argued above, the M293 clade provides evidence of a migration independent of the initial Bantu expansion. If indeed M293 is indicative of the spread of pastoralism, then the Y-chromosome does not support the model of Bantu-speaking agropastoralists initially introducing sheep to southern Africa. Instead, haplogroup E3b1f-M293 points toward a different source population for the immigrating pastoralists. The shared Sandawe/Kxoe and Hadza/Datog/!Kung haplotype supports a connection between radically different branches of Khoisan. However, it is also possible that a third population contributed the same haplotypes to both the Sandawe, Hadza, !Kung, and Kxoe within a relatively short period. The Hadza/Datog/!Kung haplotype sharing supports this hypothesis. More than any other East African population in our dataset, the Datog dominate the M293(DYS389I-10) diversity (Fig. 1) and overall M293 diversity (Table 1). Newman (36), in his study of the Sandawe subsistence strategies, describes one Sandawe clan, the Alagwa, which is derived from people with Barabaig heritage. Barabaig is a dialect of Datog, a Southern Nilotic language, and Barabaig individuals self-report their ethnicity as Datog. This Barabaig clan became incorporated into the Sandawe because of their purported rainmaking abilities and eventually came to occupy a dominant position within the Sandawe society (36). Ethnographic evidence and shared Y-STR haplotypes support exchange between Tanzanian click-speaking groups and Southern Nilotic-speaking groups in Tanzania (10). Given the high frequency and diversity of E3b1f-M293 in the Datog, our data provide tentative support for a Southern Nilotic linguistic affiliation of the population responsible for introducing pastoralism to southern Africa.
Although genetic data can be a powerful tool for inferring past human migrations, the additional inference of cultural processes associated with migration will always be more tenuous because culture is necessarily decoupled from the genes. Exceptions include aDNA from skeletal remains that are directly associated in situ with archaeological artifacts (6). In rare cases, natural selection for genes that have coevolved with cultural practices, such as lactose persistence, can provide more direct evidence for the adoption of agropastoralism (37). These inferences rely on the synthesis of genetic and a variety of types of nongenetic information.
Although the nongenetic evidence surrounding this migration is consistent with a pastoralist component, a nonpastoralist interpretation is also possible. As demonstrated above, the distribution of the E3b1f-M293 haplogroup is consistent with a migration from eastern to southern Africa within the last 5,000 y. Agriculture is generally linked to the appearance of iron-producing Bantu-speaking peoples in southern Africa ≈1,500 years ago (15, 32). However, the southern African M293 haplotypes are unlikely to have originated among Bantu speakers (see above). Thus, if this migration did not involve agricultural or pastoral populations, then it would have involved, minimally, male hunter-gatherer individuals moving between the regions. Historically, hunter-gatherer populations in Tanzania are best represented by the click-speaking Hadza and Sandawe, although there were almost certainly other hunter-gatherer populations in the region that did not persist as independent groups up to the present day. A connection between eastern and southern African hunter-gatherer groups, although possible, is somewhat unlikely given genetic support for isolation between the populations for >30,000 years (10).
In summary, the E3b1f-M293 lineage accounts for a large fraction of the paraphyletic set of E3b1-M35*(former) Y-chromosome lineages found in eastern and southern Africa. The discovery of even a single SNP, when well characterized in the context of a phylogeny, can illuminate previously obscure connections between southern and eastern African populations. Associated STR diversity has informed questions about both the mode of transmission of pastoralism and the population affiliation of the southern African pastoralists. These data provide evidence compatible with a model of demic diffusion accompanying the spread of pastoralism to southern Africa, and suggest an eastern African Southern Nilotic-speaking population as the most likely source for pastoralism in south-central Africa. Genetic estimates indicate that gene flow between eastern and southern populations most likely occurred between 1,200 (credible region bounded by 40–5,000 ya) and 2,700 years ago (SE bounded by 1,600–3,800 ya). The combination of the two dating methods indicates that gene flow between eastern and southern populations most likely occurred ≈2,000 years ago (bounded by 40–5,000 ya). With more extensive population sampling, E3b1f-M293 lineages may also yield insights into the migration of the Khoekhoe pastoralists within southern Africa, where they eventually reached as far south as the Cape of Good Hope (28, 29).
