Abstract
The Epstein–Barr virus latent membrane protein 1 (LMP1) is essential for the transformation of B lymphocytes into lymphoblastoid cell lines. Previous data are consistent with a model that LMP1 is a constitutively activated receptor that transduces signals for transformation through its carboxyl-terminal cytoplasmic tail. One transformation effector site (TES1), located within the membrane proximal 45 residues of the cytoplasmic tail, constitutively engages tumor necrosis factor receptor-associated factors. Signals from TES1 are sufficient to drive initial proliferation of infected resting B lymphocytes, but most lymphoblastoid cells infected with a virus that does not express the 155 residues beyond TES1 fail to grow as long-term cell lines. We now find that mutating two tyrosines to an isoleucine at the carboxyl end of the cytoplasmic tail cripples the ability of EBV to cause lymphoblastoid cell outgrowth, thereby marking a second transformation effector site, TES2. A yeast two-hybrid screen identified TES2 interacting proteins, including the tumor necrosis factor receptor-associated death domain protein (TRADD). TRADD was the only protein that interacted with wild-type TES2 and not with isoleucine-mutated TES2. TRADD associated with wild-type LMP1 but not with isoleucine-mutated LMP1 in mammalian cells, and TRADD constitutively associated with LMP1 in EBV-transformed cells. In transfection assays, TRADD and TES2 synergistically mediated high-level NF-κB activation. These results indicate that LMP1 appropriates TRADD to enable efficient long-term lymphoblastoid cell outgrowth. High-level NF-κB activation also appears to be a critical component of long-term outgrowth.
Epstein–Barr virus (EBV) infection of B lymphocytes is nonpermissive for virus replication. Instead, EBV expresses two latent membrane proteins (LMPs) and six nuclear proteins. These proteins efficiently transform resting B lymphocytes into continuously proliferating lymphoblastoid cell lines (LCLs) (reviewed in ref. 1).
LMP1 is a key effector of EBV-mediated transformation. LMP1 has oncogene-like activity in rodent fibroblast cell lines (2–4). In non-EBV-infected B lymphoma cells, LMP1 activates NF-κB and induces most of the changes associated with EBV infection, including up-regulated expression of activation markers, adhesion molecules, and Bcl-2 (5–7). In epithelial cells, LMP1 causes hypertrophy and alters differentiation (8, 9). LMP1 is essential for EBV to growth-transform B lymphocytes into LCLs (10) and is expressed in most malignancies associated with EBV infection, including lymphoproliferative disease, Hodgkin disease, and nasopharyngeal carcinoma (reviewed in ref. 11).
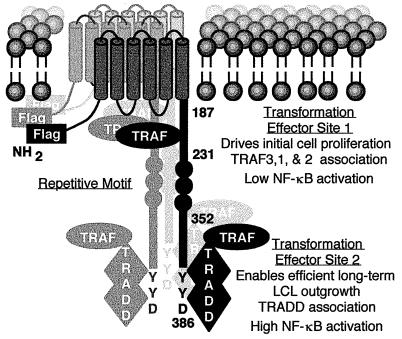
Recombinant EBV genetic and biochemical analyses are consistent with a model that LMP1 is a constitutively activated growth factor receptor that signals through its cytoplasmic carboxyl terminus (Fig. 1). The six transmembrane domains constitutively aggregate LMP1 in the plasma membrane without exogenous ligand. Aggregation is essential for B lymphocyte transformation (10, 12). The six transmembrane domains are also essential for oncogene-like activity in rodent fibroblast cell lines and phenotypic changes in B lymphoma cells (6, 13, 14). The importance of the amino and carboxyl-terminal cytoplasmic tails has been difficult to assess in rodent fibroblast transformation assays. Some assays indicate the amino terminus is important for transformation (13) whereas others implicate the carboxyl-terminal tail (4). The cytoplasmic amino terminus is not an effector of B lymphocyte transformation but is important for tethering the first transmembrane domain to the cytoplasm (12). By contrast, the cytoplasmic carboxyl terminus is essential for B lymphocyte growth transformation and therefore is implicated in signaling (15).
Figure 1.
Diagram of LMP1. The Flag epitope was introduced at the amino terminus (NH2). LMP1 residues 187, 231, 352, and 386 are noted. LMP1 constitutively aggregates in the plasma membrane. TES1 engages TRAFs to drive initial B lymphocyte proliferation and induce low-level NF-κB activation. TES2 engages TRADD to enable efficient long-term lymphoblastoid cell outgrowth and mediate high-level NF-κB activation through TRAF2.
One B lymphocyte transformation effector site (TES1) in the cytoplasmic carboxyl terminus interacts with tumor necrosis factor (TNF) receptor (TNFR) signaling pathways. The first evidence for TES1 was from EBV recombinant genetic experiments in which a mutated LMP1 MS231 was sufficient for transforming primary B lymphocytes into LCLs that required fibroblast feeder cells for long-term outgrowth. MS231 consists of the amino terminus, six transmembrane domains, and membrane proximal 45 residues of the cytoplasmic tail (15). Further studies of MS231 recombinant virus revealed that infected cells proliferate for several weeks, but thereafter most of these cells replicate progressively more slowly and then die. In contrast, nearly all wild-type EBV-transformed LCLs are immortal (K.M. Kaye, K.I., E. Johanssen, and E.K., unpublished work). TES1 was delineated as a potential signaling motif with the finding that residues within the membrane proximal 45 amino acids of the cytoplasmic tail engage TNFR-associated factors (TRAFs) (16). Deletion of the TRAF binding site results in a nontransforming EBV (17). In LCLs, LMP1 is constitutively associated with TRAF3 and TRAF1 and, to a lesser extent, with TRAF2 (18). In contrast, TRAFs are recruited to TNFR2 or to TNFR family members such as CD40 and LTβ receptor in response to ligand binding and receptor aggregation (19–21). Moreover, mimicry of an activated TNFR is concordant with the similar effects of LMP1 and activated CD40 on B lymphocyte growth and gene activation (18, 21–26).
