Abstract
The nuclear receptor steroidogenic factor 1 (SF-1) plays essential roles in the development and function of the ventromedial hypothalamic nucleus (VMH). Considerable evidence links the VMH and SF-1 with the regulation of energy homeostasis. Here, we demonstrate that SF-1 colocalizes in VMH neurons with the cannabinoid receptor 1 (CB1R) and that a specific CB1R agonist modulates electrical activity of SF-1 neurons in hypothalamic slice preparations. We further show that SF-1 directly regulates CB1R gene expression via a SF-1-responsive element at −101 in its 5′-flanking region. Finally, we show that knockout mice with selective inactivation of SF-1 in the brain have decreased expression of CB1R in the region of the VMH and exhibit a blunted response to systemically administered CB1R agonists. These studies suggest that SF-1 directly regulates the expression of CB1R, which has been implicated in the regulation of energy homeostasis and anxiety-like behavior.
STEROIDOGENIC FACTOR 1 (SF-1, officially designated NR5A1) is a nuclear receptor that plays multiple roles throughout the hypothalamic-pituitary-steroidogenic organ axis. A number of target genes of SF-1 have been identified in the gonads, adrenal cortex, and pituitary gonadotropes (1,2). Within the brain, SF-1 is expressed only in the ventromedial nucleus of the hypothalamus (VMH). The molecular basis for this highly restricted expression and the roles of SF-1 in the VMH and its potential hypothalamic target genes remain incompletely understood.
The VMH has been implicated as an important brain region for the regulation of appetite (3), energy balance (4,5,6), anxiety (7), sexual behavior (8,9), and thermogenesis (10). The nucleus serves as a satiety center that provides excitatory input to proopiomelanocortin neurons in the arcuate nucleus, thereby helping to activate anorexigenic neuronal pathways (4). Many genes that regulate energy balance are expressed in the VMH, including membrane receptors, transporters and neuropeptides (11,12).
SF-1 is a key transcription factor in establishing the cytoarchitecture of the VMH. The SF-1 neurons in the VMH have been implicated in energy homeostasis and contribute to the sensing of important regulators of energy balance such as leptin (13,14) and glucose (15). Global SF-1 knockout (KO) mice exhibited delayed-onset obesity (6), further suggesting important roles for SF-1 in the regulation of energy homeostasis in the VMH. In mice with central nervous system (CNS)-specific KO of SF-1, the VMH structure was again disrupted and the mice exhibited marked increases in anxiety-like behavior (6). As a result of this disruption, the mice had decreased hypothalamic expression of brain-derived neurotrophic factor (BDNF) and the type 2 receptor for CRH.
One neurotransmitter/receptor system that has recently been implicated in the regulation of energy homeostasis is the endogenous cannabinoid/cannabinoid receptor system. Endogenous ligands for this system are derived from arachidonic acid and interact with a G protein-coupled receptor designated cannabinoid receptor 1 (CB1R). The cannabinoid system was initially characterized because tetrahydrocannabinol, the active species in marijuana, interacts with the CBRs. More recently, genetic KO models that specifically inactivate components of the cannabinoid system, specific agonists and antagonists, and drugs that inhibit enzymes that degrade the endogenous cannabinoids have provided the tools to dissect the functions of the cannabinoid system at the whole-animal, cellular, and molecular level, implicating it in complex behaviors that include feeding and anxiety-like behavior.
Based on the relatively high level of CB1R expression in the VMH and the association of CB1R with hypothalamic regulation of energy homeostasis and other complex behaviors, we extended our studies of the VMH disruption caused by conditional inactivation of SF-1. Experiments described below show that the absence of SF-1 is associated with markedly decreased expression of CB1R and with diminished response to systemic administration of drugs that affect CB1R function.
RESULTS
SF-1 Is Required for CB1R Expression in the VMH
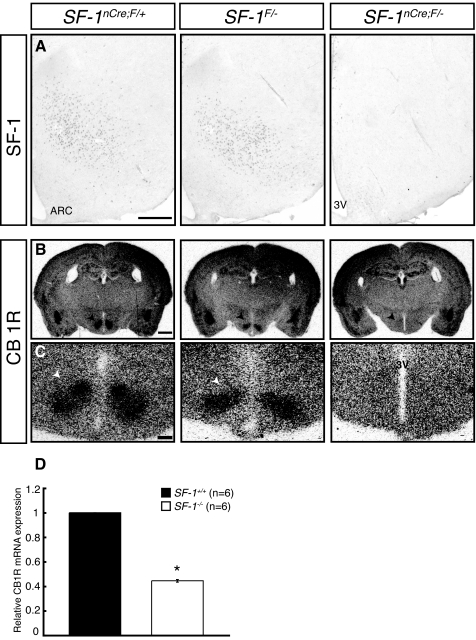
We used CNS-specific SF-1 KO mice to explore potential regulatory roles of SF-1 in CB1R expression in the VMH. As shown in Fig. 1, the effects of this conditional inactivation to abolish SF-1 immunoreactivity and perturb VMH structure (Fig. 1A) were consistent with our previous findings (7). As described (16,17), CB1R transcripts in wild-type (WT) mice were expressed in many brain regions, including the cerebral cortex, amygdala, hippocampus, and hypothalamus (Fig. 1, B and C). CB1R expression was generally preserved in the CNS-specific SF-1 KO mice (Fig. 1B) but was decreased considerably in the region where the VMH normally resides (Fig. 1, B and C). These results indicate that the CNS-specific KO of SF-1 selectively impairs CB1R expression in the VMH, suggesting that it may be a direct target of SF-1 in the VMH neurons.
Figure 1.
SF-1 Is Necessary for CB1R Expression in the VMH
A, Immunohistochemical analysis showing SF-1 expression in mice of the indicated genotypes. Relative to WT (SF-1nCre; F/+) and heterozygous (SF-1F/−) mice, SF-1 expression in the VMH is markedly diminished in CNS-specific SF-1 KO mice (SF-1nCre;F/−). Scale bar, 200 μm. B, Lower power magnification of CB1R mRNA expression in brains from mice of the indicated genotypes. ISH revealed CB1R transcripts in the cerebral cortex, hippocampus, amygdala, and hypothalamus. CB1R mRNA expression in the VMH was specifically abrogated in CNS-specific SF-1 KO mice. Arrowheads indicate the VMH. Scale bar, 500 μm. C, Higher magnification view of CB1R mRNA expression in the mediobasal hypothalamus. CB1R is expressed at high levels in the VMH, where its expression is decreased considerably in the CNS-specific SF-1 KO mice. 3V, Third ventricle. Scale bar, 200 μm. D, The effect of SF-1 on the expression of CB1R in eGFP-positive neurons isolated from WT and SF-1 KO mice. RNA was isolated from fluorescence-activated cell sorting-purified SF-1/eGFP-positive neurons from WT and SF-1 KO mice and analyzed by real-time PCR assays as described in Materials and Methods.
Electrophysiological Regulation of SF-1 Neurons by CB1R
To demonstrate colocalization of CB1R and SF-1 in VMH neurons, we first used quantitative RT-PCR assays with RNA samples prepared from neurons that express a SF-1/enhanced green fluorescent protein (eGFP) transgene (18). CB1R was expressed in these SF-1 neurons, and the level of CB1R expression was decreased by more than 50% in eGFP-positive cells isolated from SF-1 KO mice (Fig. 1D). These studies demonstrate that SF-1 neurons express CB1R and further suggest that SF-1 may regulate their expression of CB1R. The finding that some CB1R expression persists in the absence of SF-1 suggests that other transcription factors also activate CB1R expression in VMH neurons; it further suggests that the decreased levels of CB1R transcripts detected by in situ hybridization (ISH) (Fig. 1) may reflect both decreased transcript levels and greater dispersion of SF-1 neurons within the mediobasal hypothalamus, as previously described (6).