Materials and Methods
Here, we report a Y-chromosome SNP, M293, originally ascertained by P.S. and P.J.O. in a South African Bantu individual, that defines a haplogroup, E3b1f, independent of the others (Fig. S1). The E3b1-M35* designation used here refers to Y chromosomes that display the derived allele for M35, but retain ancestral alleles for M78, M81, M123, M281, V6, and M293 binary polymorphisms. The previous definition of E3b1-M35* (uninformed by M293) will be referred to as “E3b1-M35*(former)” throughout the present article.
We considered Y-chromosome data for 454 individuals from 13 populations of eastern and southern Africa. DNA samples were collected from the following Tanzanian populations by S.A.T.: Hadza, Sandawe, Burunge, Datog, Turu, Wafiome, Mbugwe, and Sukuma in the Arusha and Dodoma provinces of Tanzania. Individuals were grouped according to self-identified ethnicity, and only samples from unrelated individuals who could trace ancestry to the same ethnic group as far back as the grandparents were included in the study. Written informed consent was obtained from all donors and Institutional Review Board approval and permits from Commission for Science and Technology (COSTECH) and National Institute for Medical Research (NIMR) in Tanzania were obtained before sample collection. The Kenyan Bantu and South African Bantu samples were obtained from the HDGP–Centre d'Etude du Polymorphisme Humain collection. The Khwe (Kxoe) and !Kung samples were collected, with appropriate consent, in Kimberley, Northern Cape Province, South Africa (38). However, these individuals are recent immigrants to the area. The Kxoe were relocated from the Caprivi strip in Namibia, and the !Kung are from southern Angola. Ethiopian samples are described in Seielstad et al. (39) and Underhill et al. (19) and were collected with informed consent.
Of these sampled populations, 87 individuals had previously been reported as E3b1-M35*(former) (Table 1, Dataset S1). All such E3b1-M35*(former) individuals were subsequently genotyped at the M293 locus, using denaturing high performance liquid chromatography (DHPLC) methodology. The specifications for marker M293 (rs9341316 in dbSNP) are a T to G transversion at nucleotide position 130 within a 338-bp fragment amplified using PCR primer sequences: forward 5′-3′ = gatattagtattgaagaaaccag and reverse 5′-3′ = gctggctaatacttccacagag. The results showed that haplogroup E3b1f-M293 accounts for the majority of the former E3b1-M35* samples in eastern and southern Africa. Specifically, a total of 76 individuals carried the derived G allele at M293. Using the Promega PowerPlex Y System, we then determined Y-STR haplotypes for each M293 derived individual (Dataset S1). Y-STR loci include: DYS19, 389I and II, 439, 390, 391, 392, and 393. In addition, two more Y-STR loci, DYS388 and A7.2, were typed as described in ref. 40. The DYS389II repeat allele number was determined by subtracting the DYS389I repeat number (41). The DYS389I locus was of particular interest.
Approximately half of the M293+ individuals carry the DYS389I-10 repeat allele, which is unusually rare in the Y-STR Haplotype Reference Database (www.yhrd.org). Only 0.004% of African individuals (12 of n = 3,087) in the database carry the DYS389I-10 repeat allele; all of them were from South Africa or Mozambique or identified as African American. The other half of our M293+ individuals carry an allele with 13 or more repeats, suggestive of a three-step mutational event. To determine whether the DYS389I-10 mutation was a unique event, we examined 407 samples from a diverse set of African populations for which Y-SNP and DYS389I information was available. All individuals carrying the DYS389I-10 allele (38 individuals, all from this study) were uniquely positive for the M293 derived allele and no intermediate 11 or 12 repeat-size alleles where observed for the 76 haplogroup E3b1f chromosomes. We concluded that generation of the DYS389I-10 allele was a rare, and possibly unique, multistep event, and that individuals positive for the M35 mutation that also carry the DYS389I-10 allele are also very likely to be derived for the M293 the mutation. In one case only, derived M293 status was inferred for a M35 derived Datog individual with the DYS389I-10 allele because DNA was exhausted: an assumption bolstered by the overall 10 locus Y-STR profile identical to a known M293 derived Datog individual.
