Abstract
Plasma high density lipoprotein (HDL), which protects against atherosclerosis, is thought to remove cholesterol from peripheral tissues and to deliver cholesteryl esters via a selective uptake pathway to the liver (reverse cholesterol transport) and steroidogenic tissues (e.g., adrenal gland for storage and hormone synthesis). Despite its physiologic and pathophysiologic importance, the cellular metabolism of HDL has not been well defined. The class B, type I scavenger receptor (SR-BI) has been proposed to play an important role in HDL metabolism because (i) it is a cell surface HDL receptor which mediates selective cholesterol uptake in cultured cells, (ii) its physiologically regulated expression is most abundant in the liver and steroidogenic tissues, and (iii) hepatic overexpression dramatically lowers plasma HDL. To test directly the normal role of SR-BI in HDL metabolism, we generated mice with a targeted null mutation in the SR-BI gene. In heterozygous and homozygous mutants relative to wild-type controls, plasma cholesterol concentrations were increased by ≈31% and 125%, respectively, because of the formation of large, apolipoprotein A-I (apoA-I)-containing particles, and adrenal gland cholesterol content decreased by 42% and 72%, respectively. The plasma concentration of apoA-I, the major protein in HDL, was unchanged in the mutants. This, in conjunction with the increased lipoprotein size, suggests that the increased plasma cholesterol in the mutants was due to decreased selective cholesterol uptake. These results provide strong support for the proposal that in mice the gene encoding SR-BI plays a key role in determining the levels of plasma lipoprotein cholesterol (primarily HDL) and the accumulation of cholesterol stores in the adrenal gland. If it has a similar role in controlling plasma HDL in humans, SR-BI may influence the development and progression of atherosclerosis and may be an attractive candidate for therapeutic intervention in this disease.
High density lipoproteins (HDLs) play a critical role in cholesterol metabolism (1, 2) and their plasma concentrations are inversely correlated with risk for atherosclerosis (3). HDL appears to have an important role in removing cholesterol from peripheral tissues (4–7) and in directly delivering cholesteryl esters to other lipoproteins (8) and to tissues (9, 10). An important mechanism for the direct delivery of HDL cholesterol to cells is called selective lipid uptake (11, 12). This pathway is fundamentally different from the low density lipoprotein (LDL) receptor pathway (13) in that cell surface binding of HDL is not associated with endocytosis and lysosomal degradation of the lipoprotein particle. Instead, after cholesteryl esters are transferred from HDL to cells, lipid-depleted HDL is released into the extracellular fluid. Selective lipid uptake appears to be especially important for transport of cholesterol to steroidogenic tissues for steroid hormone synthesis (9) and to the liver where HDL-derived cholesterol is either secreted into the bile, used for bile acid synthesis, or packaged and secreted in newly synthesized lipoproteins (10, 14). The HDL-mediated transport of cholesterol from extrahepatic tissues to the liver, called reverse cholesterol transport (15), is believed to play a critical role in cholesterol homeostasis and may account for some of the protective effects of HDL against atherosclerosis (2, 4–8, 16).
Recently, we discovered that the class B, type I scavenger receptor, SR-BI (17), is a cell surface HDL receptor (ref. 18, reviewed in ref. 19) that can recognize the apolipoproteins on the surface of the HDL particle (20), including the major component apolipoprotein A-I (apoA-I). In cultured cells, murine SR-BI (mSR-BI), which is an ≈82-kDa glycoprotein that appears to be clustered in caveolae (21), can mediate selective HDL lipid uptake (18). In vivo, SR-BI is most abundantly expressed in liver and steroidogenic tissues (18, 22, 23), which are those tissues most actively engaged in selective HDL lipid uptake (refs. 11, 12, and 24, reviewed in ref. 19). SR-BI expression in rodent steroidogenic tissues is coordinately regulated with steroid synthesis by trophic hormones (22, 25, 26). Hormonal regulation of SR-BI protein and mRNA also has been observed in cultured steroidogenic cells (25, 26), and SR-BI mRNA levels have been reported to be altered by mutations in apoA-I and lecithin:cholesterol acyltransferase (LCAT) (26, 27). Hepatic overexpression of mSR-BI in mice dramatically reduces plasma HDL and increases biliary cholesterol levels (28). Taken together, these studies provide strong, although indirect, evidence that SR-BI may normally be a physiologically relevant HDL receptor.
To determine directly if SR-BI normally plays an important role in HDL metabolism in vivo and to establish an experimental system to examine the role of SR-BI in pathologic states, we have generated mice containing a targeted null mutation in the gene encoding SR-BI. Analysis of the mutants provides strong proof that SR-BI is a physiologically relevant HDL receptor that plays an important role in the accumulation of cholesterol in the adrenal gland and in regulating plasma HDL cholesterol levels.
MATERIALS AND METHODS
Generation of SR-BI Mutant Mice.