To explore possible functional effects of cannabinoid signaling in SF-1 neurons, we used the same SF-1/eGFP transgene described above to mark SF-1-positive and SF-1-negative VMH neurons. As described in Materials and Methods, presynaptic influences were eliminated by preincubation with a cocktail of inhibitors of glutamate, GABAA, and glycine receptors and whole-cell patch-clamp recordings were collected in slice preparations that included the VMH. The average action potential frequency in SF-1 neurons was 6.2 ± 1 Hz (Fig. 2).
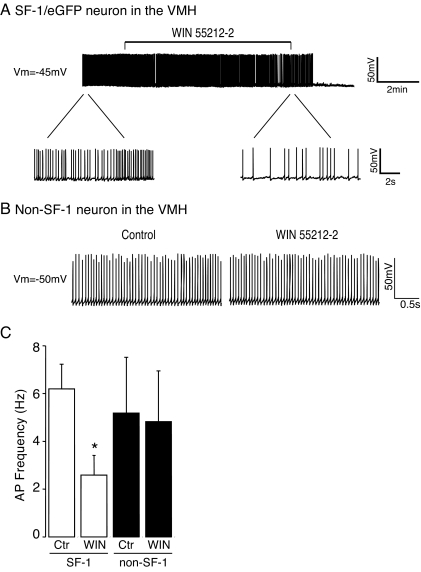
Figure 2.
Activation of CB1R Decreases Action Potential Frequency in SF-1 Neurons
A, Sample traces from a SF-1/eGFP-positive neuron recorded before, during, and after application of the CB1R agonist WIN 55212-2 (1 μm) in current clamp configuration. Treatment with WIN 55212-2 markedly decreased action potential frequency of SF-1 neuron. (Top panel: scale bar: 50 mV, 2 min). The bottom panel shows activity of the same SF-1 neuron on an expanded time scale before and immediately after application of WIN 55212-2. Scale bar: 50 mV, 2 sec. Vm = −45 mV. B, Sample traces from a SF-1/eGFP-negative VMH neuron recorded before and after application of WIN 55212-2 (1 μm) in current clamp configuration. The CB1R agonist did not alter the firing rate of SF-1/eGFP-negative neurons. Scale bar: 50 mV, 0.5 sec. Vm = −50 mV. C, Pooled data of the frequency of action potentials in SF-1/eGFP-positive and -negative VMH neurons before and after treatment with WIN 55212-2 (9 and 7 neurons, respectively; *, P < 0.05). Error bars represent sem.
We first examined the direct effect of cannabinoids on the firing rate of these SF-1/eGFP-positive neurons. After at least 10 min of stable recording of action potentials, the CB1R receptor agonist WIN 55212-2 (1 μm) was added to the bath medium. WIN 55212-2 reduced the frequency of action potentials in approximately 60% of SF-1/eGFP-positive neurons tested (9 of 14 neurons; Fig. 2A). In the affected neurons, treatment with WIN 55212-2 significantly decreased the firing rate of SF-1 neurons from 6.2 ± 1 Hz to 2.6 ± 0.8 Hz (Fig. 2C), which was associated with a statistically insignificant hyperpolarization of membrane potential (control: −45 ± 1.5 mV, plus WIN: −47 ± 1.7 mV; n = 9). The effect of WIN 55212-2 to decrease excitability of SF-1 neurons was blocked by the CB1R antagonist, AM251, suggesting that the effect is specific for CB1R (data not shown). In contrast to the direct effect of cannabinoids on SF-1 neurons, the action potential frequency of SF-1/eGFP-negative VMH neurons in our slice preparations was not significantly altered by treatment with WIN 55212-2 (firing rate: control: 5.2 ± 2 Hz, plus WIN 55212-2: 4.8 ± 2 Hz; membrane potential: control, −48 ± 2.5 mV; plus WIN, −43 ± 1.8 mV; Fig. 2, B and C). These results thus show that functional CB1R is expressed in SF-1 neurons whose activity can be inhibited by a CB1R agonist.
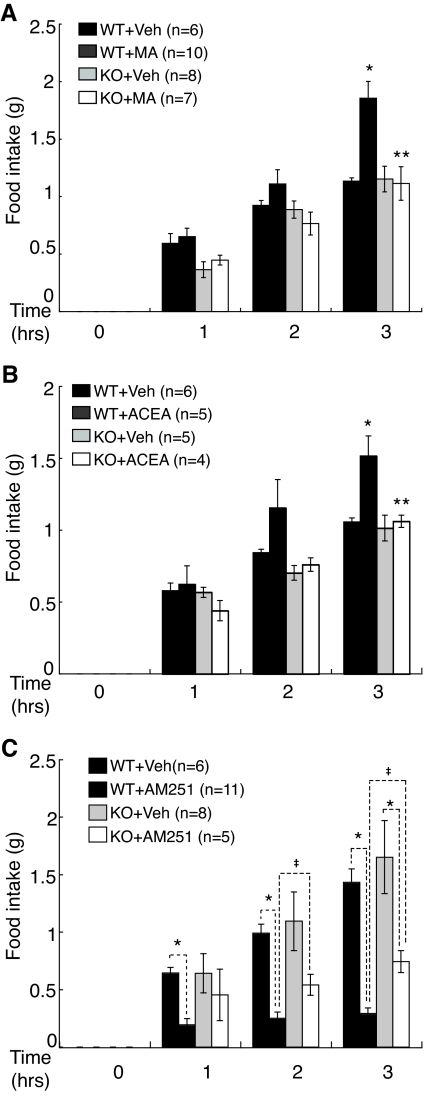
Based on the effects of the SF-1 KO on CB1R expression in the VMH and of the CB1R agonist on action potential frequency of SF-1 neurons, we examined the impact of the CNS-specific SF-1 KO—and the resulting perturbation of VMH structure and CB1R expression—on the in vivo response to systemic administration of drugs that affect the cannabinoid system. Because the effects of cannabinoids on food intake involve hypothalamic action (3) and the VMH is a prominent site of CB1R expression (16,19), we examined CB1 agonist and/or antagonist effects on nocturnal food intake in SF-1nCre;F/+ (WT) and SF-1nCre;F/− (KO) mice. After 24 h of fasting, mice were systemically administered either of two different CB1R agonists, methanandamide (MA) and arachidonyl-2′-chloroethylamide hydrate (ACEA), or the antagonist AM251, and then nocturnal food intake was measured over 3 h as described in Materials and Methods. Consistent with a previous report that rats given MA ate significantly more food in a partial satiety state (20), administration of MA or ACEA to WT mice significantly increased food intake (Fig. 3, A and B). This increased food intake was significantly diminished in CNS-specific SF-1 KO mice (Fig. 3, A and B). Consistent with the apparent preservation of VMH structure and SF-1 immunoreactivity (Fig. 1), mice heterozygous for the SF-1 KO (SF-1F/−) responded to treatment with the CB1R agonists in a manner indistinguishable from WT mice (Kim, K., unpublished observation).
Figure 3.
CNS Expression of SF-1 Contributes to CB1R-Dependent Effects on Nocturnal Food Intake and Body Weight
Effects of MA (A), ACEA (B), and AM251 (C) on nocturnal food intake in WT (SF-1nCre; F/+) and CNS-specific SF-1 KO (SF-1nCre; F/−) mice. After 24 h of fasting, mice were injected with either vehicle (veh), the CB1R agonists MA (1 mg/kg) or ACEA (3 mg/kg), or the CB1R antagonist AM251 (5 mg/kg), and nocturnal food intake was measured hourly thereafter for 3 h as described in Materials and Methods. Injection of MA or ACEA significantly stimulated nocturnal food intake in WT mice (*, P < 0.01, t test); their effects on appetite were insignificant in CNS-specific SF-1 KO (**, P > 0.1, t test). Treatment with the CB1R antagonist AM251 significantly inhibited food intake in both WT and SF-1 KO mice (*, P < 0.01, t test), but the effect was significantly greater in WT mice (‡, P < 0.05, t test). Graphs represent means ± sem and data were analyzed by mutifactorial ANOVA with repeated measure (strain × drug × time, P < 0.05).