Because EBV recombinants that express only TES1 are functionally impaired in causing LCL outgrowth, the objective of these experiments was to define the role of the membrane distal 155 residues beyond TES1 in enabling efficient, long-term LCL outgrowth. Potentially relevant to the role of these residues in transformation are previous findings that TES1 mediates only 25% of LMP1-mediated NF-κB activation whereas 75% maps to the terminal 55 or 35 residues (27, 28). NF-κB activation correlates with lymphocyte activation, and high-level NF-kB activation could be important in EBV-mediated transformation (29, 30). TRAF1 and TRAF2 dimers are implicated in NF-κB activation from TES1, and NF-κB activation by TES1 was more than 75% blocked by a dominant negative TRAF2 (18, 26). Yet, despite similarities to a TRAF binding site within the terminal 55 residues of LMP1, TRAF 1, 2, or 3 did not bind to the carboxyl-terminal 155 residues, and a dominant negative TRAF2 only partially blocked NF-κB activation by the last 55 residues (17, 18, 31). Thus, neither the significance nor the mechanism of high-level NF-κB activation was evident at the start of these studies.
MATERIALS AND METHODS
Viruses, Cells, and DNA Clones.
Viruses and cell lines were grown as described (12, 17). Vectors pRK-mycTRADD and pRK5 were gifts from David Goeddel (Tularik, South San Francisco, CA) (32). F-LMP1-ID and F-LMP1-FFD DNAs and expression vectors are derived by endonuclease deletion or codon insertions into F-LMP1 as described (17).
Recombinant EBV Construction and Characterization.
EBV recombinants were generated by second-site homologous recombination in P3HR-1 cells, tested in B lymphocyte transformation assays, and characterized as described (12, 17, 33–35).
Yeast Two-Hybrid Assay.
The methods of yeast culture, transformation, and β-galactosidase detection were as described (36). GAL-DBD-LMP1 fusions are derived from F-LMP1 clones (above) or as before (17).
Coimmune Precipitation Analyses.
Precipitations with M2 affinity gel (Kodak) were as described except that TRADD antisera (Santa Cruz Biotechnology) or 9E10 antibody to myc were used (17, 18).
NF-κB Activation.
3×-κB-L luciferase reporter and mut-κB-L control were gifts from Bill Sugden (University of Wisconsin at Madison) (27). Methods were as described (17, 18).
RESULTS
The Last Three Residues of LMP1 Are Critical for Primary B Lymphocyte Growth Transformation.
To identify a site in the carboxyl-terminal 155 residues that is critical in enabling efficient, long-term LCL outgrowth, two point mutations in the last three carboxyl-terminal residues and a series of incremental deletions were constructed in an LMP1 gene that has a Flag epitope insertion at the amino terminus (F-LMP1, Fig. 1) (17). The last three carboxyl-terminal amino acids of LMP1 are YYD, and the two point mutations were YYD to FFD or YYD to ID.
The method of P3HR-1 second site homologous recombination was used to introduce the point mutations and series of incremental deletions into complete EBV genomes (17, 33, 34, 37–39). The P3HR-1 EBV is replication competent but transformation defective because of a deletion. This deletion can be rescued by transfecting P3HR-1 EBV-infected cells with DNA that spans the deletion site. Recombinants then can be recovered because of their ability to growth transform B lymphocytes into LCLs. Cotransfecting P3HR-1-infected cells with the rescuing DNA fragment and a second EBV DNA fragment results in incorporation of the second DNA fragment into 10% of the rescued EBV genomes. Consistent with predicted results, F-LMP1 recombinants were recovered in 14% (31 of 233) of the LCLs, and F-LMP1-FFD recombinants were recovered in 8% (20 of 243) of the LCLs. In contrast, F-LMP1-ID recombinants were recovered in only 3% (13 of 458) of the LCLs. Recombinants with more severe carboxyl-terminal deletions also were recovered in fewer LCLs than the predicted 10%. All LCLs except for one infected with an F-LMP1-FFD recombinant were coinfected with P3HR-1 EBV. Because the low percentage of F-LMP1-ID-infected LCLs appeared to define a site important in transformation, we put aside the series of deletion mutations and focused on characterizing the transformation phenotypes of F-LMP-ID and F-LMP-FFD recombinants by comparing them with control F-LMP1 recombinants.
