Abstract
γ-Secretase is an unusual and ubiquitous aspartyl protease with an intramembrane catalytic site that cleaves many type-I integral membrane proteins, most notably APP and Notch. Several reports suggest that cleavage of APP to produce the Aβ peptide is regulated in part by lipids. As γ-secretase is a multipass protein complex with 19 transmembrane domains, it is likely that the local lipid composition of the membrane can regulate γ-activity. To determine the direct contribution of the lipid microenvironment to γ-secretase activity, we purified the human protease from overexpressing mammalian cells, reconstituted it in vesicles of varying lipid composition, and examined the effects of individual phospholipids, sphingolipids, cholesterol, and complex lipid mixtures on substrate cleavage. A conventional γ-activity assay was modified to include a detergent-removal step to facilitate proteoliposome formation, and this increased baseline activity over 2-fold. Proteoliposomes containing sphingolipids significantly increased γ-secretase activity over a phosphatidylcholine-only baseline, whereas the addition of phosphatidylinositol significantly decreased activity. Addition of soluble cholesterol in the presence of phospholipids and sphingolipids robustly increased the cleavage of APP- and Notch-like substrates in a dose-dependent manner. Reconstitution of γ-secretase in complex lipid mixtures revealed that a lipid raft-like composition supported the highest level of activity compared with other membrane compositions. Taken together, these results demonstrate that membrane lipid composition is a direct and potent modulator of γ-secretase and that cholesterol, in particular, plays a major regulatory role.
Alzheimer disease (AD)3 is a neurodegenerative disorder marked by progressive memory loss and cognitive failure. A major pathogenic feature of AD is the accumulation of amyloid β-protein (Aβ) in brain regions serving memory and cognition (1). Aβ is generated by sequential proteolytic cleavages of the β-amyloid precursor protein (APP) by β- and γ-secretases. γ-Secretase is a unique aspartyl protease that comprises a multiprotein complex composed of presenilin, nicastrin, Aph-1, and Pen-2 (2–4). The novel intramembranous catalytic site of γ-secretase is now known to be required for the processing of a wide range of type-I transmembrane proteins, most notably APP and the Notch receptor, and the list is rapidly expanding (for reviews, see Refs. 5 and 6). Therefore, it is of great interest to gain a better understanding of factors that can regulate γ-secretase cleavage activity and substrate specificity.
Although scientific attention has focused on identifying and characterizing proteins that affect γ-secretase activity or trafficking, there has been far less study of the lipid requirements and lipid modulation of the activity of γ-secretase. Because β-secretase, γ-secretase, and APP are all integral membrane proteins implicated in AD, it is highly likely that their functions are sensitive to the surrounding lipid environment. Several reports have described changes in lipid composition between brain samples of control and AD patients (7–13). Moreover, the activity of cellular γ-secretase has been shown to vary among different subcellular compartments with different lipid compositions (14–18). It has been proposed that even the cleavage specificity at the Aβ40 and Aβ42 residues of APP may be sensitive to bilayer thickness (14). Recent data suggest that γ-secretase activity is enriched in lipid raft subdomains, which are rich in cholesterol and sphingolipids (19, 20).
Semi-purified and purified γ-secretase preparations have been found to be inactive unless lipids, generally phosphatidylcholine (PC) and phosphatidylethanolamine (PE), are added to the reaction mixture (21, 22). The dependence of γ-secretase activity on cholesterol has been observed in several studies conducted in cultured cells or rodent models, further supporting the notion that γ-secretase activity can be regulated by its surrounding lipid microenvironment (8, 21, 22). The contribution of individual lipid species has not been thoroughly investigated and the direct effect of lipid composition on γ-secretase activity remains elusive. Although some cellular studies have suggested that particular lipids regulate γ-secretase activity, the complexity of these assays, including potential trafficking/localization issues of the enzyme or substrate due to lipid modulation, pleiotropic effects of the inhibitors used to modulate cellular lipids, and interference of other proteins, make it difficult to determine the direct effect of lipid modulation on γ-secretase. Moreover, in vitro γ-secretase assays to date almost always include detergents, most commonly CHAPSO or digitonin, each of which is compatible with γ-secretase activity but generally incompatible with proteoliposome formation at the concentrations used. It is known that the presence of detergents in activity assays of membrane proteins can interfere with liposome formation and integrity and overall enzymatic activity (23), and lipid specificity profiles can be drastically altered in the presence or absence of detergent (24). Therefore, in an effort to better understand the role of the lipid microenvironment on γ-secretase activity in a system that supports lipid structure, we have assessed the effect of detergent removal on the activity of purified mammalian γ-secretase and then used a reductionist approach to systematically examine the effects of individual lipids and lipid mixtures directly on proteolytic activity.
EXPERIMENTAL PROCEDURES
Reagents—Lipids were purchased from Sigma (PC, PE, and sphingomyelin (SM) from bovine brain) or Avanti Polar Lipids (phosphatidylserine (PS), gangliosides (GS), and cerebrosides (CS) from porcine brain; phosphatidylinositol (PI) from bovine liver, phosphatidic acid (PA) from egg, and complex lipid mixtures from porcine brain, bovine liver, bovine heart, Escherichia coli, and soybean). Methyl-β-cyclodextrin-caged water-soluble cholesterol was purchased from Sigma.
Purification of γ-Secretase and Recombinant Substrates—The multistep procedure for the high grade purification of human γ-secretase from the S-1 Chinese hamster ovary cell line (co-expressing human PS1, FLAG-Pen-2, Aph1α2-HA, and NCT-GST) and the purification of recombinant C100FLAG and N100FLAG substrates were performed as described previously (21, 25).
Reconstitution of γ-Secretase to Form Proteoliposomes—Lipids in chloroform solvent were dried under a stream of nitrogen gas and then hydrated in 50 mm HEPES buffer, pH 7.0, 150 mm NaCl. Lipid hydration was carried out at or above the Tm, with occasional vortexing, and the lipid mixtures were sonicated in a water bath for 10 min. For each reaction, lipids were diluted to 1.25 mg/ml in HEPES buffer, and CHAPSO was added at a final concentration of 4 mm, the concentration that yielded complete liposome solubilization as determined by the nadir of turbidity measurements at 540 nm. The solubilization reaction took place for 10 min at room temperature, at which time we added purified γ-secretase, at a 10-fold dilution from stock (stock = the M2 anti-FLAG-eluted fraction in the purification protocol from S-1 cells (21)). The mixture was incubated at 32 °C for 30 min and then at room temperature for 10 min. To remove detergent and facilitate proteoliposome formation, Bio-Beads SM-2 (Bio-Rad) were added at a concentration of 25× lipid weight to the protein/lipid mixture, and this mixture was incubated at room temperature for 1 h with gentle rocking. Thereafter, the Bio-Beads were sedimented, and the supernatant was collected and used as the starting material in the following assays. We refer to these assay conditions as “detergent-free” in the article, although addition of substrate (see below) will add trace amounts of detergent to the reaction mixture, i.e. 0.02% Nonidet P-40 final concentration, which has no effect on turbidity measurements.
In Vitro γ-Secretase Activity Assays—γ-Secretase/lipid mixtures post-Bio-Bead treatment were incubated with either the C100FLAG or N100FLAG substrates at a final concentration of 1 μm for 4 h at 37 °C. Samples incubated with C100FLAG were assayed for Aβ40 or Aβ42 by ELISA (Invitrogen) or by Western blot for AICD, whereas samples incubated with N100FLAG were assayed by Western blot to detect ICD production.
Western Blotting and Antibodies—For Western blot analysis of the purified γ-secretase complex from S-1 cells comprising PS1-NTF, PS1-CTF, Aph1α2-HA, FLAG-Pen-2, and NCT-GST, the samples were run on a 4–20% Tris glycine polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, and probed with Ab14 (for PS1-NTF, 1:2000; a gift of S. Gandy), 13A11 (for PS1-CTF, 5 μg/ml; a gift of Elan Pharmaceuticals), 3F10 (for Aph1α2-HA, 50 ng/ml; Roche Applied Science), anti-FLAG M2 (for FLAG-Pen-2, 1:1000; Sigma), or α-GST antibodies (for NCT-GST, 1:3000; Sigma). The biotinylated γ-secretase sample (below) was detected with an antibiotin antibody (1:2000; Sigma). Samples from the γ-secretase activity assays were run on 10–20% Tris-Tricine gels and transferred to polyvinylidene difluoride membranes to detect AICD-FLAG with M2 anti-FLAG antibody. Samples from γ-secretase activity assays with the N100FLAG substrate were run on 4–20% Tris glycine gels and transferred to polyvinylidene difluoride membranes to detect NICD-FLAG with the Notch Ab1744 antibody (1:1000, Cell Signaling Technology), which is selective for the free N terminus of the NICD. Levels of ICD-FLAG were estimated by densitometry using the Scion Image Program.