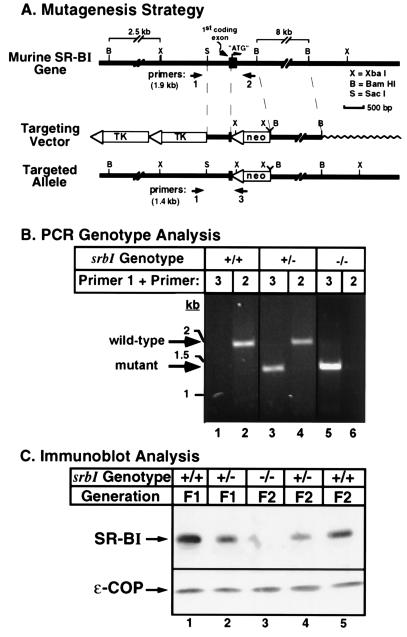
SR-BI genomic DNA was isolated from a mouse strain 129 DNA library (Genome Systems, St. Louis) screened by PCR amplification using primer pairs corresponding to the 5′ and 3′ ends of the mSR-BI cDNA (18). From one clone we identified a 12-kb XbaI fragment containing the first coding exon. A replacement-type targeting vector (see Fig. 1A), containing 0.75 kb and 9 kb short and long homology regions and the pol2sneobpA (29) and herpes simplex virus thymidine kinase (30) cassettes, was constructed using standard methods (31, 32). The vector was linearized and 100 μg were transfected by electroporation (240 V, 500 μF) into 112 × 106 murine D3 embryonic stem cells (31, 33), which were then plated onto irradiated mouse embryonic fibroblast feeder layers. After G418/gancyclovir positive/negative selection for 7–8 days as described (31), 492 of the ≈5,800 surviving colonies were picked and screened by PCR analysis using primers specific for the targeted allele (Fig. 1B, primer 1, 5′-TGAAGGTGGTCTTCAAGAGCAGTCCT-3′; and primer 3, 5′-GATTGGGAAGACAATAGCAGGCATGC-3′; all oligonucleotide primers were synthesized by Research Genetics, Huntsville, AL) as described (32). The presence of the targeted allele (amplification of a 1.4-kb band) was confirmed by Southern blot analysis of XbaI-digested genomic DNA using probes that yielded either the predicted 12-kb fragment characteristic of the wild-type allele or the predicted 2.5-kb and 9-kb fragments from the targeted mutant allele (data not shown). BamHI digested genomic DNA was also probed with a 0.9-kb fragment derived by PstI digestion of the neomycin resistance gene cassette to confirm the presence of a single neo gene in the mutant cells (data not shown). Embryonic stem cell clones containing a disrupted SR-BI allele were injected into C57BL/6 blastocysts, which were implanted into recipient females (31). The resulting chimeric mice were crossed to C57BL/6 female mice to generate F1 wild-type (srbI+/+) and heterozygous (srbI+/−) mice on an identical 129 (agouti)/C57BL/6 background. F1 heterozygotes were crossed to generate F2 wild-type (srbI+/+), heterozygous mutant (srbI+/−), and homozygous mutant (srbI−/−) progeny. The presence of the targeted or wild-type SR-BI alleles in DNA extracted from tail biopsies (34) was detected by PCR amplification using primer 1 in combination with either primer 3 (mutant-specific) or primer 2 (wild-type-specific, 5′-TATCCTCGGCAGACCTGAGTCGTGT-3′). Genotypes were confirmed by Southern blot analysis. Mice were housed in microisolator cages and were fed ad libitum a regular rodent chow diet (Prolab 3000, PMI Feeds, St. Louis).
Figure 1.
Strategy for targeted disruption of the SR-BI locus in the mouse (A) and confirmation of the expected null mutation by PCR genotype analysis (B) and immunoblot analysis of liver membranes (C). (A) Restriction map of the genomic DNA surrounding the first coding exon of the murine gene encoding SR-BI. The targeting vector and the predicted structure of the targeted (mutant) allele are shown and described in the text. The locations of the sequences for the PCR primers used to specifically detect either the wild-type (primers 1 and 2) or targeted mutant (primers 1 and 3) alleles are indicated along with the predicted PCR product lengths. TK, herpes simplex thymidine kinase; neo, pol2sneobpA expression cassette, X, XbaI; B, BamHI; S, SacI; “ATG”, codon for the initiator methionine. (B) PCR genotype analysis. Two sets of primer pairs specific for the wild-type (primers 1 and 2) or targeted mutant (primers 1 and 3) alleles (see A) were used to screen genomic DNA by PCR as described. Representative results from individual F1 wild-type (+/+, lanes 1 and 2), F1 heterozygous (+/−, lanes 3 and 4), and F2 homozygous mutant (−/−, lanes 5 and 6) animals are shown. (C) Immunoblot analysis of hepatic membranes (50 μg protein/lane) from unfasted wild-type (F1 and F2 generations, lanes 1 and 5), heterozygous (F1 and F2 generations, lanes 2 and 4), and homozygous mutant (F2 generation, lane 3) male mice were performed using polyclonal antipeptide antibodies to SR-BI (≈82 kDa, Upper) or the internal control ɛ-COP (≈36 kDa, Lower), as described. Essentially identical results were obtained using specimens from female mice (data not shown).
Analysis of Animal Tissues.
Samples were obtained from fasted (4–8 hr) or nonfasted mice that were approximately 8–12 weeks old (F1 generation) or 5–11 weeks old (F2 generation).
Immunoblot analysis.