Virus from four independently derived LCLs that were coinfected with an F-LMP1-ID recombinant and P3HR-1 and from two independently derived LCLs that were coinfected with an F-LMP1 recombinant and P3HR-1 was passaged onto primary B lymphocytes. Virus from the LCL infected with an F-LMP1-FFD recombinant without P3HR-1 also was passaged to verify that this recombinant is wild type for replication and growth transformation. By PCR analysis, virus preparations from the respective coinfected LCLs contained nearly equimolar amounts of F-LMP1-ID DNA and P3HR-1 LMP1 DNA or nearly equimolar amounts of F-LMP1 DNA and P3HR-1 LMP1 DNA (data not shown). Hundreds of B lymphocytes were transformed with these virus preparations, and these LCLs were analyzed for EBV genotypes. The results presented in Table 1 were that F-LMP1-ID recombinants were recovered in 116 LCLs, and in every instance the LCLs were coinfected with P3HR-1 EBV. Infection of B lymphocytes with less input virus reduces the number of LCLs and increases the probability of clonally segregating a transforming recombinant from P3HR-1 EBV, but none of the 41 LCLs that were obtained after infection with less input virus were infected with an F-LMP1-ID recombinant only. Overall, more LCLs (157) were transformed by secondary recombinants that have complete genomes with the P3HR-1 LMP1 gene in place of the F-LMP1-ID gene. Given the nearly equal molarity of the F-LMP1-ID DNA and P3HR-1 LMP1 DNA in the virus preparations, these data indicate a strong preference for the P3HR-1 LMP1 gene in B lymphocyte growth transformation. Clonal transformation assay of virus preparations containing F-LMP1 recombinant and P3HR-1 EBV produced contrasting results. More than half of the resultant LCLs (27 of 51) were infected with an F-LMP1 EBV recombinant without P3HR-1 EBV, 17 of 51 were infected with a secondary recombinant that had the P3HR-1 LMP1 in place of F-LMP1, and 7 of 51 were coinfected with an F-LMP1 recombinant and P3HR-1 EBV. Because no P3HR-1 EBV was in the F-LMP1-FFD recombinant virus preparation, 48 of 48 LCLs were transformed by F-LMP1-FFD recombinant. The F-LMP1-infected LCLs that lacked P3HR-1 LMP1 on initial PCR screening and all of the F-LMP1-FFD-infected LCLs were retested for the presence of P3HR-1 EBV by using a PCR method that specifically detects P3HR-1 LMP1 DNA (12). These LCLs had less than the assay limit of four copies of P3HR-1 LMP1 DNA per 10,000 cells (data not shown). To summarize these results, F-LMP1 and F-LMP1-FFD recombinants are able to transform B lymphocytes into LCLs without P3HR-1 EBV whereas F-LMP1-ID recombinants cannot. Thus, the YYD to ID mutation defines a second effector site that is critical for transforming B lymphocytes into LCLs capable of long-term outgrowth.
Table 1.
B lymphocyte transformation by EBV recombinants
| Original LCL source of virus | Viral genomes in original LCLs | % infected wells with LCLs | Number of resultant LCLs with:
|
||
|---|---|---|---|---|---|
| F-LMP1 alone | F-LMP1 & P3HR-1 LMP1 | P3HR-1 LMP1 alone | |||
| F-LMP1-1 | F-LMP1 & P3HR-1 | 17 | 8 | 7 | 16 |
| 1 | 1 | 0 | 1 | ||
| F-LMP1-2 | F-LMP1 & P3HR-1 | 9 | 17 | 0 | 0 |
| 1 | 1 | 0 | 0 | ||
| F-LMP1-ID-1 | F-LMP1-ID & P3HR-1 | 19 | 0 | 86 | 8 |
| 2 | 0 | 19 | 2 | ||
| F-LMP1-ID-2 | F-LMP1-ID & P3HR-1 | 100 | 0 | 10 | 85 |
| 5 | 0 | 1 | 19 | ||
| F-LMP1-ID-3 | F-LMP1-ID & P3HR-1 | 11 | 0 | 0 | 14 |
| F-LMP1-ID-4 | F-LMP1-ID & P3HR-1 | 15 | 0 | 0 | 29 |
| F-LMP1-FFD-1 | F-LMP1-FFD | 38 | 48 | 0 | 0 |
Virus replication was induced in EBV-infected LCLs (column 1). B lymphocytes were infected with dilutions of the resultant viruses (column 2) and seeded into 96-well plates. If the percentage of wells with LCLs (column 3) is less than 40%, P3HR-1 usually is segregated from a transforming recombinant. LCLs that are infected with a recombinant alone (column 4) are positive for F-LMP1 DNA and negative for P3HR-1 LMP1 DNA. LCLs coinfected with a recombinant and P3HR-1 (column 5) are positive for both F-LMP1 and P3HR-1 LMP1 DNA. Secondary recombinant transformed LCLs (column 6) are infected with a transforming EBV that has only P3HR-1 LMP1 DNA.
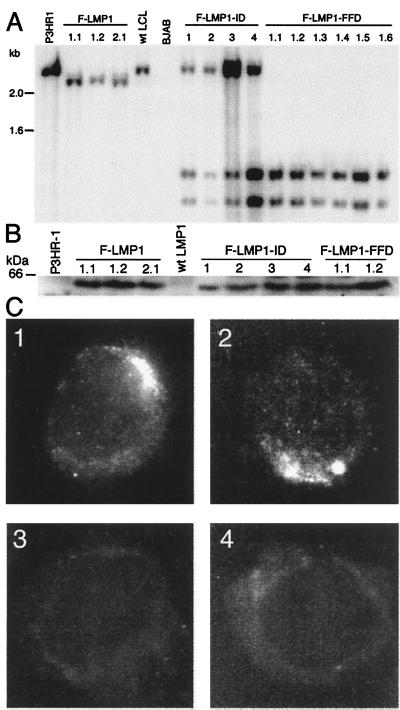
The LMP1 genes in LCLs coinfected with F-LMP1-ID recombinant and P3HR-1 and in LCLs infected with F-LMP1 or F-LMP1-FFD recombinants were confirmed to have recombined homologously by Southern blot (Fig. 2A) and PCR analyses (12, 17). DNA extracted from LCLs was cut with MluI and SacI and Southern blot-hybridized with a wild-type 2.4-kb LMP1 DNA probe. P3HR-1 LMP1 DNA was identified by a characteristic 2.4-kb DNA, F-LMP1 recombinant DNA by a 2.3-kb DNA because of a SacI site at the Flag codon insertion, and F-LMP1-FFD or F-LMP1-ID recombinant DNA by 1.2- and 1.1-kb DNAs because of a second SacI site near the site of the FFD or ID mutations. In data not shown, PCR with one primer specific for LMP1 exon 1 and a second primer anchored in the EBV terminal repeat but ending in the adjacent unique DNA confirmed that the F-LMP1, F-LMP1-ID, F-LMP1-FFD, and P3HR-1 LMP1 DNAs were separated by the expected number of base pairs from the right end terminal repeat. P3HR1 EBV LMP1 DNA yielded an 880-bp DNA that was not cut with SacI whereas F-LMP1, F-LMP1-ID, and F-LMP1-FFD recombinant DNA produced 908-bp DNAs that were cut with SacI to yield the expected 189- and 719-bp DNAs.