The CB1R antagonist AM251 significantly inhibited food intake in WT mice, but its anorexic effect was significantly reduced in CNS-specific SF-1 KO mice relative to WT mice (Fig. 3C). Taken together, these results argue that SF-1 regulation of CB1R expression in the VMH plays an important role in cannabinoid-induced effects on food intake.
SF-1 Directly Regulates CB1R Promoter Activity
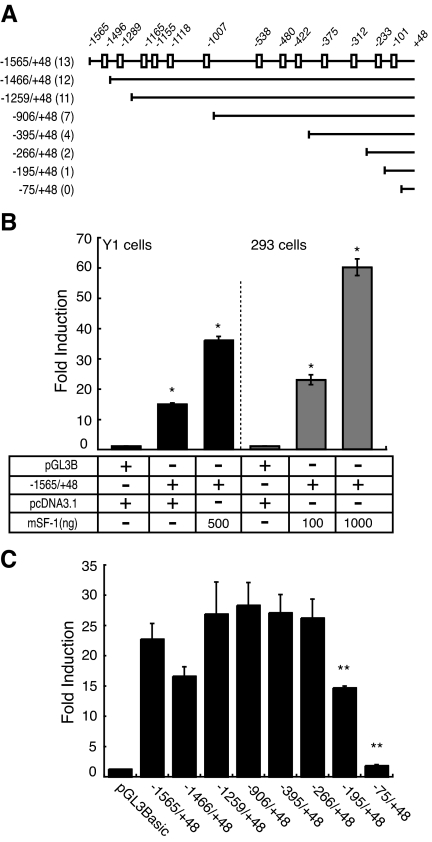
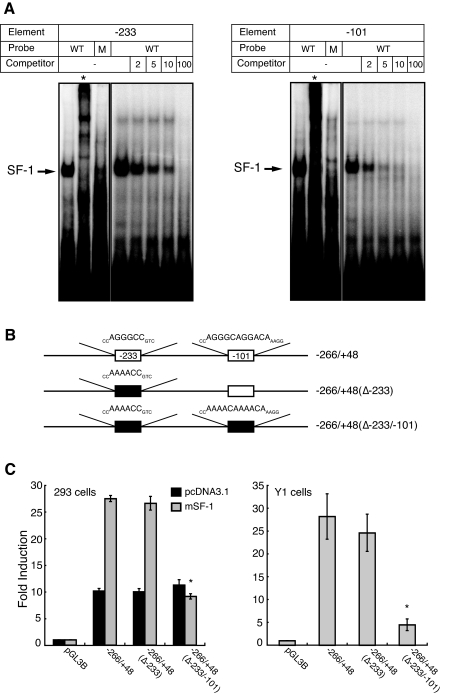
We next asked whether CB1R is a direct target of SF-1 transcriptional regulation. Inspection of the 5′-flanking sequence of CB1R identified 16 potential SF-1-binding sites (21). In EMSAs, 13 of these 16 potential sites formed specific SF-1-related complexes that were supershifted by the addition of an anti-SF-1 antiserum (data not shown). The boxes in Fig. 4A depict the 13 sites in the CB1R 5-flanking region that bound SF-1.
Figure 4.
SF-1 Directly Regulates CB1R Expression
A, Schematic diagram of the 5′-flanking region of the mouse Cb1r gene, including the location of potential SF-1 binding sites. Shown below are the 5′-deletion constructs. B, CB1R promoter activity is significantly stimulated by SF-1 in mouse Y1 adrenocortical cells and human HEK 293 kidney cells in a dose-dependent manner (*, P < 0.001, t test). C, 5′-Deletion analysis of SF-1 binding sites that regulate the CB1R promoter. The 293 cells were cotransfected with SF-1 and the deletion constructs shown in Fig. 4A. Deletion of putative SF-1 binding sites in the −246 and −108 constructs significantly decreased promoter activity (**, P < 0.001, one-way ANOVA).
We next performed transient transfection analyses to examine whether SF-1 could stimulate CB1R promoter activity. As shown in Fig. 4, B and C, SF-1 stimulated activity of CB1R sequences spanning −1565 to +48 (−1565/+48) in both Y1 cells and 293 cells. These data suggest that SF-1 can directly stimulate CB1R promoter activity.
To identify which, if any, of the 13 putative SF-1-responsive elements might mediate this induction, we made 5′-deletion constructs that included CB1R sequences from −1466 to +48 (−1466/+48), −1259/+48, −906/+48, −395/+48, −266/+48, −195/+48, and −75/+48 (Fig. 4A). We then analyzed promoter activities of these constructs via transient transfection in 293 cells (Fig. 4C). Deletion of 5′-flanking sequences from −1565 to −266 did not significantly affect SF-1 stimulated CB1R promoter activity. In contrast, deletion of sequences from −266 to −75 completely abolished SF-1 activation of promoter activity, implying that the sites at −233 and −101 may mediate SF-1 regulation (Fig. 4C).
To examine directly the roles of these two CB1R promoter sites in SF-1 regulation, we mutated the respective sequences in the context of the promoter region from −266/+48. Oligonucleotides that incorporated these mutations failed to bind SF-1 in gel mobility shift assays (Fig. 5A), whereas oligonucleotides containing native sequences at −233 and −101 specifically inhibited the SF-1 complex in a dose-dependent manner (Fig. 5A).
Figure 5.
The SF-1-Responsive Element at −101 Is Required for SF-1 Regulation of CB1R Promoter Activity
A, EMSAs with the potential SF-1 binding sites at −233 and −101 in the CB1R promoter. Oligonucleotides comprising the CB1R promoter elements at −233 (left panel) and −101 (right panel) formed specific complexes with SF-1, whereas probes with mutation of the SF-1 core motif (M) failed to form this complex. *, Reactions that included 1 μl of an anti-SF-1 antiserum. Competition analysis revealed that the complexes formed with the −233 and −101 elements were specific. Unlabeled oligonucleotide competitors at the indicated molar excesses (2, 5, 10, and 100×) specifically inhibited SF-1 binding to the −233 and −101 elements, with complete abrogation of binding at the 100× molar excess. Oligonucleotides with mutations in the SF-1 binding motif (see Materials and Methods) did not inhibit the SF-1 complex. M, Mutated probe. B, Schematic diagram of promoter constructs examined by transient transfection analyses. Empty squares indicate putative SF-1 binding sites and filled squares represent sites that were mutated. C, The SF-1 binding sites at −101 is crucial for SF-1 regulation. The indicated CB1R promoter constructs were transfected with or without SF-1 in 293 cells (left panel) and without SF-1 in Y1 cells (right panel). Mutation of the −233 site alone did not significantly impair SF-1 induction, whereas mutation of both the −233 and the −101 sites abolished SF-1 induction in 293 cells and markedly impaired promoter activity in Y1 cells (*, P < 0.01, one-way ANOVA).
To further define the roles of the sites at −233 and −101 in SF-1 regulation of CB1R promoter activity, we examined the promoter activity and regulation by SF-1 using various mutated constructs (Fig. 5B). Mutation of the site at −233 alone did not significantly impair SF-1 induction of CB1R promoter activity, whereas mutation of both elements virtually abolished the response to SF-1. These results suggest that SF-1 directly stimulates CB1R promoter activity, predominantly via the element at −101 (Fig. 5C).