Animals were sacrificed and livers and adrenal glands were removed and immediately frozen. Membranes from homogenates were prepared as described (22, 25, 35). A total of 50 μg of protein per specimen were analyzed by SDS/polyacrylamide (8%) gel electrophoresis (36) and immunoblotting (37) with chemiluminescence detection as described using rabbit antipeptide polyclonal antibodies that specifically recognize either the ≈82-kDa mSR-BI protein (anti-mSR-BI495; ref. 18) or the ≈36-kDa ɛ-COP control cytoplasmic protein (anti-ɛCOP; ref. 38). The data shown in Fig. 1C are from a single gel so that the staining for ɛ-COP serves as a control for equal sample loading.
Plasma and adrenal cholesterol analysis.
Plasma total cholesterol (unesterified plus esterified, mg/dl) was measured using an enzymatic kit (Sigma). Adrenal glands were homogenized as described above. Protein concentrations in the homogenates were measured using the method of Lowry et al. (39). Duplicate samples of homogenates (30–70 μl each) were extracted with 2 ml of hexane/isopropanol (2:1) for 1 hr at room temperature, back-washed with 1 ml of water, and phases separated by centrifugation at 800 × g for 5 min. The upper organic phase was recovered and evaporated at 37°C in a Speedvac concentrator, and cholesterol was measured in the dried pellet using an enzymatic kit (Sigma). Cholesterol values were corrected based on the recovery of a [3H]cholesteryl ester internal standard added prior to lipid extraction. Total cholesterol content was expressed as μg of cholesterol/mg total protein.
Lipoprotein Analysis.
Pooled plasma (150 μl total from two to six animals) was diluted with an equal volume of elution buffer (154 mM NaCl/1 mM EDTA, pH 8) and subjected to fast protein liquid chromatography (FPLC) using two Superose 6 columns (Pharmacia) connected in series (40, 41). Proteins were eluted at 0.25 ml/min. Forty-seven fractions (0.5 ml) were collected after the first 14 ml were eluted and total cholesterol in each fraction was determined as described above. Immunoblotting of the FPLC fractions was performed (as described above) with specific anti-apoA-I (42), anti-apoA-II (43), or anti-apoE (42) antibodies on independent samples or by sequential labeling of a single membrane to permit simultaneous visualization of all three proteins.
Statistical Analysis.
Results are expressed as the arithmetic mean ± SD. The statistical significance of the differences of the mean between groups was evaluated using the Student’s t test for unpaired comparisons. The χ2 test was used for genotype distribution analysis. P values < 0.05 are considered to be statistically significant.
RESULTS AND DISCUSSION
The SR-BI gene was inactivated in embryonic stem cells by standard homologous recombination methods as outlined in Fig. 1 (also see Materials and Methods). The segments replaced in the recombined mutant (targeted allele) include the entire coding region of the first coding exon [28 bp of 5′ untranslated sequence and 126 bp, 42 amino acids, encoding a short N-terminal cytoplasmic domain, and a portion of the N-terminal putative transmembrane domain that probably also functions as an uncleaved leader sequence for insertion into the endoplasmic reticulum during biogenesis (21)] and an additional 554 bases of the adjacent downstream intron (Fig. 1A). The mutated locus is expected to encode a transcript that would not be translated or would be translated into nonfunctional, nonmembranous, and presumably unstable, protein.
Three independently derived embryonic stem cell clones containing the targeted allele were injected into C57BL/6 blastocysts and two produced 24 male chimeras, of which 11 gave germ-line transmission of the targeted SR-BI allele when crossed to C57BL/6 females. F1 offspring were either homozygous (+/+) for the wild type allele (Fig. 1B, lane 2, 1.9-kb amplified product from primers 1 and 2, no mutant allele specific product in lane 1) or heterozygous (+/−) with both mutant (lane 3, 1.4-kb product from primers 1 and 3) and wild-type (lane 4) PCR products. F1 heterozygotes should be isogenic with the F1 wild-type controls except at the SR-BI locus (44). Wild-type, heterozygous, and homozygous mutant F2 generation offspring (Fig. 1B, lanes 5 and 6 and data not shown), whose phenotypes are subject to genetic background variability (44), were generated from F1 intercrosses. In the F2 progeny analyzed to date (n = 317), the observed ratios of wild-type:heterozygous mutant:homozygous mutant offspring were 1.0:1.7:0.5, values significantly different from the expected Mendelian ratio of 1:2:1 (P = 0.003). Thus, there may be partially penetrant effects of the mutation either on neonatal survival or on embryonic development, which would be consistent with the distribution of SR-BI on the maternal surfaces of cells in the placenta and yolk sac during embryonic development (A. Hatzopoulos, A.R., R. Rosenberg, and M.K., unpublished results).
All of the mutants looked normal (weight, general appearance and behavior) and the males were fertile (female fertility is currently being evaluated). Immunoblot analysis of liver membranes from F1 (+/+, +/−) and F2 (+/+, +/−, −/−) mice using antipeptide antibodies which recognize the C terminus of the SR-BI protein (anti-mSR-BI495, Fig. 1C), or a segment of the putative extracellular loop (anti-mSR-BI230) (ref. 18, and data not shown), revealed that there was about half as much mSR-BI protein in the heterozygous mutants (lanes 2 and 4) as in the wild-type controls (lanes 1 and 5) and no detectable SR-BI in the homozygous mutants (lane 3). No fragments or other variants (45) of the full-length protein were detected in any of the samples. In contrast, no significant differences were observed in the levels of the control protein, ɛ-COP (Fig. 1C) (38). Similar results were observed using adrenal tissue (data not shown). Thus, the mutated SR-BI gene is a functionally null allele.