Figure 2.
Characterization of LMP1 in infected LCLs. (A) Southern blot analysis of EBV recombinant infected LCLs. DNA cut with SacI and MluI was Southern blot-probed with EBV MluI DNA (nucleotide 167,129 to 169,560), which comprises LMP1 DNA. A 2.4-kb DNA from P3HR-1 cells and a wild-type EBV-transformed LCL is hybridized by the probe. In LCLs F-LMP1 1.1, 1.2, and 2.1, which are infected with an F-LMP1 recombinant from F-LMP1–1 or F-LMP1–2 LCL, a SacI site near the Flag codons results in a 2.3-kb DNA whereas the 0.16-kb DNA ran off. In coinfected LCLs F-LMP1-ID 1–4 or in singly infected LCLs F-LMP1-FFD 1.1–1.6, which are infected with an F-LMP1-FFD recombinant from F-LMP1-FFD-1 LCL, F-LMP1-ID, and F-LMP1-FFD DNAs, have a second SacI site near the last codon, resulting in 1.2- and 1.1-kb DNAs whereas the 0.16-kb DNA ran off. The 2.4-kb DNA in LCLs F-LMP1-ID 1–4 is P3HR-1 LMP1 DNA. Markers in kb are at left. (B) Immunoblot analysis of LMP1. Proteins from 5 × 104 cells were size-separated, blotted to filters, and probed with antibody to Flag. A 60-kDa band in LCLs F-LMP1 1.1, 1.2 and 2.1, F-LMP1-ID 1–4, and F-LMP1-FFD 1.1 and 1.2 is Flag-LMP1 (wild type or mutant). A standard in kDa is noted at left. (C) Immunofluorescent staining of cells with antibody to Flag. (C1) F-LMP1-ID-1, an LCL coinfected with P3HR-1 and F-LMP1-ID recombinant. (C2) F-LMP1 1.1, an LCL infected with F-LMP1 recombinant only. (C3) P3HR-1 that expresses LMP1. (C4) LCL infected with wild-type EBV that expresses LMP1 without Flag.
F-LMP1-ID recombinant and P3HR-1 coinfected LCLs also were confirmed to express F-LMP1-ID by immunoblot and by immunofluorescence with antibody to Flag. In Fig. 2B, Western immunoblots revealed similar levels of a 60-kDa Flag-LMP1 (wild type and mutant) in lysates from LCLs infected with F-LMP1, F-LMP1-FFD, or F-LMP1-ID EBV recombinants. In Fig. 2C, immunofluorescent staining with antibody to Flag revealed F-LMP1-ID (C1) and F-LMP1 (C2) were in plasma membrane aggregates indistinguishable from wild-type LMP1. Flag antibody does not detect LMP1 in P3HR-1 cells (C3) or in a wild-type EBV-transformed LCL (C4). As expected, S12 antibody to the LMP1 repetitive motif detected LMP1 in plasma membrane aggregates in all four LCLs (data not shown; ref. 40). These data indicate that the inability of F-LMP1-ID recombinants to growth transform B lymphocytes is not caused by protein instability or aberrant localization. In data not shown, F-LMP1-ID aggregated in the plasma membrane in transfected B lymphoma cells, indicating that its aggregation is not dependent on wild-type LMP1. These results indicate that the failure of F-LMP1-ID recombinants to transform cells without wild-type LMP1 likely is caused by mutation of an effector site rather than a global effect on protein stability or localization. We therefore designate the two tyrosines to be a principal component of TES2.
TRADD Is Unique Among Interacting Proteins in Specificity for Wild-Type TES2.
We used a yeast two-hybrid screen to search for a cell protein(s) that interacts with F-LMP1 but not with F-LMP1-ID. LMP1 codons 355–386 fused to the GAL4 DNA binding domain (GAL4-DBD) were used as bait to trap cDNA library prey fused to the GAL4 activating domain (GAL4-AD) (36). From 17 million yeast transformed with GAL4-DBD-LMP1(355–386) bait and GAL4-AD-cDNA prey, 19 interacting cDNAs were identified by activation of GAL4 responsive histidine biosynthesis and β-galactosidase production. The cDNAs were the aspartate transcarbamalase component of dihydroorotase (four times), glucosidase I, an X-linked nuclear protein, an E1 ubiquitin activating enzyme, heat shock protein hsp70, an HLA-B associated transcript, a protease, an ORF within an intron of ABL, an ion channel protein, a senescent fibroblast cDNA, and TRADD (32). The others were novel cDNAs. All of the cDNA-encoded proteins interacted with full-length LMP1 carboxyl terminus GAL4-DBD-LMP1(182–386). When tested for specificity for wild-type TES2, one of the cDNAs failed to interact with isoleucine-mutated LMP1 fusion proteins GAL4-DBD-LMP1(355–383ID) or GAL4-DBD-LMP1(182–383ID). That cDNA encoded the TRADD (residues 195–312). Because of its unique specificity for wild-type TES2, TRADD is the most likely cellular mediator of TES2 effects.