Electron Microscopy—For EM labeling studies, purified γ-secretase was biotinylated with EZ-Link Sulfo-NHS-LC-Biotin (Pierce) according to the manufacturer's protocol. Excess biotin was removed with a desalting column, and the biotinylated γ-secretase preparation was bound to M2 resin, washed extensively with 0.1% digitonin in Tris-buffered saline, and eluted with 200 μg/ml FLAG peptide. The M2-elute 1 protein preparation was used as the starting material in the immunogold EM studies. The γ-secretase sample was adsorbed to a glow-discharged formvar/carbon-coated copper grid for 1 min. Excess liquid was blotted off on a filter paper, and the grid was floated on 1% bovine serum albumin/phosphate-buffered saline for 5 min (blocking). For labeling, an anti-biotin antibody was diluted in 1% bovine serum albumin/phosphate-buffered saline followed by 10 nm protein A-gold (G. Posthuma, Utrecht, the Netherlands). After labeling, the sample was washed in phosphate-buffered saline and water and stained with uranyl acetate. Images were taken with an AMT 2k CCD camera on a “Tecnai G2 Spirit Bio Twin” transmission electron microscope (FEI).
Immunoprecipitation/Mass Spectrometry Analysis of Aβ—Aβ peptides generated in the in vitro γ-secretase assays were immunoprecipitated using monoclonal anti-Aβ antibody 4G8 (Senetek, Maryland Heights, MO) and protein A/G plus-agarose beads (Oncogene) and subjected to MALDI-TOF mass spectrometry using a Voyager-DE STR mass spectrometer (Applied Biosystems), as described (26). The molecular masses were accurately measured and searched against the amino acid sequence of human APP C99 carrying a methionine residue at the N terminus and a FLAG tag sequence at the C terminus (C100FLAG).
Statistical Analysis—All quantified data represent an average of at least three independent experiments. Error bars represent mean ± S.E. Data comparing differences between two groups were statistically analyzed using the unpaired, 2-tailed Student's t test. The Bonferroni correction was used when more than two groups were present. In experiments in which three or more groups were compared against each other, data were statistically analyzed by one-way analysis of variance with Tukey-Kramer post-testing.
RESULTS
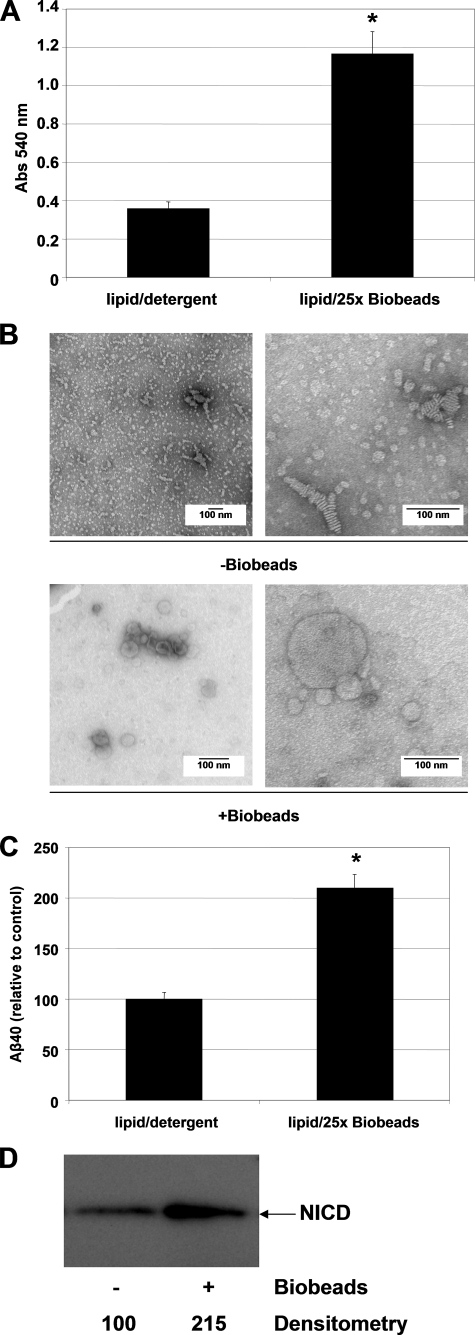
γ-Secretase is a complex of four membrane proteins containing a total of 19 transmembrane domains that cleaves type-I membrane proteins within their transmembrane domains in the lipid bilayer. To date, γ-secretase activity assays with isolated enzyme have been carried out in the presence of detergent, generally CHAPSO or digitonin, both of which are compatible with detectable cleavage activity of the enzyme (21). However, there is strong evidence that the presence of detergent can affect enzymatic activity, alter the lipid specificity profile in activity assays (24), interfere with the process of proteoliposome formation, and increase proteoliposome permeability (23, 27). Therefore, we examined the effect of detergent removal on activity assays carried out with purified γ-secretase in the presence of 0.1% PC and 0.025% PE (w/v), a lipid composition that was previously reported to allow robust γ-secretase cleavage in vitro (21). We used Bio-Beads, polystyrene beads that remove detergent through hydrophobic adsorption, as a very efficient tool for proteoliposome reconstitution (27, 28). Bio-Beads were added to reaction mixtures containing lipids and the purified enzyme prior to substrate addition. Inclusion of this detergent-removal step led to a marked increase in the turbidity of the γ-secretase reaction mixture (measured at 540 nm), which is consistent with the formation of lipid vesicles (23) (Fig. 1A). To visualize the effect of the detergent-removal step on lipid vesicle structure, we performed negative stain electron microscopy (EM) on these samples. In the samples containing 0.25% CHAPSO (i.e. before Bio-Bead treatment), we observed small, irregularly shaped lipid vesicles and, in some cases, what appeared to be irregularly shaped multilamellar liposomes (Fig. 1B, top panel). After detergent removal with Bio-Beads, the structure of the lipids changed drastically, and now numerous spherical liposomes were observed; these ranged from ∼50 to 500 nm in diameter (Fig. 1B, bottom panel). These results indicate that inclusion of a detergent-removal step in the γ-secretase assay protocol allows the formation of variously sized lipid vesicles.
FIGURE 1.
Effect of Bio-Bead-mediated detergent removal on the in vitro γ-secretase activity assay. A, effect of Bio-Bead detergent removal on the turbidity (measured at 540 nm) of the γ-secretase reaction mixture containing 0.1% PC and 0.025% PE (w/v) in 0.25% CHAPSO-HEPES. Incubation with and then removal of Bio-Beads led to a 3.1-fold increase in turbidity. Error bars represent mean ± S.E. Data were statistically analyzed using the unpaired, two-tailed Student's t test. * indicates statistical significance of p < 0.0001. B, negative stain EM of the γ-secretase reaction mixture before versus after Bio-Bead-mediated detergent removal. Liposomes of 50–500 nm were only observed upon detergent removal. C, effect of Bio-Bead detergent removal on the cleavage efficiency of the C100FLAG substrate by purified γ-secretase. Aβ40 levels were measured by ELISA (see “Experimental Procedures”). Maximum potentiation of Aβ40 production occurred at a 25× Bio-Bead to lipid ratio (w/w). Error bars represent mean ± S.E. Data were statistically analyzed using the unpaired, two-tailed Student's t test. * indicates statistical significance of p < 0.0001. D, effect of Bio-Bead detergent removal on the cleavage efficiency of the N100FLAG substrate by purified γ-secretase. NICD was probed with the 1744 antibody (Cell Signaling Technologies).
Next, we examined the effect of this detergent removal and subsequent liposome formation on γ-secretase activity. We determined that the optimal amount of Bio-Beads to add to the reaction mixture was 25× Bio-Beads/lipids (w/w) (data not shown). γ-Secretase activity assays performed with the C100FLAG substrate after detergent removal/liposome formation had 2.1-fold higher Aβ40 production than the detergent-containing controls (Fig. 1C). Likewise, cleavage of the N100FLAG substrate was also increased 2.2-fold under these new conditions (Fig. 1D). Therefore, modifying the γ-secretase activity assay protocol by including a detergent-removal step yields higher activity than that achieved under the standard published conditions, for both C100FLAG and N100FLAG substrates (23, 27).