To determine how decreased SR-BI protein expression influenced lipoprotein metabolism, we compared the plasma cholesterol levels in male and female wild-type and mutant mice (Table 1). Because there were no statistically significant differences between the data from animals derived from the two independent embryonic stem cell clones (data not shown), data from these two independent sets of animals were pooled. Relative to wild-type controls there were statistically significant increases in the plasma total cholesterol concentrations of ≈30–40% in F1 and F2 heterozygotes and 2.2-fold in F2 homozygous mutants. In contrast to the increased plasma cholesterol in the mutants, there was no statistically significant change in the levels of plasma apoA-I (Table 1). These findings are consistent with the suggestion (18, 19, 28) that hepatic SR-BI plays a key role in selective removal of cholesterol from circulating HDL: lower levels of hepatic SR-BI were expected to increase plasma HDL cholesterol but not directly alter apoA-I levels.
Table 1.
Effects of disruption of the gene encoding SR-BI on plasma total cholesterol and apoA-I concentrations, and adrenal gland total cholesterol content in wild-type (srbI+/+), and heterozygous (srbI+/−), and homozygous (srbI−/−) mutant mice.
| srbI genotype | Gender | F1 generation*
|
F2 generation*
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Plasma total cholesterol
|
Plasma total cholesterol
|
Plasma apoA-I†
|
Adrenal gland total cholesterol
|
||||||
| mg/dl‡§ | % of control | mg/dl‡§ | % of control | mg/dl‡§ | % of control | μg/mg protein‡§ | % of control | ||
| Male | 93 ± 8 (29) | 100 | 99 ± 12 (18) | 100 | — | — | — | — | |
| +/+ | Female | 80 ± 7 (13) | 100 | 94 ± 20 (27) | 100 | — | — | — | — |
| Both | 89 ± 10 (42) | 100 | 96 ± 17 (45) | 100 | 25 ± 3 (10) | 100 | 128 ± 28 (5) | 100 | |
| Male | 126 ± 10 (21)¶ | 135¶ | 137 ± 21 (29)¶ | 138¶ | — | — | — | — | |
| +/− | Female | 112 ± 9 (23)¶ | 140¶ | 118 ± 19 (49)¶ | 126¶ | — | — | — | — |
| Both | 119 ± 12 (44)¶ | 134¶ | 126 ± 22 (78)¶ | 131¶ | 28 ± 2 (12)‖ | 112‖ | 74 ± 18 (6)¶ | 58¶ | |
| Male | — | — | 220 ± 41 (10)¶ | 222¶ | — | — | — | — | |
| −/− | Female | — | — | 209 ± 32 (7)¶ | 222¶ | — | — | — | — |
| Both | — | — | 216 ± 37 (17)¶ | 225¶ | 27 ± 3 (11)‖ | 108‖ | 36 ± 7 (5)¶ | 28¶ | |
F1 generation animals were not fasted. F2 generation animals were not fasted prior to analysis of adrenal gland cholesterol levels but were fasted for 4–8 h prior to analysis of plasma.
Values determined with an Autoanalyzer and human apoA-I standards.
Values represent mean ± SD.
Values in parenthesis represent the numbers of animals analyzed.
Statistically significantly different from wild-type controls, P < 0.005 or better.
Not statistically significantly different from wild-type controls, independently confirmed by quantitative evaluation of immunoblot analysis.
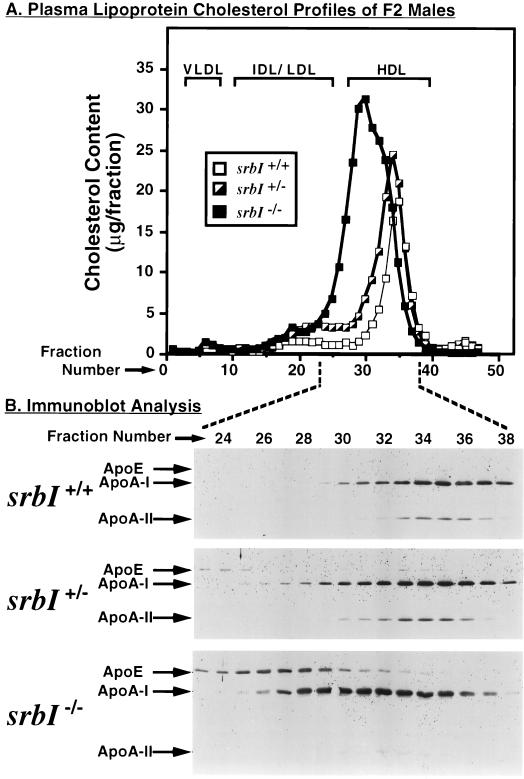
To determine if the elevated levels of plasma cholesterol in the mutants were due to changes in HDL, pooled plasma samples from F1 male and female (data not shown) and F2 male (Fig. 2) animals were subjected to FPLC and the total cholesterol content (Fig. 2A) as well as the relative amounts of apoA-I, apoA-II, and apoE (Fig. 2B) in each fraction were measured. As previously reported (40, 41, 43), for wild-type mice (srbI+/+) most of the cholesterol (Fig. 2A, open squares), apoA-I and apoA-II (Fig. 2B Top) were in the HDL fraction, with small or undetectable amounts in the very low density lipoprotein (VLDL) and intermediate density lipoprotein (IDL)/LDL fractions. There was an apparently low level of apoE which both comigrated with the HDL (centered around fraction 35) and with a small cholesterol peak in the IDL/LDL region (around fraction 20, very faint bands in Fig. 2B Top and data not shown). The cholesterol and apolipoprotein profiles of the heterozygous mutants (Fig. 2, half-solid squares in A; Fig. 2B Middle) were similar to those of the wild-type controls, except that there was an increase in the amount of cholesterol in the HDL fractions and there was a tendency of the HDL peak (cholesterol and/or apolipoproteins) to be broader than that of wild-type and shifted slightly to the left (lower fraction number, greater apparent size), which may represent large HDL particles (46, 47). This result suggested that there might be a difference in the average sizes of the HDL particles because of the inactivation of one of the SR-BI alleles; however, this shift was not observed in all specimens.