TRADD Associates Specifically with Wild-Type TES2 in 293 Cells and LCLs.
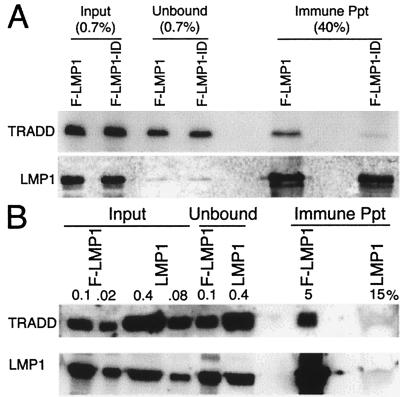
To evaluate the association of TRADD with wild-type TES2 in human cells, antibody to Flag was used to immune precipitate F-LMP1 or F-LMP1-ID from 293 cells cotransfected with myc-tagged TRADD vector DNA and F-LMP1 or F-LMP1-ID vector DNAs (Fig. 3A). In the unfractionated input cell lysates, myc-TRADD was expressed at similar levels in the respective cotransfected cells. F-LMP1 and F-LMP1-ID also were expressed at similar levels in the respective cotransfected cells and were immune-precipitated with similar efficiency. TRADD, however, coprecipitated with F-LMP1 but not with F-LMP1-ID. In data not shown, F-LMP1-FFD and an F-LMP1 with a proline to alanine mutation within a site near TES2 that resembles a TRAF binding motif (see below) also coimmune-precipitate TRADD. Because F-LMP1 and F-LMP1-FFD are transforming LMP1 genes whereas F-LMP1-ID is not, these results further indicate TRADD is the most likely cellular mediator of TES2 effects.
Figure 3.
(A) Specific coprecipitation of TRADD with F-LMP1. 293 cells were electroporated with pRK5-myc-TRADD and pSG5-based F-LMP1 or F-LMP1-ID vector DNAs. Nonidet P-40-solubilized proteins were incubated with M2 antibody to Flag coupled to beads (Kodak). M2 beads were washed, and precipitated proteins were solubilized and immunoblot-analyzed with 9E10 antibody to myc-TRADD or S12 antibody to LMP1. Unfractionated cell proteins are inputs, and proteins not precipitated with M2 beads are unbound. Percentages indicate fraction of total sample. (B) Coprecipitation of TRADD with F-LMP1. LCLs (1.5 × 108 cells) infected with an F-LMP1 EBV recombinant or with a wild-type LMP1 EBV recombinant were solubilized in buffer containing Brij 58 and incubated with M2 beads. Precipitated proteins were solubilized and Western blot-analyzed with antisera to TRADD (Santa Cruz Biotechnology) or S12 antibody to LMP1. Unfractionated cell proteins are inputs and proteins not precipitated with M2 beads are unbound. Percentages indicate fraction of total sample.
The association of TRADD with F-LMP1 in EBV recombinant-transformed LCLs also was evaluated by immune-precipitating F-LMP1 with Flag antibody and analyzing precipitates by Western blot with antibodies to LMP1 or TRADD (Fig. 3B). An LCL infected with a secondary recombinant that has a complete EBV genome and LMP1 without Flag was analyzed as control. In the unfractionated input cell lysates, TRADD was present at similar levels in the two LCLs. F-LMP1 and LMP1 were also present at similar levels in input lysates from the respective LCLs. Flag antibody specifically immune-precipitates F-LMP1 and coimmune precipitates TRADD with F-LMP1. Taking into account the efficiency of F-LMP1 precipitation (25%), 4% of TRADD is constitutively associated with F-LMP1 in LCLs transformed by F-LMP1 recombinant EBV.
NF-κB Is Activated by TES2 and TRADD Mediates NF-κB Activation from TES2.
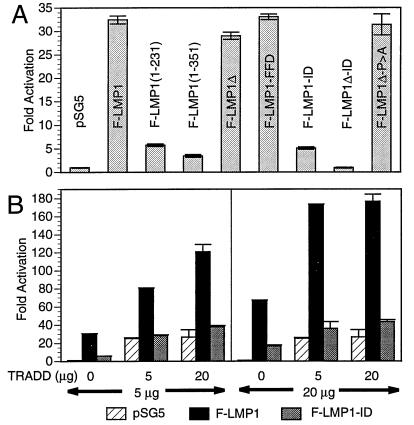
Because TES2 maps at the end of carboxyl terminus in a region previously implicated in high-level NF-κB activation (27, 28) and TRADD mediates NF-κB activation from TNFR1 (32), we investigated the role of TRADD and TES2 in mediating NF-κB activation (Fig. 4A). In 293 cells, F-LMP1 induces high-level NF-κB activation whereas a Flag-LMP1-TES1 expression vector [F-LMP1(1–231), see Fig. 1] induces low-level NF-κB activity. The addition of residues that include the repetitive motif to TES1 in vector F-LMP1(1–351) contributes little to TES1-mediated NF-κB activation whereas F-LMP1 deleted for TES1 and the repetitive motif (F-LMP1Δ) was nearly equal to F-LMP1. Further, F-LMP1-FFD was similar to F-LMP1 whereas F-LMP1-ID was similar to the low levels mediated by TES1. Most strikingly, high-level NF-κB activation by Flag-LMP1-TES2 expression vector F-LMP1Δ was completely ablated by the YYD to ID mutation (F-LMP1Δ vs. F-LMP1Δ-ID). Further, despite resemblance of TES2 adjacent residues P379XQXS383 to the core element of a TRAF binding site (18, 31), mutation of P379 to A had no effect on NF-κB activation (F-LMP1Δ-P > A). These results indicate that the two tyrosines are also principal elements of a high-level NF-κB activation effector site, and taken together with the coimmune precipitation results are consistent with an effector role for TES2 interaction with TRADD in mediating transformation and activating NF-κB.