SF-1 Regulates CB1R-Mediated ERK Activation Both in Cultured Cells and in Vivo
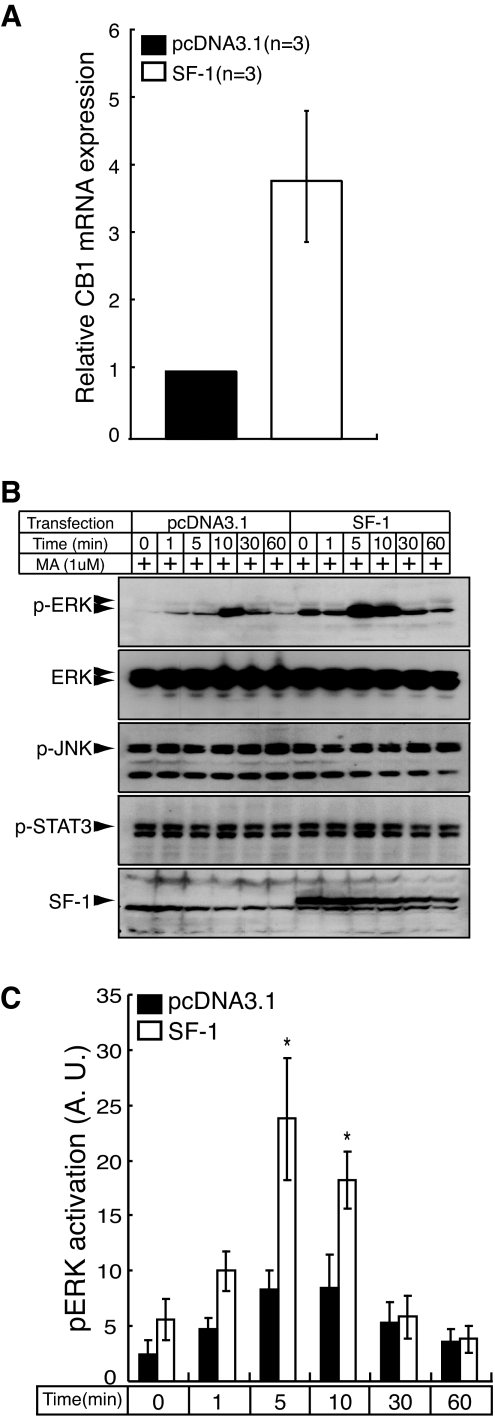
CB1R is a G protein-coupled receptor that signals via Gαi/o to activate cellular responses, including c-Jun N-terminal kinase (JNK), signal transducer and activator of transcription (STAT) 3, and the MAPK pathway (22,23,24). If CB1R is a bona fide target of SF-1, it is plausible that modulation of SF-1 could increase its expression in a manner that affected one of these CB1R-mediated signaling pathways. To address SF-1 regulation of CB1R-mediated cellular responses, we used mouse neuroblastoma Neuro2A cells, which endogenously express CB1R but do not express SF-1. First, we used real-time PCR assays to examine whether transfection of Neuro2A cells with SF-1 induces their expression of CB1R transcripts. These studies showed that introduction of SF-1 in the Neuro2A cells increased CB1R transcripts by approximately 4-fold (Fig. 6A).
Figure 6.
SF-1 Stimulates CB1R Expression and CB1R-Mediated ERK Phosphorylation in Neuro2A Neuroblastoma Cells
A, Transfection with SF-1 stimulates endogenous expression of CB1R transcripts in Neuro2A cells. Neuro2A cells were transiently transfected with either pcDNA3.1 or with the same vector expressing SF-1, and levels of CB1R transcripts were determined by real-time PCR assays as described in Materials and Methods. B, Transfection of Neuro2A cells with an expression plasmid for SF-1 selectively stimulates CB1R-mediated induction of ERK phosphorylation in Neuro2A cells. Immunoblot analyses showed that transfection of Neuro2A cells with an SF-1 expression plasmid dramatically induced MA-induced ERK phosphorylation at 5 and 10 min without affecting the total level of immunodetectable ERK, p-JNK, or p-STAT3. C, Time course quantitating the SF-1 stimulation of MA-mediated ERK phosphorylation in Neuro2A cells shown in B. Black bars indicate cells transfected with pcDNA3.1 and white bars represent cells transfected with SF-1. SF-1 expression significantly stimulated ERK phosphorylation (2.9-fold) at 5 and 10 min after MA treatment (*, P < 0.05, n = 5, t test). The data were expressed as mean ± sem and analyzed by repeated two-way ANOVA (SF-1 transfection × time, P < 0.01). A.U., Arbritrary units.
Treatment of Neuro2A cells with MA (1 μm) increased phosphorylation of ERK, and this increase was significantly potentiated by SF-1 transfection (Fig. 6, B and C). Unlike the dynamic change of ERK phosphorylation by MA treatment, the levels of p-JNK and p-STAT3 were not affected by MA treatment (Fig. 6B). To determine whether Gαi/o mediates the potentiation of ERK phosphorylation by SF-1 overexpression, we pretreated cells with pertussis toxin, which inhibits Gαi/o-mediated signaling, or AM251, a CB1R antagonist. The potentiation of ERK phosphorylation by SF-1 overexpression was completely abolished by pretreatment with either pertussis toxin or AM251 (data not shown), indicating that the potentiation of ERK activation by SF-1 overexpression is CB1R agonist-dependent and requires the Gαi/o-mediated pathway.
To further ascertain that the potentiation of ERK activation by SF-1 is directly mediated by CB1R in the Neuro2A cells, we showed that this effect was abolished after introduction of multiple siRNAs that specifically targeted CB1R (Kim, K., unpublished observation). These data collectively establish that pERK potentiation by SF-1 is directly mediated by CB1R.
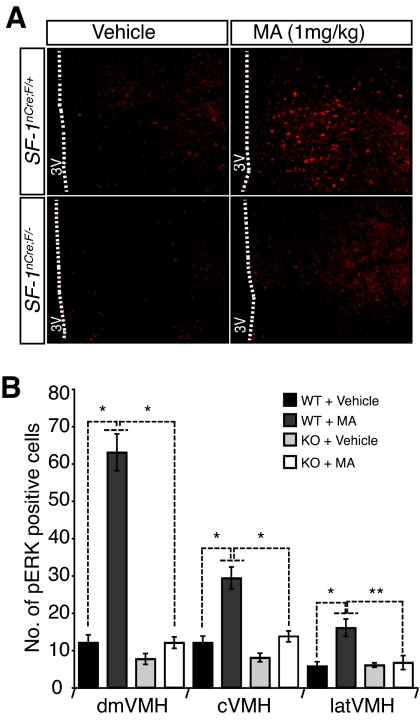
Based on the potentiation of CB1R-mediated ERK phosphorylation by SF-1 in cultured cells, we wondered whether a similar effect also took place in vivo. Indeed, systemic administration of MA to WT (SF-1nCre; F/+) mice significantly increased p-ERK immunoreactivity in the VMH. Consistent with the ISH studies (Fig. 1, B and C), MA-induced phosphorylation of ERK in the VMH was markedly impaired in CNS-specific SF-1 KO mice (Fig. 7).
Figure 7.
Administration of MA to WT But Not SF-1 KO Mice Significantly Stimulates ERK Phosphorylation in the VMH
WT and SF-1 KO mice were treated with MA and cells that contained phospho-ERK were quantified in sections of the mediobasal hypothalamus as residing in the dorsomedial (dm), central (c), or lateral (lat) regions of the VMH. A, Representative sections of immunoreactive phospho-ERK in vehicle- and MA-treated WT and CNS-specific SF-1 KO mice. B, The ERK phosphorylation induced by MA injection was significantly diminished in CNS-specific SF-1 KO mice. Bars represent means ± sem (*, P < 0.01; **, P < 0.05, n = 3, t test).
DISCUSSION
Results presented here show that SF-1 directly regulates CB1R via its 5′-flanking region and that the CNS-specific KO of SF-1 has significant effects on the response of food intake to exogenously administered CB1R agonist and antagonists. Specifically, we have demonstrated that CB1R expression was impaired in the VMH when we selectively removed SF-1 in the CNS, implicating SF-1 in CB1R expression in the VMH. We further found that CB1R colocalized with SF-1 expression in VMH neurons and that cannabinoids decreased excitability of SF-1 neurons but had no effect on non-SF-1 neurons. Recent evidence suggests that cannabinoid and leptin signals are integrated in lateral hypothalamic neurons (25). A similar integration may also occur in SF-1 neurons in the VMH, whose action potential frequency is inhibited by CB1R agonists (Fig. 2) but increased by leptin (14). These results suggest that integration of cannabinoid and leptin signals may regulate feeding behavior in the VMH.