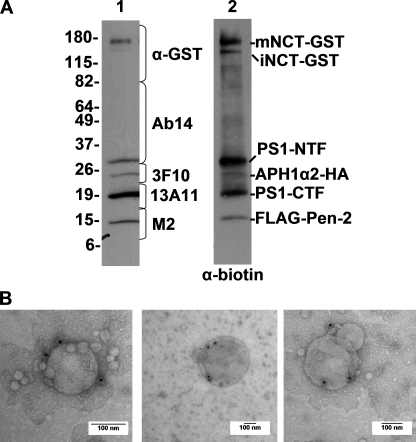
To confirm that the liposomes that formed after detergent removal were in fact proteoliposomes containing γ-secretase, we used immunoelectron microscopy to label γ-secretase-containing liposomes. Because we were working with low concentrations of purified enzyme relative to the concentration of lipid, we wanted to increase the labeling efficiency of γ-secretase within the proteoliposome. To that end, we biotinylated a small sample of a purified γ-secretase preparation, which resulted in specific biotin labeling of all of the γ-secretase components, as seen by Western blotting with an antibody to biotin (Fig. 2A, lane 2). Importantly, the α-biotin blots revealed only the known proteins of the γ-secretase complex, confirming the purity of our γ-secretase preparation. When this biotinylated γ-secretase preparation was used in our lipid reconstitution protocol (see above) and then examined by immuno-EM, the lipid vesicles were specifically labeled with the α-biotin antibody. Although a large percentage of vesicles did not contain γ-secretase due to the low concentration of enzyme used in the reconstitution, the great majority of immunodetected γ-secretase was membrane-associated, with minimal labeling outside vesicles (Fig. 2B). Taken together, our results indicate that whereas standard γ-secretase reaction conditions using 0.25% CHAPSO are compatible with γ-secretase activity, removal of the detergent substantially increases baseline activity and enables proteoliposome formation, which provides a more physiological environment for studying membrane proteins and their regulation by lipids.
FIGURE 2.
Labeling of γ-secretase within the proteoliposomes. A, Western blot of purified γ-secretase run on a 4–20% Tris glycine gel and probed with γ-secretase antibodies specific for NCT-GST, PS1-NTF, APH1α2-HA, PS1-CTF, and FLAG-Pen-2, before in vitro biotinylation (lane 1) and then probed with anti-biotin after biotinylation (lane 2). B, immunogold EM of proteoliposomes probed with anti-biotin antibody plus 10-nm conjugated Protein A-gold, and counterstained with uranyl acetate.
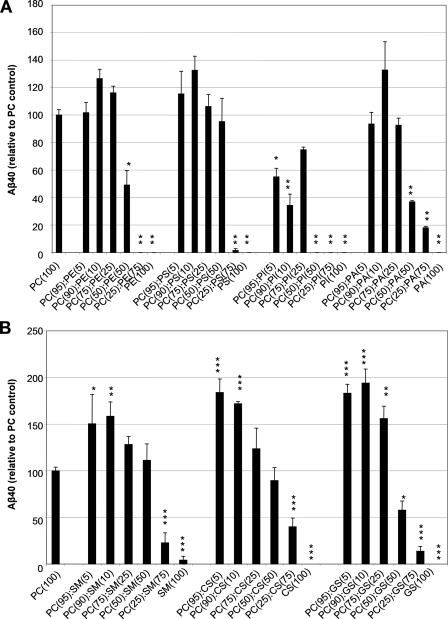
Phospholipid Dependence of Aβ Formation by Purified γ-Secretase—Our laboratory has previously demonstrated that PC and PE are necessary to reconstitute activity of purified γ-secretase (21). Despite the huge diversity of lipid species in biological membranes, the effects of individual phospholipids other than PC and PE on γ-secretase activity have not been described heretofore. To understand the roles of individual lipid species on the activity of γ-secretase in our detergent-free system that supports proteoliposome formation, we examined the effect of various concentrations of PC, PE, PS, PI, and PA on the cleavage efficiency of the C100FLAG substrate by measuring Aβ40 production by ELISA. With the exception of PC, none of these phospholipid species alone supported measurable cleavage activity, as measured by Aβ40 production (Fig. 3A). As PC is generally the most abundant phospholipid in cellular membranes and is well suited to serve as the bulk structural element of membranes, we assessed the effect of various ratios of PC to other membrane phospholipids on the efficiency of C100FLAG cleavage in our detergent-free assay. We found that vesicles containing 90% PC plus 10% of PE, PS, or PA had the greatest increase in Aβ40 production relative to the PC-only control, with a 1.3-fold increase in activity for each of the lipid mixtures (Fig. 3A). When PC was present in the reaction mixtures at 50% (w/w) or less versus PE, PS, or PA, Aβ40 production was reduced compared with the PC-only control, most likely because these lipid compositions may not form functional proteoliposomes, as the bulk structural element, PC, is markedly reduced. Interestingly, we found that vesicles containing PI at any concentration led to a marked decrease in Aβ40 production compared with the PC only control. The results indicate that significant changes in the efficiency of cleavage of APP substrate by the purified γ-secretase complex are induced by altering the ratios of certain membrane phospholipids.
FIGURE 3.
Effects of individual phospholipids and sphingolipids on the cleavage efficiency of C100FLAG substrate by purified γ-secretase reconstituted in detergent-free proteoliposomes. A, effect of increasing concentrations of PC, PE, PS, PI, or PA on the cleavage efficiency of the C100FLAG substrate, as measured by Aβ40 ELISA. Total lipid concentration in the mixtures was 1.25 mg/ml under all conditions. Error bars represent mean ± S.E. Data were statistically analyzed using the unpaired, two-tailed Student's t test with Bonferroni correction. Statistical significance of p < 0.01 (*) and p < 0.0001 (**) versus the PC(100) control is indicated. B, effect of increasing concentrations of SM, CS, or GS on the cleavage efficiency of the C100FLAG substrate, as measured by Aβ40 ELISA. Total lipid concentration was 1.25 mg/ml under all conditions. Error bars represent mean ± S.E. Data were statistically analyzed using the unpaired, two-tailed Student's t test with Bonferroni correction. Statistical significance of p < 0.05 (*), p < 0.001 (**), and p < 0.0001 (***) versus the PC(100) control is indicated.
Sphingolipid Dependence of Aβ Formation by Purified γ-Secretase—γ-Secretase activity has been reported to be enriched in lipid raft microdomains, which are rich in sphingolipids (19, 20). To determine the effect of sphingomyelin, cerebrosides, or gangliosides on the activity of purified γ-secretase in our detergent-free proteoliposome assay, we assessed varying ratios of PC to these sphingolipids as regards the efficiency of C100FLAG cleavage. SM, which is the most abundant sphingolipid in animal tissues and has structural similarities to phosphatidylcholine, enhanced γ-secretase activity when it comprised 5–25% of the total lipids in the PC-containing vesicles (Fig. 3B). The maximum increase in Aβ40 production occurred in vesicles consisting of 90% PC and 10% SM, which yielded 1.6-fold more activity than the vesicles containing PC alone. CS, glycosphingolipids that are abundant in cell membranes of the brain and other neural tissues, also enhanced Aβ40 production over the PC-only control when they comprised 5–25% (w/w) of the PC-containing vesicles (Fig. 3B). The maximum increase in Aβ40 production occurred in vesicles consisting of 95% PC and 5% CS, which enhanced γ-secretase activity 1.8-fold over the PC-only control. GS, complex glycosphingolipids found predominantly in central nervous system tissues and in lipid raft subdomains, also markedly increased Aβ40 production compared with the PC-only control when they comprised 5–25% (w/w) of the PC-containing vesicles (Fig. 3B). The maximum increase in Aβ40 production was observed in vesicles containing 90% PC and 10% GS, in which γ-activity increased a striking 1.9-fold over control conditions. Again, Aβ40 production dropped sharply when PC levels fell below 50% (w/w) of the total lipid, presumably because functional proteoliposomes could not form under these lipid conditions. Therefore, the activity of γ-secretase is increased under lipid conditions that are rich in sphingolipids, consistent with reports demonstrating increased activity in lipid raft environments, which are known to be enriched in this lipid class (19, 20).