Figure 4.
(A) F-LMP1 expression in 293 cells activates NF-κB. Five million cells were electroporated with 5 μg of pSG5 or pSG5-LMP1 vector DNA, 2.5 μg of 3×-κB-L luciferase reporter DNA, and 2.5 μg of glucokinase promoter/β-galactosidase reporter DNA as control. Residues of LMP1 are indicated within parenthesis, Δ indicates deletion of residues 187–351, FFD or ID indicates replacement of Y384YD386, and P > A indicates P379 replaced with A within a potential TRAF binding site. Cell lysates were analyzed for luciferase (Promega) and β-galactosidase (Tropix, Bedford, MA) on an Optocomp I luminometer. Immunoblots probed with M5 antibody to Flag (data not shown) indicated that LMP1 expressions levels were similar in the transfected cells. (B) TRADD and F-LMP1 synergize to activate NF-κB. 293 cells were electroporated with the indicated amounts of pSG5 or pSG5-F-LMP1 or F-LMP-ID vector DNA and with pRK5 or pRK5-mycTRADD vector DNA, 2.5 μg of 3×-κB-L luciferase reporter DNA, and 2.5 μg of glucokinase promoter/β-galactosidase reporter DNA as control. Lysates were analyzed for luciferase (Promega) and β-galactosidase (Tropix) on an Optocomp I luminometer. Immunoblots probed with M5 antibody to Flag (data not shown) indicated that LMP1 expressions levels were similar for the indicated amounts of DNA in the respective transfected 293 cells.
We further investigated the role of TRADD as a mediator of TES2- induced NF-κB activation (Fig. 4B). In a representative 293 cell transfection experiment, 5 μg of F-LMP1 or TRADD vector DNAs activated NF-κB 30-fold or 25-fold, respectively. Four times as much F-LMP1 DNA activated NF-κB 70-fold whereas four times as much TRADD DNA did not increase NF-κB activation. Cotransfecting 5 μg of F-LMP1 and 5 μg of TRADD vector DNAs activated NF-κB 80-fold, which is greater than the 55-fold sum of F-LMP1 plus TRADD activation. Even more striking, 20 μg of F-LMP1 with 5 μg of TRADD DNA activated NF-κB 175-fold, which is substantially greater than the 95-fold expected for an additive effect. In contrast, F-LMP1-ID had less than 25% of the activity of F-LMP1, and F-LMP1-ID did not synergize with TRADD. TRADD interaction with F-LMP1, TRADD synergy with F-LMP1 in NF-κB activation, and the lack of interaction or synergy with F-LMP1-ID are biochemical, physiologic, and genetic evidence that TRADD is a mediator of TES2-induced NF-κB activation and B lymphocyte growth transformation.
Because TRAF2 is a downstream effector of TRADD-mediated NF-κB activation from TNFR1 (25, 32), the effect of a dominant negative TRAF2 (25, 26) on TRADD and TES2 was investigated. In DNT/293 cells, a 293 cell line derivative that expresses eight times more dominant negative TRAF2Δ6–86 than TRAF2, TRADD activated NF-κB 10-fold or 44% less than in 293 cells where TRADD activated NF-κB 18-fold (mean of three experiments). This result is consistent with the previously observed 40% inhibition by dominant negative TRAF2Δ6–86 on LMP1Δ187–332 mediated NF-κB activation (26). Significantly, TES2 and TRADD synergistic NF-κB activation in DNT/293 cells (mean 24-fold) was also 48% less than in 293 cells (mean 46-fold). Thus, a dominant negative TRAF2 has similar effects on TES2, TRADD, or TES2 and TRADD-mediated NF-κB activation. These results taken together with the coimmune precipitation results are further evidence TES2 mediates NF-κB activation by interaction with TRADD and TRADD association with TRAFs.
DISCUSSION
The definition of a second LMP1 transformation effector site (TES2) involving Y384Y385 and the finding of its biochemical and physiologic interaction with TRADD adds a surprising dimension to the extent to which LMP1 mimics a constitutively activated TNFR to mediate B lymphocyte growth transformation. This model (Fig. 1) is based on the findings that the amino terminus tethers the first transmembrane domain to the cytoplasm, the six transmembrane domains constitutively aggregate LMP1 in the plasma membrane, and aggregation enables TES1 to constitutively engage TRAF3, 1, and 2 to mediate initial growth transformation of B lymphocytes into LCLs (refs. 10, 12, and 15–18; K. M. Kaye, K.I., E. Johanssen, and E.K., unpublished work). In contrast, TNFR2 or lymphotoxin β receptor aggregation, TRAF recruitment, and subsequent TRAF-mediated effects require ligand binding (20, 25). We now find that TES2 mimics TNFR1 by appropriating TRADD as its signaling adapter. Similar to TES1 association with TRAFs, LMP1 aggregation enables TES2 to constitutively engage TRADD to enable efficient long-term LCL outgrowth. In contrast, TNFR1 aggregation, TRADD recruitment, and TRADD-mediated effects require TNF (19, 32, 41, 42).