Figure 2.
FPLC profiles of plasma lipoprotein cholesterol (A) and apolipoproteins (B) for wild-type (srbI+/+) and heterozygous (srbI+/−) and homozygous (srbI−/−) mutant F2 male mice. (A) The chromatograms represent single analyses of pooled samples (150 μl of plasma from three animals per sample) from 4 to 8 h fasted wild-type (srbI+/+, open squares), and heterozygous (srbI+/−, partly filled squares) and homozygous (srbI−/−, filled squares) mutant mice and are representative of multiple, independent determinations. Approximate positions of VLDL, IDL/LDL, and HDL elution are indicated by brackets and were determined both by analysis of human lipoprotein standards and by previous analysis of lipoproteins in murine plasma (28). (B) Combined immunoblot analysis of fractions 23–38 from the chromatograms shown in A were performed with polyclonal antibodies to apoE, apoA-I, and apoA-II as described. Additional analysis of these and independent chromatograms (data not shown) established that there were no additional peaks containing apoA-I in fractions containing larger lipoproteins (fractions 1–22) and that the only other peak containing a small amount of apoE was in fraction 6, which corresponds to VLDL.
In the F2 homozygous mutant animals (srbI−/−, Fig. 2, filled squares in A; Fig. 2B Bottom) the cholesterol was found in a large, somewhat heterogeneous peak in the HDL range, but shifted to the left (larger apparent size) of the wild-type HDL peak. The amount of cholesterol in the IDL/LDL fraction (around fraction 20) varied between samples (the size of this peak in Fig. 2A is about midway between the lowest and highest values observed). The distributions of apoA-I and apoA-II were similar to that of cholesterol, although unlike the case for apoA-I there was a notable reduction in the amount of apoA-II relative to that seen in wild-type and heterozygous mutant animals. Conversely, in the homozygous mutants there was a substantial increase in the amount of apoE, whose distribution profile (larger particles, centered around fractions 26–28) differed from, but overlapped, those of apoA-I and apoA-II. Detailed analyses of the production, composition and structure, and turnover of lipoproteins in these mutants will help further define the mechanism by which SR-BI affects HDL metabolism and will be the subject of future studies. It is noteworthy that a significant increase in apoE associated with HDL particles in homozygous mutants in the gene for the SR-BI ligand apoA-I (48, 49) has also been reported, although it is not clear if the mechanisms responsible for the apoE increase in SR-BI and apoA-I mutants are related.
These results with the mutant animals, in which the changes in SR-BI expression are in the physiologic range (22, 25), are complementary to and consistent with the recent observation that transient adenovirus-mediated hepatic SR-BI overexpression results in dramatically decreased levels of HDL cholesterol and increased delivery of HDL-associated lipid to hepatocytes and the bile (28). In rodents, most of the plasma HDL cholesterol appears to be removed by the liver via selective uptake (refs. 11, 12, and 24; reviewed in ref. 19) and the liver appears to be the site of the highest total amount of SR-BI protein expression (18, 22, 23). It seems likely that buildup of large, cholesterol-enriched lipoprotein particles in the circulation of SR-BI mutants was primarily due to decreased hepatic selective HDL cholesterol uptake. Thus, it appears that murine plasma HDL cholesterol levels are particularly sensitive to physiologically relevant changes in the levels of hepatic SR-BI protein expression (e.g., ≈50% reduction in heterozygotes, Fig. 1C). The effect of the null mutation in SR-BI on total plasma cholesterol levels was quantitatively similar to that of a null mutation in the LDL receptor (50). For both sets of mutants, total plasma cholesterol levels were ≈36% above wild-type controls for heterozygotes and ≈114% for homozygotes. It is important to emphasize that although the magnitudes of the effects on total plasma cholesterol of these distinct mutations (SR-BI vs. LDL receptor) are similar, the mechanistic consequences on lipoprotein metabolism (e.g., effects on the various lipoproteins) differ.