Supplementary Material
Acknowledgments.
We thank Antonel Olkers (North-West University, Potchefstroom, South Africa) for collecting the !Kung and Khwe samples from South Africa. We thank R. Klein and K. A. Horsburgh for helpful discussion. P.J.O. was supported by BayGene (the Bavarian Genome Network). S.A.T.'s funding sources for field work/sample collection included: National Science Foundation (NSF) grants BCS-0196183 and BCS-0552486 and Wenner Gren Foundation grants and a L.S.B. Leakey Foundation grant. J.L.M.'s funding sources included: NSF grant BCS-9905574 and National Institutes of Health Grant GM28428 and a L.S.B. Leakey Foundation grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the Single Nucleotide Polymorphism (dbSNP) database, www.ncbi.nlm.nih.gov/projects/SNP (accession no. rs9341316).
This article contains supporting information online at www.pnas.org/cgi/content/full/0801184105/DCSupplemental.
References
- 1.Barbujani G, Bertorelle G. Genetics and the population history of Europe. Proc Natl Acad Sci USA. 2001;98:22–25. doi: 10.1073/pnas.98.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavalli-Sforza LL, Menozzi P, Piazza A. Demic expansions and human evolution. Science. 1993;259:639–646. doi: 10.1126/science.8430313. [DOI] [PubMed] [Google Scholar]
- 3.Chikhi L, et al. Clines of nuclear DNA markers suggest a largely neolithic ancestry of the European gene pool. Proc Natl Acad Sci USA. 1998;95:9053–9058. doi: 10.1073/pnas.95.15.9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richards M, et al. Tracing European founder lineages in the near eastern mtDNA pool. Am J Hum Genet. 2000;67:1251–1276. [PMC free article] [PubMed] [Google Scholar]
- 5.Dupanloup I, Bertorelle G, Chikhi L, Barbujani G. Estimating the impact of prehistoric admixture on the genome of Europeans. Mol Biol Evol. 2004;21:1361–1372. doi: 10.1093/molbev/msh135. [DOI] [PubMed] [Google Scholar]
- 6.Haak W, et al. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science. 2005;310:1016–1018. doi: 10.1126/science.1118725. [DOI] [PubMed] [Google Scholar]
- 7.Arredi B, et al. A predominantly neolithic origin for Y-chromosomal DNA variation in North Africa. Am J Hum Genet. 2004;75:338–345. doi: 10.1086/423147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruciani F, et al. Phylogeographic analysis of haplogroup E3b (E-M215) y chromosomes reveals multiple migratory events within and out of Africa. Am J Hum Genet. 2004;74:1014–1022. doi: 10.1086/386294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luis J, et al. The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am J Hum Genet. 2004;74:532–544. doi: 10.1086/382286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tishkoff S, et al. History of Click-Speaking Populations of Africa Inferred from mtDNA and Y Chromosome Genetic Variation. Mol Biol Evol. 2007;24:2180–2195. doi: 10.1093/molbev/msm155. [DOI] [PubMed] [Google Scholar]
- 11.Wood E, et al. Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes. Eur J Hum Genet. 2005;13:867–876. doi: 10.1038/sj.ejhg.5201408. [DOI] [PubMed] [Google Scholar]
- 12.Semino O, et al. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet. 2004;74:1023–1034. doi: 10.1086/386295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knight A, et al. African Y chromosome and mtDNA divergence provides insight into the history of click languages. Curr Biol. 2003;13:464–473. doi: 10.1016/s0960-9822(03)00130-1. [DOI] [PubMed] [Google Scholar]
- 14.Semino O, et al. Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am J Hum Genet. 2002;70:265–268. doi: 10.1086/338306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillipson DW. African Archaeology. Cambridge, UK: Cambridge Univ Press; 2005. [Google Scholar]