Although SF-1 clearly is a key factor in VMH development, its specific roles in VMH function remain incompletely understood. We recently identified the CRH (Crh) receptor 2 as a direct target of SF-1, providing a plausible molecular basis for the increased anxiety-like behavior seen in CNS-specific SF-1 KO mice (7). Although CB1R in the brain also has been implicated in the regulation of anxiety, the key regions for emotional regulation apparently are the amygdala, hippocampus, and cortex. These sites do not express SF-1, and thus it cannot regulate their expression of CB1R. However, impairment of CB1R expression by genetic ablation of SF-1 may also contribute to the increased anxiety-like behavior because the VMH is known to be a critical brain region for defensive responses to predators (26,27). Moreover, the acute administration of the CB1 antagonist SR141716A induced anxiety-like responses in certain behavioral tests (28), and global CB1R KO showed increased anxiety-like behavior (29,30).
Both N-methyl-d-aspartate subunit (31) and BDNF (32) have been identified as direct targets of SF-1 in the VMH. A recent report expanded considerably the list of SF-1 targets in the VMH, including several cell adhesion molecules such as Amigo2, Cdh4, Sema3a, Slit3, and Netrin3 and other genes that are highly expressed in the VMH, such as Fezf1, Nptx2, Nkx2-2, and A2bp1 (11). Our results show that SF-1 can regulate CB1R expression directly, expanding the group of target genes to this G protein-coupled receptor, which has become an exciting target for drug development (33).
Given that CB1R activation by cannabinoids leads to phosphorylation of STAT3, JNK, and ERK (22,23,34), we sought to determine whether SF-1 has a role in signaling mediated by CB1R activation. As shown in Fig. 6, transfection of Neuro2A cells with SF-1 induced CB1R expression and potentiated CB1R-mediated ERK phosphorylation. Activation of STAT3 or JNK by CB1R has been implicated in neurite outgrowth (22,23,24), but the precise effects of CB1R-dependent ERK phosphorylation after CB1R activation are not fully understood. Activation of ERK has been related to the regulation of proliferation and differentiation (35), and this pathway, like SF-1 itself, may play important roles in the differentiation/migration of VMH neurons. Studies presented here show that systemic administration of a CB1R agonist is associated with increased levels of immunoreactive phospho-ERK in VMH neurons, suggesting that CB1R also couples to the MAPK pathway in the VMH (Fig. 7).
Although behaviors modulated by cannabinoids are regulated by complex interactions among structures such as the ventral tegmental area, medial forebrain bundle, nucleus accumbens, and amygdala (36), the hypothalamic nuclei surrounding the third ventricle are central to CB1R-mediated energy balance (3,37,38,39). For example, direct administration of the CB1 agonist anandamide (ANA) into the VMH significantly increased food intake, whereas the CB1R antagonist SR141716 inhibited the AEA induction of food intake (3) and infusion of ANA into the nucleus accumbens shell showed hyperphagic effects with marked activation of hypothalamic nuclei involved in feeding behaviors (39). These results implicate hypothalamic CB1R in orexigenic actions of cannabinoids. In agreement with these studies, we observed that administration of CB1R agonists (e.g. MA and ACEA) increased food intake in WT mice (Fig. 3, A and B); this increased food intake was blunted significantly in mice with CNS-specific KO of SF-1 (Fig. 3, A and B). Likewise, administration of the specific CB1R antagonist AM251 markedly suppressed food intake in WT mice, but suppression was significantly impaired in CNS-specific SF-1 KO mice (Fig. 3C). These data imply that the lack of CB1R expression in VMH neurons may be a factor in the behavioral effects exhibited by CNS-specific SF-1 KO mice (e.g. increased anxiety-like behavior and adiposity) (6,7).
A number of reported interactions between the cannabinoid system and neuropeptides involved in energy homeostasis provide insights into potential roles of the cannabinoid system on food intake and hypothalamic regulation. For example, SF-1 neurons in the VMH express leptin receptor and leptin signaling in SF-1 neurons is required for normal energy homeostasis (13,14); similarly, these SF-1 neurons express CB1R and SF-1 directly regulates the CB1R promoter (this study). Leptin negatively regulates endocannabinoid production (37) and modulates food intake by inhibiting the orexigenic neuropeptide Y (NPY) in arcuate nucleus (40,41). There are conflicting data regarding the cannabinoid system and neuropeptide NPY regulation. Stimulation of the cannabinoid system with ANA and CP55,940 reportedly augmented NPY release in the rat hypothalamus (42), suggesting that activation of the cannabinoid system in the hypothalamus generated orexigenic effects by augmenting NPY levels. In contrast, others have proposed that cannabinoids modulate energy balance by NPY-independent mechanisms (37). Consistent with the latter model, hypothalamic NPY expression was not affected by activation of the cannabinoid system by the CB1R agonist MA (data not shown) or by the perturbed VMH expression of CB1R in the VMH in CNS-specific SF-1 KO mice. Rather, the orexigenic effect of cannabinoids may involve other neuropeptides such as CRH (43), cocaine-amphetamine regulated peptide (44), proopiomelanocortin (45), and ghrelin (46).
In summary, our data identify CB1R as a novel target of SF-1 in the VMH, providing clear evidence that SF-1 plays important roles in the regulation of CB1R expression as well as in CB1R-mediated regulation of food intake. Furthermore, the excitability of SF-1 neurons is directly inhibited by CB1R agonists, which may be an important factor in homeostatic regulation by the VMH. Future studies that identify downstream modulators whose activity is controlled by cannabinoid signaling in SF-1 neurons will provide new insights into how SF-1 maintains CB1R-mediated energy balance in the VMH and will hopefully identify other genes that determine how the VMH and other hypothalamic nuclei regulate various physiological functions.
MATERIALS AND METHODS
Animal Care
All mouse care and experimental procedures were approved by the Institutional Animal Care Research Advisory Committee at UT Southwestern or at Albert Einstein Medical Center. Mice were housed at room temperature with a 12-h light, 12-h dark cycle (lights on at 0600 h) with regular mouse chow (Teklad mouse/rat diet 7001; 3.82 kcal/g, gross energy, 4.25% kcal from fat) and water provided ad libitum. The generation of CNS-specific SF-1 KO (SF-1nCre; F/−) mice was described previously (7).
Drugs
The CB1 agonists, MA and ACEA, and the CB1 antagonist AM251 were purchased from Sigma-Aldrich (St. Louis, MO). MA was dissolved in saline solution containing 2% polyoxyethylenesorbitan monoleate (Tween 80; Sigma) and 20% ethanol, ACEA was suspended in saline solution with 1% Tween 80, and AM251 was dissolved in saline containing 2% Tween 80 and 10% dimethylsulfoxide. All drugs were injected ip in a volume of 1 μl/g body weight.
Nocturnal Food Intake and Body Weight Studies
The WT (SF-1nCre; F/+), heterozygous (SF-1F/−), and CNS-specific SF-1 KO (SF-1nCre; F/−) mice were individually housed beginning at 4 wk of age. Body weight and food intake were monitored three or four times per week until 9 wk of age. Matched littermates with comparable food intake and weights (21–23 g for males and 18–19 g for females) were used for these studies. At d 65, food was removed from the cages at 1800 h and mice were fasted for 24 h; thereafter, they were administered the indicated drugs via ip injection and food intake was monitored over 3 h.