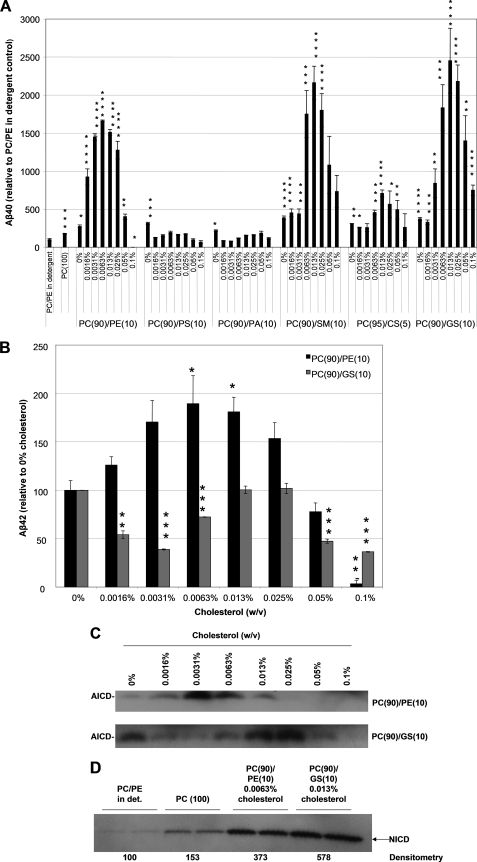
Cholesterol Dependence of ICD and Aβ Formation by Purified γ-Secretase—Cholesterol has long been implicated in AD pathogenesis by cell biological (8, 29, 30), animal modeling (31), and even epidemiological (12) data, and several mechanisms for its role have been proposed, including effects on γ-secretase activity. We therefore examined the effect of cholesterol on the activity of the purified protease reconstituted in the more physiological, detergent-free lipid microenvironment described above. Because of the poor solubility of cholesterol described in our previous publication (21), we used water-soluble, methyl-β-cyclodextrin (MβCD)-caged cholesterol. We investigated the effect of a range of cholesterol concentrations on Aβ40 production in each of the optimal PC-phospholipid/sphingolipid mixtures from Fig. 3 that gave an increase over the PC(100) control: PC(90)/ PE(10), PC(90)/PS(10), PC(90)/PA(10), PC(90)/SM (10), PC(95)/CS(5), PC(90)/GS (10). Increasing concentrations of water-soluble cholesterol from 0 to 0.0063% in the PC(90)/PE(10) reaction mixture increased production of Aβ40 by a striking ∼17-fold over the PC/PE detergent-containing control, whereas higher concentrations (≥0.013%) promoted a large decrease of the peptide, yielding a bell-shaped dose response (Fig. 4A). Interestingly, cholesterol did not stimulate Aβ40 production in either of the other phospholipid combinations, PC(90)/PS(10) or PC(90)/PA(10) at any concentration tested, but rather decreased Aβ40 relative to the 0% cholesterol treatment (Fig. 4A). Increasing concentrations of water-soluble cholesterol from 0 to 0.013% in all of the detergent-free PC/sphingolipid mixtures increased Aβ production markedly over the PC/PE detergent-containing control. At 0.013% cholesterol, the PC(95)/CS(5), PC(90)/SM(10), and PC(90)/GS(10) reaction mixtures markedly increased Aβ40 production 7-, 22-, and 25-fold over the PC/PE detergent-containing control, respectively (Fig. 4A). Higher concentrations (≥0.025%) promoted a large decrease of the Aβ40 peptide, yielding bell-shaped dose-response curves.
FIGURE 4.
Effect of cholesterol on the cleavage efficiency of purified γ-secretase in detergent-free proteoliposomes. A, the effect of various concentrations of cholesterol on Aβ40 production in proteoliposomes of optimal phospholipid or sphingolipid compositions from Fig. 3 (PC(90)/PE(10), PC(90)/PS(10), PC(90)/PA(10), PC(90)/SM(10), PC(95)/CS(5), PC(90)/GS(10)) was examined. Error bars represent mean ± S.E. Data were statistically analyzed using the unpaired, two-tailed Student's t test with Bonferroni correction. Statistical significance of p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), and p < 0.0001 (****) versus the PC/PE-detergent-containing control is indicated. B, the effect of various concentrations of cholesterol on Aβ42 production in proteoliposomes (PC(90)/PE(10) and PC(90)/GS(10)) was examined. Error bars represent mean ± S.E. Data were statistically analyzed using the unpaired, two-tailed Student's t test with Bonferroni correction. Statistical significance of p < 0.05 (*), p < 0.001 (**), and p < 0.0001 (***) versus the 0% cholesterol control is indicated. C, AICD levels of samples from the C100FLAG assay in A and B were analyzed by Western blot. AICD fragments were detected with the M2 FLAG antibody. D, the effect of various concentrations of cholesterol on NICD production in proteoliposomes of PC/PE in detergent, PC(100), PC(90)/PE(10)/0.0063% cholesterol, and PC(90)/GS(10)/0.013% cholesterol was examined. NICD levels were analyzed by Western blot and the NICD was detected with the 1744 antibody (Cell Signaling Technologies).
Next, we examined the effect of cholesterol on Aβ42 production in our optimal phospholipid (PC(90)/PE(10)/0.0063% cholesterol) and sphingolipid (PC(90)/GS(10)/0.013% cholesterol) conditions relative to the 0% cholesterol control. Increasing concentrations of water-soluble cholesterol from 0 to 0.0063% in the PC(90)/PE(10) reaction mixture increased production of Aβ42 by ∼1.9-fold over the 0% cholesterol control, whereas higher concentrations (≥0.013%) promoted a large decrease of the peptide, yielding a bell-shaped dose-response (Fig. 4B). This result indicates that the optimal cholesterol concentration for Aβ42 production was the same as that for Aβ40 production in this reaction mixture. Interestingly, in PC(90)/GS(10) proteoliposomes, cholesterol did not have the same stimulatory effect on Aβ42 production as it did on Aβ40 production; rather Aβ42 production decreased from 0 to 0.0031% cholesterol, increased from 0.0063%, to its peak level at both 0.013 and 0.025% cholesterol, and then decreased at higher concentrations (≥0.05%) (Fig. 4B). At the peak of the dose-response (0.013 and 0.025% cholesterol), Aβ42 levels never exceeded the levels of the 0% cholesterol control. These results indicate that lipid composition and cholesterol levels have a direct and strongly differential effect on the Aβ peptide species generated by γ-secretase, further implicating the importance of the lipid microenvironment in regulating γ-secretase activity.
We next examined the effect of cholesterol on AICD production in our PC(90)/PE(10) and PC(90)/GS(10) reaction mixtures. Increasing the concentration of water-soluble cholesterol from 0 to 0.0031% induced peak AICD production in the PC(90)/PE(10) reaction mixture, whereas higher concentrations (≥0.0063%) led to a decrease of the fragment, yielding a bell-shaped dose-response (Fig. 4C, top panel). In the PC(90)/GS(10) reaction mixture, AICD production decreased relative to the 0% control from 0.0016 to 0.0063% cholesterol, then increased above the control baseline levels at 0.013 and 0.025% cholesterol, and decreased at higher concentrations (≥0.05%) (Fig. 4C, bottom panel). Just as with Aβ production, maximal AICD production occurred at a higher cholesterol concentration in the PC(90)/GS(10) reaction mixture than in the PC(90)/PE(10) reaction mixture, perhaps because the more ordered acyl chains of sphingolipids favor the intercalation of the planar cholesterol molecule (32).
To see if the strong modulatory effect of cholesterol on in vitro cleavage by the purified protease was specific to the APP-based substrate, we examined the cleavage of a Notch-derived “C100-like” substrate (designated N100FLAG) in the optimized phospholipid or sphingolipid/cholesterol mixtures (PC(90)/PE(10), 0.0063% cholesterol and PC(90)/GS(10)/0.013% cholesterol), in comparison to the PC/PE detergent-containing and the PC(100)-detergent-free (i.e. Bio-Bead-treated) controls. Cleavage activity on N100FLAG rose upon reconstitution in proteoliposomes with optimal phospholipid or sphingolipid compositions, with PC(90)/PE(10)/0.0063% cholesterol and PC(90)/GS(10)/0.013% cholesterol yielding a 3.7- or 5.8-fold increase in NICD production, respectively (Fig. 4D). Our results suggest that in purified γ-secretase, where the many other cellular effects of altering cholesterol and/or lipid levels are obviated, there are direct and potent effects on the proteolytic processing of APP and Notch substrates.