In addition to playing an important role in regulating plasma HDL cholesterol, SR-BI has been implicated (reviewed in ref. 19) in the delivery of HDL cholesterol to the adrenal gland and other steroidogenic tissues, both for the accumulation of esterified cholesterol stores (11, 12, 51, 52) and for steroid hormone synthesis (9, 51). To examine this, we measured the cholesterol content of adrenal glands in mutant and wild-type mice (Table 1). As predicted, cholesterol stores in the adrenal gland dropped substantially in the heterozygous and homozygous mutants to 58% and 28% of control, respectively. We also noted that the color of intact adrenal glands from homozygous mutants was brownish-red whereas that of wild-type and heterozygous animals was light yellow (data not shown) and, in preliminary studies, we observed a dramatic decrease in oil-red O staining of the adrenal cortex in the homozygous mutants relative to the wild-type mice. Thus, the total cholesterol content, color, and oil-red O staining characteristics of the adrenal glands in SR-BI homozygous mutants resembled those in their cholesterol-depleted counterparts in other murine mutants (27, 51, 53), including, as one might expect, null mutants in the SR-BI ligand apoA-I (51). This similarity with apoA-I knockouts is consistent with the possibility that the reduction in adrenal cholesterol in the SR-BI homozygotes is a direct consequence of the loss of the key receptor for selective lipid uptake; however, the data do not rule out possible indirect effects of the mutations (e.g., alteration in HDL size and composition). Recent antibody blocking experiments have provided additional support for a major role of mSR-BI in delivering HDL cholesterol to cultured adrenocortical cells for steroidogenesis (69). Based on the tissue distribution and hormonal regulation of SR-BI protein expression (18, 22, 25) and the phenotypes of apoA-I knockouts (51), it seems likely that there would also be reductions in cholesterol stores in other steroidogenic tissues (e.g., ovary, testes) in SR-BI homozygous mutants. Adrenal cholesterol deficiency in both the apoA-I and SR-BI homozygous mutants also suggests that LDL receptors in the mouse, in which there normally is little LDL in the plasma, do not normally contribute significantly to murine adrenal cholesterol accumulation. However, this may not be the case in other species in which plasma LDL levels are higher and LDL receptors may provide significant amounts of cholesterol to the adrenal gland (54). Future studies will help address these questions as well as determine if SR-BI gene inactivation alters other aspects of cholesterol metabolism and function of adrenal and other steroidogenic tissues.
Previous studies have shown that it is possible to alter plasma HDL cholesterol and/or adrenal cholesterol levels in mice by genetically manipulating the expression of a wide variety of apolipoproteins, plasma enzymes and lipid transfer proteins (refs. 27, 53, and 55–64; reviewed in refs. 16 and 65). In contrast to these studies, which involve alterations in HDL’s synthesis, secretion, or modification in the circulation, the inactivation of the SR-BI gene (this study) and its adenovirus-mediated hepatic overexpression (28) directly affect the mechanism of cellular recognition of HDL and consequent uptake of HDL cholesterol. It should be of interest to cross SR-BI mutant mice with other mice bearing different naturally occurring or experimentally induced genetic alterations to further study the role of SR-BI in lipoprotein and cholesterol metabolism and to establish if the predisposition to atherosclerosis or gallstone disease is altered in SR-BI-deficient mice.
In humans the influence of SR-BI on the levels of plasma HDL cholesterol, adrenal cholesterol, and possibly biliary cholesterol remains unknown. Human SR-BI, hSR-BI [also called CLA-1 (66)] may have physiologic activities similar to those of rodent SR-BI, because it also mediates selective cholesterol uptake and exhibits ligand binding properties and tissue specific expression (23) similar to those of rodent SR-BI (18, 22). It is possible that genetic and/or metabolic variations in the expression of SR-BI contribute to differences in HDL cholesterol levels in humans (16, 67). However, caution should be exercised in extending interpretations from one species (e.g., mice) to another (e.g., rabbits or humans) because of well-known species-dependent differences in lipoprotein metabolism (e.g., refs. 54, 62, and 68). Nevertheless, if SR-BI does have a similar role in lipoprotein metabolism in mice and humans, SR-BI may influence the development and progression of atherosclerosis or gallstone disease and may be an attractive candidate for therapeutic intervention.
For some time, a thorough understanding of cellular and systemic HDL metabolism has been hampered by the absence of a molecularly well-defined, physiologically relevant HDL receptor. The current work establishes that SR-BI is such a receptor. Additional characterization of the structure, mechanism of action, and physiologic and pathophysiologic activities of SR-BI should help provide a greater understanding of HDL metabolism and its role in health and disease.
Acknowledgments
We are grateful to R. Rosenberg, R. Hynes, D. Housman, H. Weiler, K. Kozarsky, M. Donahee, E. Vasile, A. Pearson, D. Resnick, J. Chatterton, P. Seifert, A. Hatzopoulos, P. Christie, K. Wyne, J. Horton, H. Shimano, and A. Rohlmann for advice, encouragement, and helpful discussions. We want to gratefully acknowledge Susan Acton for her contributions to the initial genomic DNA cloning stage of this project and J. Hsu and M. Cummiskey for expert technical assistance. We thank D. Rader and P. Smith for generously providing access to the autoanalyzer for the measurement of plasma apoA-I levels, and L. Castellani and A. Lusis for generously providing anti-apoA-II antibody. We also thank H. Hobbs for generous hospitality, ongoing encouragement, and helpful discussions. This work was supported by Grant HL41484 from the National Institutes of Health Heart Lung and Blood Institute. A.R. was a Howard Hughes Medical Institute Postdoctoral Fellow, B.L.T. was a Medical Research Council of Canada Postdoctoral Fellow, and J.H. was the recipient of an Established Investigator Award from the American Heart Association and Parke–Davis.