An important aspect of the interaction of TES2 with TRADD and of TES1 with TRAFs is how these two sites differentially alter cell growth. TES1 mediates low-level NF-κB activation as measured with the Igκ (28) or class I major histocompatibility complex NF-κB response elements (18, 27), which preferentially respond to NFKB1(p50)/RelA (p65) dimers. Yet, TES1 mediates initial B lymphocyte transformation and induces epidermal growth factor receptors in C33 epithelial cells whereas TES2 mediates high-level NF-κB activation but cannot by itself mediate B lymphocyte growth transformation or epidermal growth factor receptor induction (17, 43). Clearly, TES1 and TES2 are transmitting qualitatively different signals that are not fully characterized by NF-κB activation as measured by the Igκ or class 1 major histocompatibility complex NF-κB response elements. Whether TRAF1 and 2 mediate different interactions with downstream effectors of IκB degradation when engaged by TES1 than when engaged by TES2 through TRADD or whether other interactions account for these differences remains to be resolved. TRADD induction of NF-κB is only in part mediated by a direct TRADD interaction with TRAF1 and 2. TRADD also interacts with receptor interacting protein, which can recruit TRAFs and substantially augment NF-κB activation (41, 42, 44). The identification of TRAFs and TRADD as the proximal effectors of TES1 and TES2 are important steps in understanding the different biochemical and genetic effects of TES1 and TES2 in B lymphocyte transformation and establish a basis for subsequent analyses of the molecular mechanisms underlying the difference between TES1 and TES2.
The biochemical and genetic data reported here affirm a central role for high-level NF-κB activation from TES2 in enabling efficient long-term LCL outgrowth. F-LMP1-ID is clearly deficient in NF-κB activation and transformation whereas F-LMP1-FFD is similar to F-LMP1 for TRADD association, NF-κB activation, and transformation. Recently, we found that those cells that establish long-term outgrowth after infection with MS231 recombinant that has only TES1 have high-level NF-κB activation similar to wild-type EBV-transformed LCLs (K.M. Kaye, K.I., E. Johanssen, and E.K., unpublished work). Thus, high-level NF-κB activation mediated by TES2 engagement of TRADD or by underlying viral or cellular changes appear to be important in the fully transformed LCL phenotype. Such a role for NF-κB in EBV-mediated B lymphocyte transformation is consistent with the role of NF-κB in normal lymphocyte growth control, differentiation, and development (29, 30). NF-κB can alter the transcription of growth regulatory genes such as c-myc, cytokines such as interleukin 2, interleukin 6, granulocyte-colony stimulating factor, and granulocyte/macrophage-colony stimulating factor, and anti-apoptotic factors such as A20 (45). NF-κB-related proteins also have been implicated in malignancy. Retroviral v-rel induces lymphomas in chickens whereas activating chromosomal translocations of NFKB2 or bcl-3 are associated with human lymphomas. Moreover, human T-lymphotrophic virus-1 infection, tax-mediated NF-κB activation, and dysregulated lymphocyte proliferation are significant early events in the causation of adult T cell leukemia. But, high-level NF-κB activation per se is not sufficient for EBV-mediated growth transformation because LMP1 deleted of TES1 retains high-level NF-κB activation but is not sufficient for B lymphocyte transformation (17).
TRADD is a mediator of TES2 effects in NF-κB activation and growth transformation and transduces death signals from TNFR1. TNF induces TRADD to recruit fas-associated death domain protein (FADD), which effects apoptosis through its interaction and activation of caspases (42). LMP1 can have toxic effects when highly expressed (46), and this could be caused by TRADD interaction. However, TNFR1 and LMP1 only partially overlap in inducing apoptosis. In most cells, LMP1 expression at levels similar to that in LCLs activates NF-κB but does not induce apoptosis. Further, the extent to which LMP1 toxicity is attributable to apoptosis is uncertain. In contrast, apoptosis is a prominent feature of TNF/TNFR1 activation and TRADD overexpression. LMP1 and TNFR1 also differ in the biochemistry of their interaction with TRADD. The sequence around TES2 that engages TRADD does not resemble TNFR1 or a death domain. Although TNFR1 interaction with TRADD can propagate a death domain interaction with FADD, LMP1 interaction with TRADD may not. TES2 may allosterically alter TRADD and resist heteroaggregation with FADD. Alternatively, LMP1 may actively counter cell death effectors from TRADD through the anti-apoptotic effects of NF-κB, A20 (45), or Bcl-2 induction (7).
Acknowledgments
Erle Robertson, Kenneth Kaye, George Mosialos, and Ellen Cahir McFarland contributed to this manuscript. This work was supported by Public Health Service Grant CA47006–06 from the National Cancer Institute.