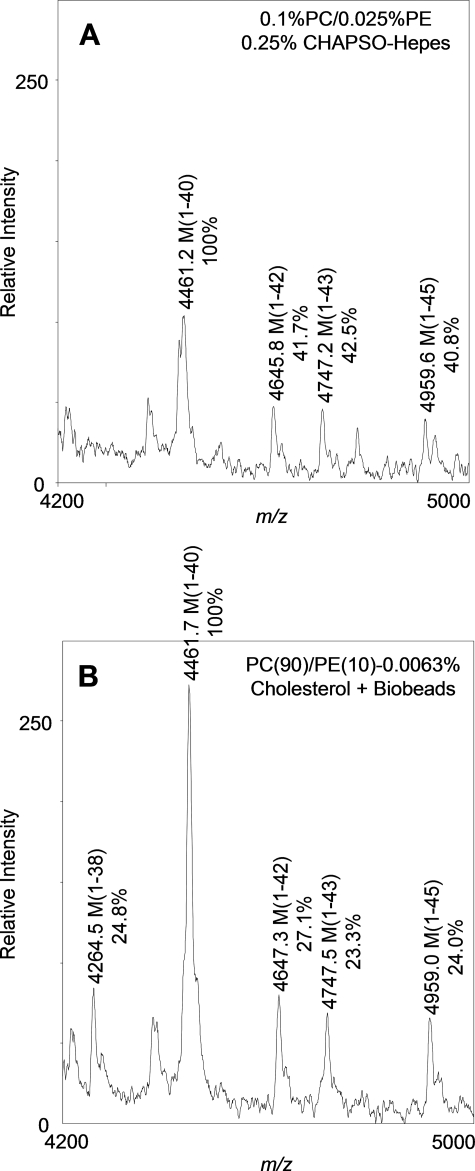
Detergent Removal/Cholesterol Addition Influences the Cleavage Specificity of Purified γ-Secretase—We next set out to determine whether γ-secretase cleavage site specificity was altered as a result of the detergent removal/cholesterol addition steps of our optimized activity assay. To examine γ-cleavage specificity, Aβ-peptides were generated in our in vitro reaction by purified γ-secretase (see “Experimental Procedures”) in the previous standard assay conditions containing 0.1% PC, 0.025% PE, and 0.25% CHAPSO or else in new conditions, i.e. post-Bio-Bead treatment plus cholesterol (PC(90)/PE(10)/0.0063% cholesterol). To detect Aβ peptides by mass spectrometry, each cleavage reaction was scaled up to a final volume of 1 ml. The Aβ cleavage products were immunoprecipitated with 4G8, a mid-region monoclonal anti-Aβ antibody that can capture all the heterogeneous Aβ species, and analyzed by MALDI-TOF mass spectrometry. The mass spectrometry spectrum revealed Aβ-(1–38), -(1–40), -(1–42), -(1–43), and -(1–45) as the principal species generated under the in vitro conditions tested, with Aβ40 being the predominant species, as expected (Fig. 5). To gain insight about how the minor Aβ species compared with the predominant Aβ40 peak between the two conditions, we measured the peak height of each corresponding Aβ species and set it as a ratio to the peak height of Aβ40. From these results, we found that removal of detergent with Bio-Beads plus addition of cholesterol subtly altered the Aβ profile in such a way that the Aβ42/Aβ40, Aβ43/Aβ40, and Aβ45/Aβ40 ratios were decreased compared with detergent-containing conditions, and the Aβ38/Aβ40 ratio was increased. These results suggest that our optimized in vitro assay protocol may promote the generation of shorter Aβ peptides-(1–38) and reduce the generation of longer Aβ peptides (1–42, 1–43, and 1–45). In addition, the intensity of the relative Aβ ion signal was increased after Bio-Bead detergent removal and cholesterol supplementation, consistent with these assay conditions having greater γ-secretase activity than detergent-containing conditions, as reported above. We conclude that alteration of the lipid microenvironment via detergent removal and cholesterol supplementation essentially gives the same variety of Aβ peptides generated under detergent-containing conditions, whereas increasing overall Aβ levels and subtly altering the cleavage specificity of γ-secretase, again indicating the importance of the lipid microenvironment in finely regulating γ-activity.
FIGURE 5.
Mass spectral analysis of Aβ peptides generated under detergent-containing conditions (0.1% PC, 0.025% PE, 0.25% CHAPSO-HEPES) (A) and post-Bio-Bead treatment plus cholesterol (PC(90)/PE(10)/0.0063% cholesterol) (B). The γ-secretase cleavage products generated after incubation with the C100FLAG substrate were captured with the 4G8 monoclonal anti-Aβ antibody and subjected to MALDI-TOF analysis using a Voyager-DE STR mass spectrometer. All Aβ-related peptides are labeled. The ratio of Aβ/Aβ40 peak heights is shown as a percent below the corresponding Aβ peak.
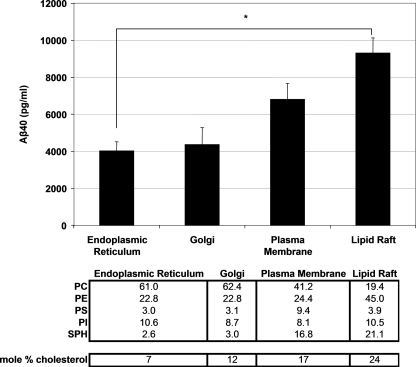
Reconstitution of γ-Secretase Activity in Lipid Environments Mimicking Those of Cellular Organelles—Some studies in intact cells have suggested that the activity of γ-secretase may be regulated by its subcellular localization, in part because different subcellular membranes have different lipid components or have similar components in different proportions (14, 16, 17). Considerable evidence suggests that lipid rafts, including those at the plasma membrane, may be a favored site for physiological γ-activity, including APP processing to Aβ (16, 17, 20, 33). To examine the effects of organelle lipid composition on the activity of γ-secretase, we reconstituted purified γ-secretase activity in complex lipid mixtures that mimicked published organelle lipid compositions and determined which composition yielded the highest γ-secretase activity (34–36). The lipid composition of the “organelle-based” vesicles is described in Fig. 6 (1.25 mg/ml final lipid concentration). Aβ40 production was readily detectable when γ-secretase was reconstituted in different lipid combinations mimicking the endoplasmic reticulum, Golgi apparatus, plasma membranes, or lipid rafts (Fig. 6). Although γ-secretase cleavage activity took place when reconstituted in lipid compositions mimicking endoplasmic reticulum and Golgi membranes, the lipid environments of the plasma membrane, and specifically the lipid raft, supported the highest levels of activity. Our results yielded the following trend in Aβ40 production: endoplasmic reticulum = Golgi < plasma membrane < lipid raft.
FIGURE 6.
Effect of organelle-like lipid compositions (see table below) on the cleavage efficiency of the C100FLAG substrate. Aβ40 production was measured by ELISA. Lipid compositions are expressed as percent of total phospholipids (w/w), except for cholesterol, which is expressed as mole percent. The final lipid concentration was 1.25 mg/ml. A representative experiment from three independent experiments is shown. Error bars represent mean ± S.E. Data were statistically analyzed using one-way analysis of variance with Tukey-Kramer post-testing. Statistical significance of p < 0.05 (*) is indicated.
Reconstitution of γ-Secretase Activity in Complex Lipid Mixtures—Next, we investigated the ability of complex lipid mixtures extracted from various biological sources to reconstitute the enzymatic activity of γ-secretase. We examined complex lipid mixtures obtained from brain, liver, heart, E. coli, or soybean to get a better understanding of the levels of individual lipids and the ratios of different lipids that best supported proteolytic activity. Compared with the other complex lipid mixtures, reconstitution of proteoliposomes with the brain lipid mixture led to the greatest Aβ40 production, followed to a much lesser extent by heart and liver (supplemental Fig. S1). E. coli and soybean lipid extracts did not support γ-secretase activity. The brain lipid composition also supported the highest cleavage activity on the Notch-like substrate (data not shown). Because these commercially available lipid mixtures were all prepared under the same conditions, i.e. a chloroform/methanol extraction followed by a partition against deionized water to remove water-soluble components (Avanti Technical Services)),4 the observed differences in activity most likely result from the distinct lipid compositions of these samples, further supporting the notion that the activity of γ-secretase is directly and potently affected by the composition of the surrounding lipid microenvironment.