Slice Preparation for Electrophysiology
Transverse brain slices were prepared from SF-1/eGFP mice at postnatal age 21–28 d. Animals were anesthetized with a mixture of ketamine and xylazine. After decapitation, the brains were transferred into a sucrose-based solution bubbled with 95% O2-5% CO2 and maintained at approximately 3 C. This solution contained (mm): sucrose 248; KCl2; MgCl2 1; KH2PO4 1.25; NaHCO3 26; sodium pyruvate 1; and glucose 10. Transverse coronal brain slices (200 μm) were prepared using a vibratome. Slices were equilibrated with an oxygenated artificial cerebrospinal fluid (aCSF) for more than 1 h at 32 C before transfer to the recording chamber. The slices were continuously superfused with aCSF at a rate of 1.5 ml/min containing (in mm): NaCl 113; KCl3; NaH2PO4 1; NaHCO3 26; CaCl2 2.5; MgCl2 1; and glucose 5 in 95% O2/5% CO2 at room temperature.
Electrophysiological Studies
Brain slices were placed on the stage of an upright, infrared-differential interference contrast microscope (Olympus BX50WI, Olympus America Inc., Center Valley, PA) mounted on a Gibraltar X-Y table (Burleigh, Olympus America Inc.) and visualized with a ×40 water immersion objective by infrared microscopy (DAGE MTI camera; DAGE, Michigan City, IN). Membrane potentials were recorded at room temperature (25–26 C) with an Axopatch 200B Patch-Clamp amplifier. CNQX (10 μm), DL-amino-phosphonovaleric acid (DL-AP-5, 50 μm), picrotoxin (100 μm) and strychnine (1 μm) were continuously present in aCSF. The internal solution contained (mm): K acetate 115; KCl 10; MgCl2 2; EGTA 0.2; HEPES 10; Na2ATP 1; Na2GTP 0.5; and phosphocreatine 5. Pipette resistance ranged from 2.5 to 4 MΩ. WIN 55212-2 and AM251 were purchased from Sigma-Aldrich and dissolved in dimethylsulfoxide at 10,000× the final concentration. Effects of the CB1R agonist compared with control were analyzed using Student’s t tests (Origin 7.0). Differences were considered significant when the P value was < 0.05. All statistical results are given as means ± sem.
Immunohistochemistry and ISH
Immunohistochemistry and ISH were performed as described (7). Briefly, floating or mounted sections from adult brain (age 12–18 wk if not specified) were used for immunohistochemistry and ISH. Images were captured with a Nikon E1000 automated microscope connected with a Nikon digital camera (DXM 1200F; Nikon, Melville, NY).
Rabbit anti-SF-1 antibody [1:1500, (47)] was used for SF-1 immunohistochemistry. Samples were incubated with a biotin-conjugated antirabbit secondary antibody (1:1000) from Vector Laboratory (Burlingame, CA; BA-1000) for 2 h, followed by incubation for 1 h in a solution of avidin-biotin complex (Vectastain Elite ABC kit, Vector Laboratory). Finally, signals were visualized with DAB substrate kit (SK-4100; Vector Laboratory).
For immunofluorescent analysis of pERK activation in the VMH, mice at 9 wk were fasted for 24 h and then fed for 1 h after MA (1 mg/kg) administration. Mice were anesthetized with tribromoethanol (Avertin, 250 mg/kg·BW) and perfused via cardiac puncture with 4% paraformaldehyde (Sigma) in PBS (pH 7.4). After cryoprotection using 30% sucrose, brains were sectioned at 20 μm on a cryostat (Leica, Bannockburn, IL; CM1900). Rabbit anti-phosphorylated (p)-ERK antibody (1:100, no. 4376; Cell Signaling Technology, Danvers, MA) was incubated overnight and followed by Cy3-conjugated goat antirabbit IgG (Jackson ImmunoResearch, West Grove, PA) as secondary antibody.
ISH for CB1R was performed on every fourth serial section from at least five brains. The mouse CB1R probe was generously provided by Dr. Joel Elmquist (UT-Southwestern, Dallas, TX) and included CB1R sequences from 953 to 1557.
Cell Culture and Transfection Analyses
Cell culture conditions were previously described (7). HEK 293 human embryonic kidney cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin-streptomycin (Invitrogen, Carlsbad, CA). Neuro2A mouse neuroblastoma cells were cultured using MEM (Invitrogen) containing nonessential amino acids, 10% FBS, and 100 U/ml penicillin-streptomycin. Y1 mouse adrenocortical tumor cells were grown in Ham’s F10 medium (Mediatech, Herndon, VA) supplemented with 15% horse serum, 5% FBS, and 100 U/ml penicillin-streptomycin.
For immunoblotting analyses, Neuro2A cells were plated in six-well culture plates at a density of 5 × 105 cells per well and cultured in the growth media for 20–24 h before transfection. Cells were transfected with Fugene 6 (Roche Diagnostics, Indianapolis, IN) according to the manufacturer’s instructions. After 24 h, cells were treated with MA for differing time periods. Cells were then lysed in 100 μl of lysis buffer containing protease and phosphatase cocktails (Roche Diagnostics) and followed by gentle shaking on ice for 10 min. Supernatants were collected and resolved on 10–12% sodium dodecyl sulfate-polyacrylamide gels for 2 h and then transferred to nitrocellulose membrane (Bio-Rad, Hercules, CA). Commercially available antibodies from Cell Signaling Technology were used; rabbit anti-pERK (1:1000; no. 4376), rabbit anti-ERK (1:2000; no. 9102), rabbit anti-pJNK (1:2500; no. 9251), and rabbit anti-pStat3 (1:1000; no. 9131) for primary reaction. Signals were detected by horseradish peroxidase-conjuated secondary antisera. The chemiluminescence was visualized with Lumi-Light Western Blotting Substrate (Roche Diagnostics), and the images were detected by Kodak Biomax MR film. The bands were quantified with the ImageJ program (National Institutes of Health; http://rsb.info.nih.gov/ij/).
EMSAs
Duplex oligonucleotides containing the putative SF-1 binding sites at −233 and −101 were labeled with 32P by fill-in with Klenow fragment (New England Biolabs, Newton, MA) and used as probes for EMSAs as described (7). Oligonucleotide probes were 5′-GGGCGCCGAGCACCAGGGCCGTCCCTCCTAG-3′ for the site −233 (WT, top strand), 5′-GCTAGGAGGGACGGCCCTGGCGCTCGGCGCCCACT-3′ for the site −233 (WT, bottom strand), 5′-GGGCGCCGAGCGCCAAAACCGTCCCTCCTAG-3′ for site −233 (M, top strand), 5′-GCTAGGAGGGACGGTTTTGGCGCTCGGCGCCCACT-3′ for the site −233 (M, bottom strand), 5′-GCGCTGCCAGGGCAGGACAAAGGCTCATT-3′ for site −101 (WT, top strand), 5′-GAATGAGCCTTTGTCCTGCCCTGGCAGCG-3′ for site −101 (WT, bottom strand), 5′-GCGCTGCCAAAACAAAACAAAAACTCATT-3′ for site −101 (M, top strand), 5′-GAATGAGTTTTTGTTTTGTTTTGGCAGCG-3′ for site −101 (M, bottom strand). Boldface represents SF-1 binding sites and mutated sequences. Unlabeled duplex oligonucleotides were used for competition experiment at 2-, 5-, 10-, and 100-fold molar excesses. Where indicated, a rabbit antimouse SF-1 antiserum (1 μl) was included in the binding reaction. Nuclear extract was prepared from mouse Y1 adrenocortical tumor cells, which endogenously express SF-1, as described (7).