ABBREVIATIONS
- HDL
high density lipoprotein
- LDL
low density lipoprotein
- VLDL
very low density lipoprotein
- IDL
intermediate density lipoprotein
- SR-BI
scavenger receptor class B type I
- apolipoprotein
apo
- m
murine
- FPLC
fast protein liquid chromatography
References
- 1.Eisenberg S. J Lipid Res. 1984;25:1017–1058. [PubMed] [Google Scholar]
- 2.Tall A R. J Clin Invest. 1990;86:379–384. doi: 10.1172/JCI114722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon D J, Rifkin B M. N Engl J Med. 1989;321:1311–13162. doi: 10.1056/NEJM198911093211907. [DOI] [PubMed] [Google Scholar]
- 4.Johnson W J, Mahlberg F H, Rothblat G H, Phillips M C. Biochim Biophys Acta. 1991;1085:273–298. doi: 10.1016/0005-2760(91)90132-2. [DOI] [PubMed] [Google Scholar]
- 5.Pieters M N, Schouten D, VanBerkel T J C. Biochim Biophys Acta. 1994;1225:125–134. doi: 10.1016/0925-4439(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 6.Fielding C J, Fielding P E. J Lipid Res. 1995;36:211–228. [PubMed] [Google Scholar]
- 7.Oram J F, Yokoyama S. J Lipid Res. 1996;37:2473–2491. [PubMed] [Google Scholar]
- 8.Tall A R. J Lipid Res. 1993;34:1255–1274. [PubMed] [Google Scholar]
- 9.Gwynne J T, Strauss J F. Endocr Rev. 1982;3:299–329. doi: 10.1210/edrv-3-3-299. [DOI] [PubMed] [Google Scholar]
- 10.Botham K M, Bravo E. Prog Lipid Res. 1995;34:71–97. doi: 10.1016/0163-7827(94)00007-9. [DOI] [PubMed] [Google Scholar]
- 11.Glass C, Pittman R C, Weinstein D W, Steinberg D. Proc Natl Acad Sci USA. 1983;80:5435–5439. doi: 10.1073/pnas.80.17.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass C, Pittman R C, Civen M, Steinberg D. J Biol Chem. 1985;260:744–750. [PubMed] [Google Scholar]
- 13.Goldstein J L, Brown M S, Anderson R G, Russell D W, Schneider W J. Annu Rev Cell Biol. 1985;1:1–39. doi: 10.1146/annurev.cb.01.110185.000245. [DOI] [PubMed] [Google Scholar]
- 14.Turley S D, Dietschy J M. In: The Liver: Biology and Pathobiology. Arias I M, Jakoby W B, Popper H, Schachter D, Schafritz D A, editors. New York: Raven; 1988. pp. 617–641. [Google Scholar]
- 15.Glomset J A. J Lipid Res. 1968;9:155–167. [PubMed] [Google Scholar]
- 16.Breslow J L. In: The Metabolic and Molecular Bases of Inherited Diseases. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 2031–2052. [Google Scholar]
- 17.Acton S L, Scherer P E, Lodish H F, Krieger M. J Biol Chem. 1994;269:21003–21009. [PubMed] [Google Scholar]
- 18.Acton S, Rigotti A, Landschulz K, Xu S, Hobbs H H, Krieger M. Science. 1996;271:518–520. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 19.Rigotti A, Trigatti B, Babitt J, Penman M, Xu S, Krieger M. Curr Opin Lipidol. 1997;8:181–188. doi: 10.1097/00041433-199706000-00009. [DOI] [PubMed] [Google Scholar]
- 20.Xu S, Laccotripe M, Huang X, Rigotti A, Zannis V I, Krieger M. J Lipid Res. 1997;38:1289–1298. [PubMed] [Google Scholar]
- 21.Babitt J, Trigatti B, Rigotti A, Smart E J, Anderson R G W, Xu S, Krieger M. J Biol Chem. 1997;272:13242–13249. doi: 10.1074/jbc.272.20.13242. [DOI] [PubMed] [Google Scholar]
- 22.Landschulz K T, Pathak R P, Rigotti A, Krieger M, Hobbs H H. J Clin Invest. 1996;98:984–995. doi: 10.1172/JCI118883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murao K, Terpstra V, Green S R, Kondratenko N, Steinberg D, Quehenberger O. J Biol Chem. 1997;272:17551–17557. doi: 10.1074/jbc.272.28.17551. [DOI] [PubMed] [Google Scholar]
- 24.Woollett L A, Spady D K. J Clin Invest. 1997;99:1704–1713. doi: 10.1172/JCI119334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rigotti A, Edelman E R, Seifert P, Iqbal S, DeMattos R B, Temel R E, Krieger M, Williams D. J Biol Chem. 1996;271:33545–33549. doi: 10.1074/jbc.271.52.33545. [DOI] [PubMed] [Google Scholar]
- 26.Wang N, Weng W, Breslow J L, Tall A R. J Biol Chem. 1996;271:21001–21004. doi: 10.1074/jbc.271.35.21001. [DOI] [PubMed] [Google Scholar]
- 27.Ng D S, Francone O L, Forte T M, Zhang J, Haghpassand M, Rubin E M. J Biol Chem. 1997;272:15777–15781. doi: 10.1074/jbc.272.25.15777. [DOI] [PubMed] [Google Scholar]
- 28.Kozarsky K F, Donahee M H, Rigotti A, Iqbal S N, Edelman E R, Krieger M. Nature (London) 1997;387:414–417. doi: 10.1038/387414a0. [DOI] [PubMed] [Google Scholar]