ABBREVIATIONS
- EBV
Epstein–Barr virus
- LMP
latent membrane protein
- LCLs
lymphoblastoid cell lines
- TNF
tumor necrosis factor
- TNFR
TNF receptor
- TRADD
TNF receptor-associated death domain
- TRAF
TNF receptor-associated factor
- TES
transformation effector site
References
- 1.Kieff E. In: Fields Virology. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S, editors. Philadelphia: Raven; 1996. pp. 2343–2396. [Google Scholar]
- 2.Wang D, Liebowitz D, Kieff E. Cell. 1985;43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 3.Baichwal V R, Sugden B. Oncogene. 1988;2:461–467. [PubMed] [Google Scholar]
- 4.Moorthy R K, Thorley-Lawson D. J Virol. 1993;67:1638–1646. doi: 10.1128/jvi.67.3.1638-1646.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammarskjold M L, Simurda M C. J Virol. 1992;66:6496–6501. doi: 10.1128/jvi.66.11.6496-6501.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D, Liebowitz D, Wang F, Gregory C, Rickinson A, Larson R, Springer T, Kieff E. J Virol. 1988;62:4173–4184. doi: 10.1128/jvi.62.11.4173-4184.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowe M, Peng P M, Huen D S, Hardy R, Croom C D, Lundgren E, Rickinson A B. J Virol. 1994;68:5602–5612. doi: 10.1128/jvi.68.9.5602-5612.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson J B, Weinberg W, Johnson R, Yuspa S, Levine A J. Cell. 1990;61:1315–1327. doi: 10.1016/0092-8674(90)90695-b. [DOI] [PubMed] [Google Scholar]
- 9.Dawson C W, Rickinson A B, Young L S. Nature (London) 1990;344:777–780. doi: 10.1038/344777a0. [DOI] [PubMed] [Google Scholar]
- 10.Kaye K M, Izumi K M, Kieff E. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rickinson A B, Kieff E. In: Fields Virology. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S, editors. Philadelphia: Raven; 1996. pp. 2397–2446. [Google Scholar]
- 12.Izumi K M, Kaye K M, Kieff E D. J Virol. 1994;68:4369–4376. doi: 10.1128/jvi.68.7.4369-4376.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baichwal V R, Sugden B. Oncogene. 1989;4:67–74. [PubMed] [Google Scholar]
- 14.Liebowitz D, Mannick J, Takada K, Kieff E. J Virol. 1992;66:4612–4616. doi: 10.1128/jvi.66.7.4612-4616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaye K M, Izumi K M, Mosialos G, Kieff E. J Virol. 1995;69:675–683. doi: 10.1128/jvi.69.2.675-683.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 17.Izumi K M, Kaye K M, Kieff E D. Proc Natl Acad Sci USA. 1997;94:1447–1452. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devergne O, Hatzivassiliou E, Izumi K M, Kaye K M, Kleijnen M F, Kieff E, Mosialos G. Mol Cell Biol. 1996;16:7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shu H B, Takeuchi M, Goeddel D V. Proc Natl Acad Sci USA. 1996;93:13973–13978. doi: 10.1073/pnas.93.24.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.VanArsdale T L, VanArsdale S L, Force W R, Walter B N, Mosialos G, Kieff E, Reed J C, Ware C F. Proc Natl Acad Sci USA. 1997;94:2460–2465. doi: 10.1073/pnas.94.6.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng G, Baltimore D. Genes Dev. 1996;10:963–973. doi: 10.1101/gad.10.8.963. [DOI] [PubMed] [Google Scholar]
- 22.Banchereau J, Bazan F, Blanchard D, Briere F, Galizzi J P, van Kooten C, Liu Y J, Rousset F, Saeland S. Annu Rev Immunol. 1994;12:881–922. doi: 10.1146/annurev.iy.12.040194.004313. [DOI] [PubMed] [Google Scholar]
- 23.Kehry M R. J Immunol. 1996;156:2345–2348. [PubMed] [Google Scholar]
- 24.Noelle R J. Immunity. 1996;4:415–419. doi: 10.1016/s1074-7613(00)80408-2. [DOI] [PubMed] [Google Scholar]
- 25.Rothe M, Sarma V, Dixit V M, Goeddel D V. Science. 1995;269:1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- 26.Kaye K M, Devergne O, Harada J N, Izumi K M, Yalamanchili R, Kieff E, Mosialos G. Proc Natl Acad Sci USA. 1996;93:11085–11090. doi: 10.1073/pnas.93.20.11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell T, Sugden B. J Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huen D S, Henderson S A, Croom C D, Rowe M. Oncogene. 1995;10:549–560. [PubMed] [Google Scholar]
- 29.Baeuerle P A, Henkel T. Annual Review of Immunology. Vol. 12. Inc., Palo Alto, CA: Annual Reviews; 1994. pp. 141–179. [DOI] [PubMed] [Google Scholar]
- 30.Baldwin A S. Annual Review of Immunology. Vol. 14. Inc., Palo Alto, CA: Annual Reviews; 1996. pp. 649–683. [DOI] [PubMed] [Google Scholar]
- 31.Franken M, Devergne O, Rosenzweig M, Annis B, Kieff E, Wang F. J Virol. 1996;24:1879–1885. doi: 10.1128/jvi.70.11.7819-7826.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu H, Xiong J, Goeddel D V. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 33.Tomkinson B, Kieff E. J Virol. 1992;66:2893–2903. doi: 10.1128/jvi.66.5.2893-2903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomkinson B, Kieff E. J Virol. 1992;66:780–789. doi: 10.1128/jvi.66.2.780-789.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomkinson B, Robertson E, Yalamanchili R, Longnecker R, Kieff E. J Virol. 1993;67:7298–7306. doi: 10.1128/jvi.67.12.7298-7306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durfee T, Becherer K, Chen P-L, Yeh S, Yang Y, Kilburn A E, Lee W, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 37.Cohen J I, Wang F, Mannick J, Kieff E. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hammerschmidt W, Sugden B. Nature (London) 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 39.Robertson E S, Tomkinson B, Kieff E. J Virol. 1994;68:1449–1458. doi: 10.1128/jvi.68.3.1449-1458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liebowitz D, Wang D, Kieff E. J Virol. 1986;58:233–237. doi: 10.1128/jvi.58.1.233-237.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu H, Huang J, Shu H B, Baichwal V, Goeddel D V. Immunity. 1996;4:387–396. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 42.Hsu H, Shu H B, Pan M G, Goeddel D V. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 43.Miller W, Mosialos G, Kieff E, Raab-Traub N. J Virol. 1997;71:586–594. doi: 10.1128/jvi.71.1.586-594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ting A T, Pimentel M F, Seed B. EMBO J. 1996;15:6189–6196. [PMC free article] [PubMed] [Google Scholar]
- 45.Laherty C D, Hu H M, Opipari A W, Wang F, Dixit V M. J Biol Chem. 1992;267:24157–24160. [PubMed] [Google Scholar]
- 46.Hammerschmidt W, Sugden B, Baichwal V R. J Virol. 1989;63:2469–2475. doi: 10.1128/jvi.63.6.2469-2475.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]