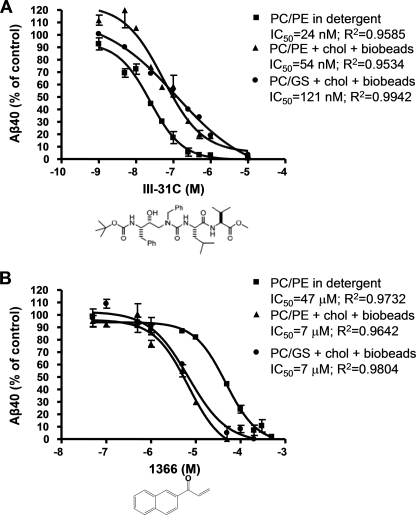
Modulation of the Potency of γ-Secretase Inhibitors by Membrane Lipids—Finally, we asked whether conditions that improve overall proteolytic activity, specifically, detergent removal and addition of cholesterol in optimized phospholipid and sphingolipid mixtures, can alter the efficacy of a transition state analogue inhibitor (III-31C (37)) or a γ-secretase modulator (1366 (38)) on the cleavage of APP by the purified enzyme. We compared the IC50 values of III-31C and 1366 under three conditions: standard detergent-containing samples (0.1% PC, 0.025% PE (w/v) in 0.25% CHAPSO), optimal phospholipid conditions (PC(90)/PE(10)/0.0063% cholesterol, Bio-Bead detergent removal), and optimal sphingolipid conditions (PC(90)/GS(10)/0.013% cholesterol, Bio-Bead detergent removal). III-31C inhibited Aβ40 generation in a concentration-dependent fashion under all assay conditions, but displayed a slightly higher potency in detergent-containing samples than in detergent-free samples supplemented with cholesterol. The IC50 values were ∼24 nm for CHAPSO-containing samples, ∼54 nm for the PC(90)/PE(10) cholesterol-containing sample, and ∼121 nm for the PC(90)/GS(10) cholesterol-containing sample (Fig. 7A). The decrease in III-31C potency under the new assay conditions may be the result of decreased accessibility of this transition state analogue, to the active site of presenilin, now buried within the lipid bilayer of a functional proteoliposome. Additionally, cholesterol, which is present in the reaction mixtures with reduced III-31C potency, has been reported to decrease bilayer permeability to small molecules. We then examined 1366, a known Janus kinase inhibitor that we previously identified as capable of selectively modulating purified γ-secretase to alter APP processing to Aβ much more than the processing of a Notch substrate (38). Here, we found that 1366 inhibited Aβ40 generation in a concentration-dependent fashion, with an IC50 of ∼47 μm under assay conditions that included 0.25% CHAPSO, a result close to our previously reported IC50 value of ∼60 μm under such conditions (Fig. 7B) (38). In contrast, altering the assay conditions via the detergent removal and cholesterol supplementation steps (above) significantly increased the potency of 1366, lowering the IC50 in both the PC(90)/PE(10)/0.0063% cholesterol and the PC(90)/GS(10)/0.013% cholesterol mixtures approximately 7-fold to ∼7 μm each. Importantly, the 1366 compound remained selective for APP versus Notch in our new assay conditions (data not shown). Because these new conditions substantially promote γ-secretase activity compared with previously published in vitro conditions and contain γ-secretase within a functional proteoliposome consisting of lipids resembling the native environment of γ-secretase, they provide a wider range for enzymatic inhibition and a more physiological vesicle for membrane-protein study, which should be useful in further inhibitor characterization studies.
FIGURE 7.
Effect of reported inhibitors on Aβ40 generation by purified γ-secretase. γ-Secretase, prepared in the standard assay conditions (0.1% PC, 0.025% PE, 0.25% CHAPSO-HEPES), in the optimal phospholipid condition (PC(90)/PE(10)/0.0063% cholesterol + Bio-Bead detergent removal), or in the optimal sphingolipid condition (PC(90)/GS(10)/0.013% cholesterol + Bio-Bead detergent removal) was incubated at 37 °C for 4 h in the presence of 1 μm C100FLAG substrate and increasing concentrations of either III-31C (A) or 1366 (B). Aβ40 was measured by ELISA.
DISCUSSION
Because γ-secretase is a four-protein complex with 19 transmembrane domains and its active site is predicted to be near the center of the bilayer, we hypothesized that its conformation and activity should be highly sensitive to membrane lipid composition. We and others have previously determined that certain lipids can affect the proteolytic activity of γ-secretase. In this study, we reconstituted the protease complex with lipids under detergent-free conditions and expanded the range of lipids and substrates examined to gain a better understanding of the lipid regulation of its activity.
Several reports have described alterations in lipid composition and lipid metabolism in the brains of AD patients compared with controls. Some studies demonstrated that levels of inositol-containing phospholipids (39), PC and PE (7, 40), and gangliosides (41) are decreased in AD brain compared with control, whereas other reports indicate that AD brain has higher PS levels than control brain (42). Increases in the activities of phospholipase A2 and phospholipase C have also been noted in AD brain (7). In addition to the measured alterations in brain phospholipids and lipases, numerous epidemiologic studies have shown that elevated cholesterol may serve as a risk factor for AD (for review, see Ref. 43). Such modifications to the composition and structure of the lipid bilayer could have profound effects on the proteins that reside there, including some of the key molecules implicated in AD pathogenesis, including APP, BACE, α-secretase, and γ-secretase.
Aβ levels have been reported to change in response to alterations in the lipid microenvironment (43, 44). BACE activity is regulated by cholesterol levels, as reported in intact cells (29, 45) and using purified enzyme (46). Interestingly, high levels of cholesterol have been found to inhibit the secretion of APPs-α, and this is thought to be due to the inhibition of α-secretase, whereas low cholesterol levels have been suggested to stimulate the α-secretase pathway (47, 48). Several studies show that reducing cholesterol levels via statin and/or cyclodextrin treatments decreases Aβ production by intact cells (29, 30, 49, 50) or in partially fractionated γ-secretase preparations (22). GM1 ganglioside was shown to promote Aβ generation, in part by increasing γ-secretase activity (51). Conversely, lipid levels themselves have been reported to be regulated by Aβ peptides, in that Aβ42 was found to down-regulate cellular sphingomyelin levels via activation of sphingomyelinase, whereas Aβ40 down-regulated cholesterol levels by inhibiting hydroxymethylglutaryl-CoA reductase activity (52).
In light of these various observations, we sought to use a reductionist approach to characterize systematically the effects of the lipid microenvironment in a controlled reconstituted system using purified γ-secretase and defined lipids. In doing so, we first wanted to ensure that our assay conditions were well suited for the study of membrane lipids. Virtually all published studies reconstitute γ-secretase activity in the presence of detergent, generally CHAPSO at 0.25–0.5% (21, 22) or digitonin at 0.01–0.1% final concentrations (21). However, as detergents are well known to permeabilize or completely solubilize liposomes and can have negative effects on overall activity of membrane-associated enzymes, we added a detergent-removal step to our assay protocol (23). We chose hydrophobic adsorption as the method of detergent removal over dialysis, dilution, or gel chromatography, due to its reproducible effects and its ease of use in small reaction volumes. Addition of a detergent-removal step led to the formation of liposomes with incorporated γ-secretase protein and resulted in more than 2-fold increases in cleavage of C100FLAG and N100FLAG substrates. We employed these improved assay conditions in all of our further analyses.
Phospholipids are the primary building blocks of cellular membranes, and many alterations in phospholipid content have been reported in AD versus control brain, as mentioned above (7, 39, 40, 42). To better understand the role of phospholipids in the regulation of γ-secretase per se, we assessed the effects of various ratios of PC to PE, PS, PI, or PA on the purified protease complex. We found that PE, PS, or PA, in the presence of PC, increased Aβ production over the PC-only control, whereas PI decreased activity over the PC-only control. These lipids may indirectly regulate γ-secretase conformation by modulating the bulk properties of the membrane (fluidity, thickness, shape, and packing properties) and/or directly as a result of electrostatic interactions of γ-secretase with negatively charged lipids, e.g. in the case of PS or PA. It was recently suggested that the increased activity of γ-secretase in budded virus particles compared with cellular membranes of Sf9 cells may be due to enhanced PS levels in the lipid composition of the budded virus envelope (53). The activity of purified BACE was also shown to be increased in proteoliposomes containing PE, PS, or PA, implying that both proteases may be regulated by the lipid microenvironment (46). Interestingly, PI, an anionic lipid, was the only phospholipid that decreased γ-activity over the PC-only control at all tested concentrations. Perhaps the bulky ring structure on the head group had a negative effect on lipid packing and membrane fluidity under these experimental conditions, accounting for the decrease in activity.