- 16.Bandelt H, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 17.Forster P, Harding R, Torroni A, Bandelt H. Origin and evolution of Native American mtDNA variation: A reappraisal. Am J Hum Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- 18.Zhivotovsky L, et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Underhill P, et al. Y chromosome sequence variation and the history of human populations. Nat Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- 20.Bosch E, et al. High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula. Am J Hum Genet. 2001;68:1019–1029. doi: 10.1086/319521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beleza S, et al. The genetic legacy of western Bantu migrations. Hum Genet. 2005;117:366–375. doi: 10.1007/s00439-005-1290-3. [DOI] [PubMed] [Google Scholar]
- 22.Cruciani F, et al. A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet. 2002;70:1197–1214. doi: 10.1086/340257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pereira L, et al. Prehistoric and historic traces in the mtDNA of Mozambique: Insights into the Bantu expansions and the slave trade. Ann Hum Genet. 2001;65:439–458. doi: 10.1017/S0003480001008855. [DOI] [PubMed] [Google Scholar]
- 24.Salas A, et al. The making of the African mtDNA landscape. Am J Hum Genet. 2002;71:1082–1111. doi: 10.1086/344348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scozzari R, et al. Combined use of biallelic and microsatellite Y-chromosome polymorphisms to infer affinities among African populations. Am J Hum Genet. 1999;65:829–846. doi: 10.1086/302538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walsh B. Estimating the time to the most recent common ancestor for the Y chromosome or mitochondrial DNA for a pair of individuals. Genetics. 2001;158:897–912. doi: 10.1093/genetics/158.2.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gifford-Gonzalez D. Animal disease challenges to the emergence of pastoralism in sub-Saharan Africa. Afr Archaeol Rev. 2000;17:95–139. [Google Scholar]
- 28.Sadr K. The First Herders at the Cape of Good Hope. Afr Archaeol Rev. 1998;15:101–132. [Google Scholar]
- 29.Smith A. The concepts of ‘Neolithic’ and ‘Neolithisation’ for Africa? Before Farming. 2005;1:1–6. [Google Scholar]
- 30.Sampson CG, Hart TJG, Wallsmith DL, Blagg JD. The ceramic sequence in the upper Seacow Valley: Problems and implications. S Afr Archaeol Bull. 1989;44:3–16. [Google Scholar]
- 31.Mitchell P. The Archaeology of Southern Africa. Cambridge, UK: Cambridge Univ Press; 2002. [Google Scholar]
- 32.Phillipson DW. The earliest South African pastoralists and the early Iron Age. Nsi. 1989;6:127–134. [Google Scholar]
- 33.Ehret C. An African Classical Age: Eastern and Southern Africa in World History, 1000 BC to AD 400. Charlottesville: University of Virginia Press; 1998. [Google Scholar]
- 34.Marshall F. Origins of specialized pastoral production in East Africa. Am Anthropol. 1990;92:873–894. [Google Scholar]
- 35.Ambrose SH. In: The Archaeological and Linguistic Reconstruction of African History. Ehret C, Posnansky M, editors. Berkeley: University of California Press; 1982. pp. 104–157. [Google Scholar]
- 36.Newman JL. The Ecological Basis for Subsistence Change Among the Sandawe of Tanzania. Washington, DC: National Academy of Sciences, National Research Council; 1970. [Google Scholar]
- 37.Tishkoff SA, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scozzari R, et al. Differential structuring of human populations for homologous X and Y microsatellite loci. Am J Hum Genet. 1997;61:719–733. doi: 10.1086/515500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seielstad M, et al. A view of modern human origins from Y chromosome microsatellite variation. Genome Res. 1999;9:558–567. [PMC free article] [PubMed] [Google Scholar]
- 40.Cinnioğlu C, et al. Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet. 2004;114:127–148. doi: 10.1007/s00439-003-1031-4. [DOI] [PubMed] [Google Scholar]
- 41.Cooper G, Amos W, Hoffman D, Rubinsztein D. Network analysis of human Y microsatellite haplotypes. Hum Mol Genet. 1996;5:1759–1766. doi: 10.1093/hmg/5.11.1759. [DOI] [PubMed] [Google Scholar]
- 42.Bandelt H, Forster P, Sykes B, Richards M. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.