Real-Time Q-PCR Analysis of RNA from Purified Hypothalamic Cells Expressing a SF-1/eGFP Transgene
Embryonic d 16.5 embryos were harvested and cells prepared as described (12). Total RNA was prepared from 10,000–15,000 fluorescence-activated cell sorting-enriched cells using TRIzol (Invitrogen), deoxyribonuclease I treatment (QIAGEN, Valencia, CA), and RNeasy columns (QIAGEN). 2.5–25 ng of RNA was then amplified using the WT-Pico Kit (NuGEN, San Carlos, CA). In a final volume of 10 μl, the real-time PCR assays contained 1 ng of amplified RNA, 150 nm of each primer, and 5 μl of SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). PCRs were performed on an Applied Biosystems 7900HT and relative mRNA levels were calculated using the comparative threshold cycle method (Applied Biosystems user bulletin no. 2). Comparisons were made to samples from cells harvested from WT embryos using cyclophilin as the reference gene. Real-time primers were designed using Primer Express software (PerkinElmer Life Sciences, Downers Grove, IL) and validated using a standard curve comparison to cyclophilin (see supplemental figure published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). Primers used were forward (F) and reverse (R) CB1R (F) 5′-GTGCCGAGGGAGCTTCTG-3′ and (R) 5′-TCTTTGATTAGGCCAGGCTCAA-3′, Cyclophilin (F) 5′-TGGAGAGCACCAAGACAGACA-3′ and (R) 5′-TGCCGGAGTCGACAATGAT-3′. Twelve different pools (three WT male, three KO male, three WT female, and three KO female) were analyzed. Because no sexual dimorphism was noted, the data were combined as WT (n = 6) or KO (n = 6).
Supplementary Material
Acknowledgments
We thank Drs. Stuart Tobet (Colorado State University, Fort Collins, CO) and Joel Elmquist (UT-Southwestern, Dallas, TX) for careful reading of manuscript and comments, and Drs. Yun-Hee Choi, Chanhee Kang, Yangsik Jeong, and Youngjae You (UT-Southwestern, Dallas, TX) for discussions.
Footnotes
This work was supported by National Institutes of Health [Grants DK54480 and 1RL1DK081185-01 (to K.L.P.) and PO1DK26687 (to S.C.C.)], Skirball Foundation to S.C.C., and a junior faculty award from the ADA (to Y.H.J.).
Disclosure Statement: The authors have nothing to disclose.
First Published Online May 29, 2008
Abbreviations: ACEA, Arachidonyl-2′-chloroethylamide hydrate; aCSF, artificial cerebrospinal fluid; ANA, anandamide; BDNF, brain-derived neurotrophic factor; CB1R, cannabinoid receptor 1; CNS, central nervous system; DMH, dorsomedial hypothalamic nucleus; eGFP, enhanced green fluorescent protein; FBS, fetal bovine serum; ISH, in situ hybridization; JNK, c-Jun N-terminal kinase; KO, knockout; MA, methanandamide; NPY, neuropeptide Y; p, phosphorylated; SF-1, steroidogenic factor 1; STAT, signal transducer and activator of transcription; VMH, ventromedial hypothalamic nucleus; WT, wild type.
References
- Parker KL, Rice DA, Lala DS, Ikeda Y, Luo X, Wong M, Bakke M, Zhao L, Frigeri C, Hanley NA, Stallings N, Schimmer BP 2002 Steroidogenic factor 1: an essential mediator of endocrine development. Recent Prog Horm Res 57:19–36 [DOI] [PubMed] [Google Scholar]
- Val P, Lefrancois-Martinez AM, Veyssiere G, Martinez 2003 A SF-1 a key player in the development and differentiation of steroidogenic tissues. Nucl Recept 1:8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshidi N, Taylor DA 2001 Anandamide administration into the ventromedial hypothalamus stimulates appetite in rats. Br J Pharmacol 134:1151–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternson SM, Shepherd GM, Friedman JM 2005 Topographic mapping of VMH –> arcuate nucleus microcircuits and their reorganization by fasting. Nat Neurosci 8:1356–1363 [DOI] [PubMed] [Google Scholar]
- Hetherington AWR, SW 1940 Hypothalamic lesions and adiposity in the rat. Anat Rec 78:149–161 [Google Scholar]
- Majdic G, Young M, Gomez-Sanchez E, Anderson P, Szczepaniak LS, Dobbins RL, McGarry JD, Parker KL 2002 Knockout mice lacking steroidogenic factor 1 are a novel genetic model of hypothalamic obesity. Endocrinology 143:607–614 [DOI] [PubMed] [Google Scholar]
- Zhao L, Kim KW, Ikeda Y, Anderson KK, Beck L, Chase S, Tobet SA, Parker KL 2008 CNS-specific knockout of steroidogenic factor 1 results in increased anxiety-like behavior. Mol Endocrinol 22:1403–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canteras NS, Simerly RB, Swanson LW 1994 Organization of projections from the ventromedial nucleus of the hypothalamus: a Phaseolus vulgaris-leucoagglutinin study in the rat. J Comp Neurol 348:41–79 [DOI] [PubMed] [Google Scholar]
- Robarts DW, Baum MJ 2007 Ventromedial hypothalamic nucleus lesions disrupt olfactory mate recognition and receptivity in female ferrets. Horm Behav 51:104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston E, Triandafillou J, Haas N 1989 Colchicine lesions of ventromedial hypothalamus: effects on regulatory thermogenesis in the rat. Pharmacol Biochem Behav 32:301–307 [DOI] [PubMed] [Google Scholar]
- Kurrasch DM, Cheung CC, Lee FY, Tran PV, Hata K, Ingraham HA 2007 The neonatal ventromedial hypothalamus transcriptome reveals novel markers with spatially distinct patterning. J Neurosci 27:13624–13634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal JP, Stallings NR, Lee CE, Zhao L, Socci N, Viale A, Harris TM, Soares MB, Childs G, Elmquist JK, Parker KL, Friedman JM 2005 Use of laser-capture microdissection for the identification of marker genes for the ventromedial hypothalamic nucleus. J Neurosci 25:4181–4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham NC, Anderson KK, Reuter AL, Stallings NR, Parker KL 2008 Selective loss of leptin receptors in the ventromedial hypothalamic nucleus results in increased adiposity and a metabolic syndrome. Endocrinology 149:2138–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon H, Zigman JM, Ye C, Lee CE, McGovern RA, Tang V, Kenny CD, Christiansen LM, White RD, Edelstein EA, Coppari R, Balthasar N, Cowley MA, Chua Jr S, Elmquist JK, Lowell BB 2006 Leptin directly activates SF1 neurons in the VMH, and this action by leptin is required for normal body-weight homeostasis. Neuron 49:191–203 [DOI] [PubMed] [Google Scholar]
- Tong Q, Ye C, McCrimmon RJ, Dhillon H, Choi B, Kramer MD, Yu J, Yang Z, Christiansen LM, Lee CE, Choi CS, Zigman JM, Shulman GI, Sherwin RS, Elmquist JK, Lowell BB 2007 Synaptic glutamate release by ventromedial hypothalamic neurons is part of the neurocircuitry that prevents hypoglycemia. Cell Metab 5:383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez S, Manzanares J, Berrendero F, Wenger T, Corchero J, Bisogno T, Romero J, Fuentes JA, Di Marzo V, Ramos JA, Fernandez-Ruiz J 1999 Identification of endocannabinoids and cannabinoid CB(1) receptor mRNA in the pituitary gland. Neuroendocrinology 70:137–145 [DOI] [PubMed] [Google Scholar]
- Mailleux P, Vanderhaeghen JJ 1992 Distribution of neuronal cannabinoid receptor in the adult rat brain: a comparative receptor binding radioautography and in situ hybridization histochemistry. Neuroscience 48:655–668 [DOI] [PubMed] [Google Scholar]