We also examined the effect of sphingolipids and found that SM, cerebrosides, or gangliosides, each in the presence of PC, increased γ-secretase activity over the PC-only control and to a greater extent than the phospholipids PE, PS, or PA. Compared with phospholipids, sphingolipids generally contain longer fatty acid chains with a higher degree of saturation, characteristics that can modulate the bulk characteristics of a lipid membrane, influencing membrane thickness and fluidity, among other qualities. Interestingly, both cerebrosides and gangliosides contain a carbohydrate moiety on their head group, which may influence bulk membrane properties or could interact with γ-secretase directly.
We found that cholesterol stimulated the activity of purified γ-secretase on C100FLAG and N100FLAG, and its effect was most pronounced in PC/sphingolipid mixtures. Our results with the purified protease are consistent with other reports that: (a) water-soluble cholesterol strongly increased Aβ production in gradient-fractionated, buoyant membrane microdomains containing PS1, whereas empty MβCD, by sequestering soluble cholesterol, did the opposite (8); (b) depletion of cholesterol from hippocampal neurons using statins ± MβCD led to a ∼2.5-fold decrease in both secreted and intracellular Aβ (30); and (c) opposing effects of water-soluble cholesterol and empty MβCD were shown on a size-separated cellular fraction containing γ-secretase (22). The variation in our lipid compositions may support vesicles of varying thickness, as sphingolipid-containing membranes are generally thicker due to longer fatty acid tails. Therefore, our results imply that proteoliposomes with thicker membranes (PC(90)/GS(10)/0.013% cholesterol) support robust Aβ40 but not Aβ42 cleavage, whereas thinner membranes (PC(90)/PE(10)/0.0063% cholesterol), do support Aβ42 production. This is consistent with the notion that thinner membranes, like those in the endoplasmic reticulum, support Aβ42 production better, whereas thicker membranes of the plasma membrane and Golgi apparatus support Aβ40 production better (14). Although other studies have reported that γ-secretase cleavage of APP is cholesterol dependent in certain membrane fractions, in semi-purified protease preparations or in intact cells (8, 29, 30), we show here for the first time a direct and differential effect of cholesterol on the activity of the purified enzyme on multiple substrates.
There is great diversity in the lipid species present in biological membranes. Although our results suggest how individual lipid species could affect γ-secretase activity, it is very difficult to predict the effect of multiple lipids on activity and on the bulk characteristics of a membrane, as the properties of individual lipids are not simply additive in a complex lipid mixture. Therefore, we examined purified γ-secretase activity after reconstituting it in complex lipid mixtures with defined compositions that mimicked certain subcellular membranes. Because the literature suggests multiple cellular locations of γ-secretase activity, we wanted to examine, in the absence of all other factors, how the lipid environment of subcellular organelles could directly affect γ-secretase activity. Although all subcellular membrane lipid mixtures supported robust cleavage activity, the lipid raft composition consistently yielded the highest γ-activity. These results support studies finding that γ-secretase activity is enriched in lipid raft microdomains (19, 33). The lipid raft mixture contained the highest level of sphingomyelin and cholesterol, two positive regulators of γ-secretase activity in our system (Figs. 3 and 4). These results suggest that not only do lipid rafts bring together the key players in Aβ production, i.e. BACE, APP, and γ-secretase, but also that the environment of the lipid raft may support the highest γ-secretase activity, thereby promoting Aβ production (19, 45).
In conclusion, we have identified new assay conditions (detergent removal) and combinations of different lipids that can strongly promote γ-secretase activity compared with previous in vitro assay protocols. As these new conditions better reflect the biological diversity of membrane lipids and enable proteoliposome formation, they provide a more physiological environment for the study of this membrane protein complex. We show that the quality and quantity of specific membrane lipid components can markedly enhance γ-activity, enabling a wider activity range in which prospective γ-secretase inhibitors and modulators can be evaluated in vitro. As our optimized assay conditions decreased the potency of an inhibitor known to target the active site of presenilin within the lipid bilayer, our results raise concerns over the current detergent-containing conditions in which γ-secretase inhibitors are initially tested, as they do not promote functional proteoliposome formation, and therefore, may not render the enzyme in its proper conformation. Moreover, our information about the mixtures of phospholipids, sphingolipids, and cholesterol that give particularly high γ-secretase activity should prove useful in current attempts to achieve two-dimensional crystals of active γ-secretase to elucidate the structure of the complex. Perhaps most importantly, emerging knowledge about the potency of lipid regulation of γ-activity can inform experiments to find new or existing small molecules that selectively modulate specific lipids as an attractive addition to protein-targeted therapeutics.
Supplementary Material
Acknowledgments
We thank Maria Ericsson of the Harvard Medical School Electron Microscopy Facility for valuable help with sample preparation and EM assistance. We also thank Georgia Dolios and Lianne Morris-Smith for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants AG00222-15 (to P. O.), AG15379 from the NIA (to D. J. S. and M. S. W.), and AG17574 and NS41355 (to M. S. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: AD, Alzheimer disease; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PI, phosphatidylinositol; PA, phosphatidic acid; SM, sphingomyelin; CS, cerebrosides; GS, gangliosides; APP, β-amyloid precursor protein; HA, hemagglutinin; ELISA, enzyme-linked immunosorbent assay; CHAPSO, 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonic acid; GST, glutathione S-transferase; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]-glycine; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; MβCD, methyl-β-cyclodextrin; AICD, APP intracellular domain; NTF, presenilin N-terminal fragment; CTF, presenilin C-terminal fragment; BACE, β-secretase; NICD, notch intracellular domain.