- Stallings NR, Hanley NA, Majdic G, Zhao L, Bakke M, Parker KL 2002 Development of a transgenic green fluorescent protein lineage marker for steroidogenic factor 1. Mol Endocrinol 16:2360–2370 [DOI] [PubMed] [Google Scholar]
- Berrendero F, Garcia-Gil L, Hernandez ML, Romero J, Cebeira M, de Miguel R, Ramos JA, Fernandez-Ruiz JJ 1998 Localization of mRNA expression and activation of signal transduction mechanisms for cannabinoid receptor in rat brain during fetal development. Development 125:3179–3188 [DOI] [PubMed] [Google Scholar]
- Chambers AP, Sharkey KA, Koopmans HS 2004 Cannabinoid (CB)1 receptor antagonist, AM 251, causes a sustained reduction of daily food intake in the rat. Physiol Behav 82:863–869 [DOI] [PubMed] [Google Scholar]
- Busygina TV, Ignatieva EV, Osadchuk AV 2003 Consensus sequence of transcription factor SF-1 binding site and putative binding site in the 5′ flanking regions of genes encoding mouse steroidogenic enzymes 3βHSDI and Cyp17. Biochemistry (Mosc) 68:377–384 [DOI] [PubMed] [Google Scholar]
- He JC, Gomes I, Nguyen T, Jayaram G, Ram PT, Devi LA, Iyengar R 2005 The G α(o/i)-coupled cannabinoid receptor-mediated neurite outgrowth involves Rap regulation of Src and Stat3. J Biol Chem 280:33426–33434 [DOI] [PubMed] [Google Scholar]
- Jordan JD, He JC, Eungdamrong NJ, Gomes I, Ali W, Nguyen T, Bivona TG, Philips MR, Devi LA, Iyengar R 2005 Cannabinoid receptor-induced neurite outgrowth is mediated by Rap1 activation through G(α)o/i-triggered proteasomal degradation of Rap1GAPII. J Biol Chem 280:11413–11421 [DOI] [PubMed] [Google Scholar]
- He JC, Neves SR, Jordan JD, Iyengar R 2006 Role of the Go/i signaling network in the regulation of neurite outgrowth. Can J Physiol Pharmacol 84:687–694 [DOI] [PubMed] [Google Scholar]
- Jo YH, Chen YJ, Chua Jr SC, Talmage DA, Role LW 2005 Integration of endocannabinoid and leptin signaling in an appetite-related neural circuit. Neuron 48:1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dielenberg RA, Hunt GE, McGregor IS 2001 “When a rat smells a cat”: the distribution of Fos immunoreactivity in rat brain following exposure to a predatory odor. Neuroscience 104:1085–1097 [DOI] [PubMed] [Google Scholar]
- Canteras NS 2002 The medial hypothalamic defensive system: hodological organization and functional implications. Pharmacol Biochem Behav 71:481–491 [DOI] [PubMed] [Google Scholar]
- Navarro M, Hernandez E, Munoz RM, del Arco I, Villanua MA, Carrera MR, Rodriguez de Fonseca F 1997 Acute administration of the CB1 cannabinoid receptor antagonist SR 141716A induces anxiety-like responses in the rat. Neuroreport 8:491–496 [DOI] [PubMed] [Google Scholar]
- Haller J, Bakos N, Szirmay M, Ledent C, Freund TF 2002 The effects of genetic and pharmacological blockade of the CB1 cannabinoid receptor on anxiety. Eur J Neurosci 16:1395–1398 [DOI] [PubMed] [Google Scholar]
- Uriguen L, Perez-Rial S, Ledent C, Palomo T, Manzanares J 2004 Impaired action of anxiolytic drugs in mice deficient in cannabinoid CB1 receptors. Neuropharmacology 46:966–973 [DOI] [PubMed] [Google Scholar]
- Pieri I, Klein M, Bayertz C, Gerspach J, van der Ploeg A, Pfizenmaier K, Eisel U 1999 Regulation of the murine NMDA-receptor-subunit NR2C promoter by Sp1 and fushi tarazu factor1 (FTZ-F1) homologues. Eur J Neurosci 11:2083–2092 [DOI] [PubMed] [Google Scholar]
- Tran PV, Akana SF, Malkovska I, Dallman MF, Parada LF, Ingraham HA 2006 Diminished hypothalamic BDNF expression and impaired VMH function are associated with reduced SF-1 gene dosage. J Comp Neurol 498:637–648 [DOI] [PubMed] [Google Scholar]
- Mackie K 2006 Cannabinoid receptors as therapeutic targets. Annu Rev Pharmacol Toxicol 46:101–122 [DOI] [PubMed] [Google Scholar]
- Eljaschewitsch E, Witting A, Mawrin C, Lee T, Schmidt PM, Wolf S, Hoertnagl H, Raine CS, Schneider-Stock R, Nitsch R, Ullrich O 2006 The endocannabinoid anandamide protects neurons during CNS inflammation by induction of MKP-1 in microglial cells. Neuron 49:67–79 [DOI] [PubMed] [Google Scholar]
- Johnson GL, Lapadat R 2002 Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298:1911–1912 [DOI] [PubMed] [Google Scholar]
- Gardner EL 2005 Endocannabinoid signaling system and brain reward: emphasis on dopamine. Pharmacol Biochem Behav 81:263–284 [DOI] [PubMed] [Google Scholar]
- Di Marzo V, Goparaju SK, Wang L, Liu J, Batkai S, Jarai Z, Fezza F, Miura GI, Palmiter RD, Sugiura T, Kunos G 2001 Leptin-regulated endocannabinoids are involved in maintaining food intake. Nature 410:822–825 [DOI] [PubMed] [Google Scholar]
- Kirkham TC, Williams CM, Fezza F, Di Marzo V 2002 Endocannabinoid levels in rat limbic forebrain and hypothalamus in relation to fasting, feeding and satiation: stimulation of eating by 2-arachidonoyl glycerol. Br J Pharmacol 136:550–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria-Gomez E, Matias I, Rueda-Orozco PE, Cisneros M, Petrosino S, Navarro L, Di Marzo V, Prospero-Garcia O 2007 Pharmacological enhancement of the endocannabinoid system in the nucleus accumbens shell stimulates food intake and increases c-Fos expression in the hypothalamus. Br J Pharmacol 151:1109–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley MA, Smart JL, Rubinstein M, Cerdan MG, Diano S, Horvath TL, Cone RD, Low MJ 2001 Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature 411:480–484 [DOI] [PubMed] [Google Scholar]
- van den Top M, Lee K, Whyment AD, Blanks AM, Spanswick D 2004 Orexigen-sensitive NPY/AgRP pacemaker neurons in the hypothalamic arcuate nucleus. Nat Neurosci 7:493–494 [DOI] [PubMed] [Google Scholar]
- Gamber KM, Macarthur H, Westfall TC 2005 Cannabinoids augment the release of neuropeptide Y in the rat hypothalamus. Neuropharmacology 49:646–652 [DOI] [PubMed] [Google Scholar]
- Cota D, Marsicano G, Tschop M, Grubler Y, Flachskamm C, Schubert M, Auer D, Yassouridis A, Thone-Reineke C, Ortmann S, Tomassoni F, Cervino C, Nisoli E, Linthorst AC, Pasquali R, Lutz B, Stalla GK, Pagotto U 2003 The endogenous cannabinoid system affects energy balance via central orexigenic drive and peripheral lipogenesis. J Clin Invest 112:423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osei-Hyiaman D, Depetrillo M, Harvey-White J, Bannon AW, Cravatt BF, Kuhar MJ, Mackie K, Palkovits M, Kunos G 2005 Cocaine- and amphetamine-related transcript is involved in the orexigenic effect of endogenous anandamide. Neuroendocrinology 81:273–282 [DOI] [PubMed] [Google Scholar]
- Ho J, Cox JM, Wagner EJ 2007 Cannabinoid-induced hyperphagia: correlation with inhibition of proopiomelanocortin neurons? Physiol Behav 92:507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kola B, Farkas I, Christ-Crain M, Wittmann G, Lolli F, Amin F, Harvey-White J, Liposits Z, Kunos G, Grossman AB, Fekete C, Korbonits M 2008 The orexigenic effect of ghrelin is mediated through central activation of the endogenous cannabinoid system. PLoS ONE 3:e1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AC, Reuter AL, Zubair M, Serecky K, Else T, Bingham NC, Lavery GG, Parker KL, Hammer GD, Targeted disruption of β-catenin in Sf-1-expressing cells impairs development and maintenance of the adrenal cortex. Development, in press [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.