P. Osenkowski, personal communalization.
References
- 1.Selkoe, D. J. (2001) Physiol. Rev. 81 741-766 [DOI] [PubMed] [Google Scholar]
- 2.Kimberly, W. T., LaVoie, M. J., Ostaszewski, B. L., Ye, W., Wolfe, M. S., and Selkoe, D. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 6382-6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edbauer, D., Winkler, E., Regula, J. T., Pesold, B., Steiner, H., and Haass, C. (2003) Nat. Cell Biol. 5 486-488 [DOI] [PubMed] [Google Scholar]
- 4.Takasugi, N., Tomita, T., Hayashi, I., Tsuruoka, M., Niimura, M., Takahashi, Y., Thinakaran, G., and Iwatsubo, T. (2003) Nature 422 438-441 [DOI] [PubMed] [Google Scholar]
- 5.Kopan, R., and Ilagan, M. X. (2004) Nat. Rev. Mol. Cell. Biol. 5 499-504 [DOI] [PubMed] [Google Scholar]
- 6.Beel, A. J., and Sanders, C. R. (2008) Cell Mol. Life Sci. [DOI] [PMC free article] [PubMed]
- 7.Farooqui, A. A., Horrocks, L. A., and Farooqui, T. (2000) Chem. Phys. Lipids 106 1-29 [DOI] [PubMed] [Google Scholar]
- 8.Wahrle, S., Das, P., Nyborg, A. C., McLendon, C., Shoji, M., Kawarabayashi, T., Younkin, L. H., Younkin, S. G., and Golde, T. E. (2002) Neurobiol. Dis. 9 11-23 [DOI] [PubMed] [Google Scholar]
- 9.Jarvik, G. P., Wijsman, E. M., Kukull, W. A., Schellenberg, G. D., Yu, C., and Larson, E. B. (1995) Neurology 45 1092-1096 [DOI] [PubMed] [Google Scholar]
- 10.Jick, H., Zornberg, G. L., Jick, S. S., Seshadri, S., and Drachman, D. A. (2000) Lancet 356 1627-1631 [DOI] [PubMed] [Google Scholar]
- 11.Notkola, I. L., Sulkava, R., Pekkanen, J., Erkinjuntti, T., Ehnholm, C., Kivinen, P., Tuomilehto, J., and Nissinen, A. (1998) Neuroepidemiology 17 14-20 [DOI] [PubMed] [Google Scholar]
- 12.Wolozin, B., Kellman, W., Ruosseau, P., Celesia, G. G., and Siegel, G. (2000) Arch. Neurol. 57 1439-1443 [DOI] [PubMed] [Google Scholar]
- 13.Kalanj, S., Kracun, I., Rosner, H., and Cosovic, C. (1991) Neurol. Croat. 40 269-281 [PubMed] [Google Scholar]
- 14.Hartmann, T., Bieger, S. C., Bruhl, B., Tienari, P. J., Ida, N., Allsop, D., Roberts, G. W., Masters, C. L., Dotti, C. G., Unsicker, K., and Beyreuther, K. (1997) Nat. Med. 3 1016-1020 [DOI] [PubMed] [Google Scholar]
- 15.Xu, H., Sweeney, D., Wang, R., Thinakaran, G., Lo, A. C., Sisodia, S. S., Greengard, P., and Gandy, S. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 3748-3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaether, C., Schmitt, S., Willem, M., and Haass, C. (2006) Traffic 7 408-415 [DOI] [PubMed] [Google Scholar]
- 17.Chyung, J. H., Raper, D. M., and Selkoe, D. J. (2005) J. Biol. Chem. 280 4383-4392 [DOI] [PubMed] [Google Scholar]
- 18.Chyung, J. H., and Selkoe, D. J. (2003) J. Biol. Chem. 278 51035-51043 [DOI] [PubMed] [Google Scholar]
- 19.Vetrivel, K. S., Cheng, H., Lin, W., Sakurai, T., Li, T., Nukina, N., Wong, P. C., Xu, H., and Thinakaran, G. (2004) J. Biol. Chem. 279 44945-44954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hur, J. Y., Welander, H., Behbahani, H., Aoki, M., Franberg, J., Winblad, B., Frykman, S., and Tjernberg, L. O. (2008) FEBS J. 275 1174-1187 [DOI] [PubMed] [Google Scholar]
- 21.Fraering, P. C., Ye, W., Strub, J. M., Dolios, G., LaVoie, M. J., Ostaszewski, B. L., van Dorsselaer, A., Wang, R., Selkoe, D. J., and Wolfe, M. S. (2004) Biochemistry 43 9774-9789 [DOI] [PubMed] [Google Scholar]
- 22.Wrigley, J. D., Schurov, I., Nunn, E. J., Martin, A. C., Clarke, E. E., Ellis, S., Bonnert, T. P., Shearman, M. S., and Beher, D. (2005) J. Biol. Chem. 280 12523-12535 [DOI] [PubMed] [Google Scholar]
- 23.Rigaud, J. L., Pitard, B., and Levy, D. (1995) Biochim. Biophys. Acta 1231 223-246 [DOI] [PubMed] [Google Scholar]
- 24.Sandermann, H., Jr. (1978) Biochim. Biophys. Acta 515 209-237 [DOI] [PubMed] [Google Scholar]
- 25.Kimberly, W. T., Esler, W. P., Ye, W., Ostaszewski, B. L., Gao, J., Diehl, T., Selkoe, D. J., and Wolfe, M. S. (2003) Biochemistry 42 137-144 [DOI] [PubMed] [Google Scholar]
- 26.Wang, R., Sweeney, D., Gandy, S. E., and Sisodia, S. S. (1996) J. Biol. Chem. 271 31894-31902 [DOI] [PubMed] [Google Scholar]
- 27.Rigaud, J. L. (2002) Braz. J. Med. Biol. Res. 35 753-766 [DOI] [PubMed] [Google Scholar]
- 28.Levy, D., Bluzat, A., Seigneuret, M., and Rigaud, J. L. (1990) Biochim. Biophys. Acta 1025 179-190 [DOI] [PubMed] [Google Scholar]
- 29.Simons, M., Keller, P., De Strooper, B., Beyreuther, K., Dotti, C. G., and Simons, K. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6460-6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fassbender, K., Simons, M., Bergmann, C., Stroick, M., Lutjohann, D., Keller, P., Runz, H., Kuhl, S., Bertsch, T., von Bergmann, K., Hennerici, M., Beyreuther, K., and Hartmann, T. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 5856-5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Refolo, L. M., Malester, B., LaFrancois, J., Bryant-Thomas, T., Wang, R., Tint, G. S., Sambamurti, K., Duff, K., and Pappolla, M. A. (2000) Neurobiol. Dis. 7 321-331 [DOI] [PubMed] [Google Scholar]
- 32.Vance, D. E., and Vance, J. E. (eds) (2002) Biochemistry of Lipids, Lipoproteins and Membranes, 4 Ed., Elsevier, Paris
- 33.Lee, S. J., Liyanage, U., Bickel, P. E., Xia, W., Lansbury, P. T., Jr., and Kosik, K. S. (1998) Nat. Med. 4 730-734 [DOI] [PubMed] [Google Scholar]
- 34.van Meer, G. (1989) Annu. Rev. Cell Biol. 5 247-275 [DOI] [PubMed] [Google Scholar]
- 35.Kadowaki, H., Grant, M. A., and Seyfried, T. N. (1994) J. Lipid Res. 35 1956-1964 [PubMed] [Google Scholar]
- 36.Pike, L. J., Han, X., Chung, K. N., and Gross, R. W. (2002) Biochemistry 41 2075-2088 [DOI] [PubMed] [Google Scholar]
- 37.Esler, W. P., Kimberly, W. T., Ostaszewski, B. L., Ye, W., Diehl, T. S., Selkoe, D. J., and Wolfe, M. S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2720-2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fraering, P. C., Ye, W., LaVoie, M. J., Ostaszewski, B. L., Selkoe, D. J., and Wolfe, M. S. (2005) J. Biol. Chem. 280 41987-41996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stokes, C. E., and Hawthorne, J. N. (1987) J. Neurochem. 48 1018-1021 [DOI] [PubMed] [Google Scholar]
- 40.Nitsch, R. M., Blusztajn, J. K., Pittas, A. G., Slack, B. E., Growdon, J. H., and Wurtman, R. J. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 1671-1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svennerholm, L., and Gottfries, C. G. (1994) J. Neurochem. 62 1039-1047 [DOI] [PubMed] [Google Scholar]
- 42.Wells, K., Farooqui, A. A., Liss, L., and Horrocks, L. A. (1995) Neurochem. Res. 20 1329-1333 [DOI] [PubMed] [Google Scholar]
- 43.Puglielli, L., Tanzi, R. E., and Kovacs, D. M. (2003) Nat. Neurosci. 6 345-351 [DOI] [PubMed] [Google Scholar]
- 44.Hartmann, T., Kuchenbecker, J., and Grimm, M. O. (2007) J. Neurochem. 103 Suppl. 1, 159-170 [DOI] [PubMed] [Google Scholar]
- 45.Ehehalt, R., Keller, P., Haass, C., Thiele, C., and Simons, K. (2003) J. Cell Biol. 160 113-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalvodova, L., Kahya, N., Schwille, P., Ehehalt, R., Verkade, P., Drechsel, D., and Simons, K. (2005) J. Biol. Chem. 280 36815-36823 [DOI] [PubMed] [Google Scholar]
- 47.Bodovitz, S., and Klein, W. L. (1996) J. Biol. Chem. 271 4436-4440 [DOI] [PubMed] [Google Scholar]
- 48.Kojro, E., Gimpl, G., Lammich, S., Marz, W., and Fahrenholz, F. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 5815-5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Runz, H., Rietdorf, J., Tomic, I., de Bernard, M., Beyreuther, K., Pepperkok, R., and Hartmann, T. (2002) J. Neurosci. 22 1679-1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burns, M., Gaynor, K., Olm, V., Mercken, M., LaFrancois, J., Wang, L., Mathews, P. M., Noble, W., Matsuoka, Y., and Duff, K. (2003) J. Neurosci. 23 5645-5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zha, Q., Ruan, Y., Hartmann, T., Beyreuther, K., and Zhang, D. (2004) Mol. Psychiatry 9 946-952 [DOI] [PubMed] [Google Scholar]
- 52.Grimm, M. O., Grimm, H. S., Patzold, A. J., Zinser, E. G., Halonen, R., Duering, M., Tschape, J. A., De Strooper, B., Muller, U., Shen, J., and Hartmann, T. (2005) Nat. Cell Biol. 7 1118-1123 [DOI] [PubMed] [Google Scholar]
- 53.Hayashi, I., Urano, Y., Fukuda, R., Isoo, N., Kodama, T., Hamakubo, T., Tomita, T., and Iwatsubo, T. (2004) J. Biol. Chem. 279 38040-38046 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.