- 29.Soriano P, Montgomery C, Geske R, Bradley A. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 30.Mansour S L, Thomas K R, Capecchi M R. Nature (London) 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 31.George E L, Hynes R O. Methods Enzymol. 1994;245:386–420. doi: 10.1016/0076-6879(94)45021-8. [DOI] [PubMed] [Google Scholar]
- 32.Willnow T E, Herz J. Methods Cell Biol. 1994;43:305–334. doi: 10.1016/s0091-679x(08)60610-x. [DOI] [PubMed] [Google Scholar]
- 33.Doetschman T C, Eistetter H, Katz M, Schmidt W, Kemler R. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- 34.Laird P W, Zijderveld A, Linders K, Rudnicki M, Jaenisch R, Berns A. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jokinen E V, Landschulz K T, Wyne K L, Ho Y K, Frykman P K, Hobbs H H. J Biol Chem. 1994;269:26411–25418. [PubMed] [Google Scholar]
- 36.Laemmli U K. Nature (London) 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Towbin H, Staehelin T, Gordon T. Proc Natl Acad Sci USA. 1979;76:4350–4355. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo Q, Penman M, Trigatti B L, Krieger M. J Biol Chem. 1996;271:11191–11196. doi: 10.1074/jbc.271.19.11191. [DOI] [PubMed] [Google Scholar]
- 39.Lowry O H, Rosebrough N J, Farr A L, Randall J R. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 40.Kozarsky K F, Jooss K, Donahee M, Strauss J F, Wilson J M. Nat Genet. 1996;13:54–62. doi: 10.1038/ng0596-54. [DOI] [PubMed] [Google Scholar]
- 41.Yokode M R, Hammer R E, Ishibashi S, Brown M S, Goldstein J L. Science. 1990;250:1273–1275. doi: 10.1126/science.2244210. [DOI] [PubMed] [Google Scholar]
- 42.Willnow T E, Sheng Z, Ishibashi S, Herz J. Science. 1994;264:1471–1474. doi: 10.1126/science.7515194. [DOI] [PubMed] [Google Scholar]
- 43.Warden C H, Hedrick C C, Qiao J-H, Castellani L W, Lusis A J. Science. 1993;261:469–472. doi: 10.1126/science.8332912. [DOI] [PubMed] [Google Scholar]
- 44.Smithies O, Maeda N. Proc Natl Acad Sci USA. 1995;92:5266–5272. doi: 10.1073/pnas.92.12.5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webb N R, de Villiers W J, Connell P M, de Beer F C, van der Westhuyzen D R. J Lipid Res. 1997;38:1490–1495. [PubMed] [Google Scholar]
- 46.Vaisman B L, Klein H-G, Rouis M, Berard A M, Kindt M R, Talley G D, Meyn S M, Hoyt R F, Jr, Marcovina S M, Albers J J, Hoeg J M, Brewer H B, Jr, Santamarina-Fojo S. J Biol Chem. 1995;270:12269–12275. doi: 10.1074/jbc.270.20.12269. [DOI] [PubMed] [Google Scholar]
- 47.DeSilva H V, Mas-Oliva J, Taylor J M, Mahley R W. J Lipid Res. 1994;35:1297–1310. [PubMed] [Google Scholar]
- 48.Li H, Reddick R L, Maeda N. Arterioscler Thromb. 1993;13:1814–1821. doi: 10.1161/01.atv.13.12.1814. [DOI] [PubMed] [Google Scholar]
- 49.Plump A S, Azrolan N, Odaka H, Wu L, Jiang X, Tall A, Eisenberg S, Breslow J. J Lipid Res. 1997;38:1033–1047. [PubMed] [Google Scholar]
- 50.Ishibashi S, Brown M S, Goldstein J L, Gerard R D, Hammer R E, Herz J. J Clin Invest. 1993;92:883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plump A S, Erickson S K, Weng W, Partin J S, Breslow J L, Williams D L. J Clin Invest. 1996;97:2660–2671. doi: 10.1172/JCI118716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reaven E, Tsai L, Azhar S. J Biol Chem. 1996;271:16208–16217. doi: 10.1074/jbc.271.27.16208. [DOI] [PubMed] [Google Scholar]
- 53.Meiner V L, Cases S, Myers H M, Sande E R, Bellosta S, Schambelan M, Pitas R E, McGuire J, Herz J, Farese R V., Jr Proc Natl Acad Sci USA. 1996;93:14041–14046. doi: 10.1073/pnas.93.24.14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown M S, Kovanen P T, Goldstein J L. Rec Prog Horm Res. 1979;35:215–257. doi: 10.1016/b978-0-12-571135-7.50009-6. [DOI] [PubMed] [Google Scholar]
- 55.Weng W, Breslow J L. Proc Natl Acad Sci USA. 1996;93:14788–14794. doi: 10.1073/pnas.93.25.14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duverger N, Tremp G, Caillaud J M, Emmanuel F, Castro G, Fruchart J C, Steinmetz A, Denefle P. Science. 1996;273:966–968. doi: 10.1126/science.273.5277.966. [DOI] [PubMed] [Google Scholar]
- 57.Farese R V, Ruland S L, Flynn L M, Stokowski R P, Young S G. Proc Natl Acad Sci USA. 1995;92:1774–1778. doi: 10.1073/pnas.92.5.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang L-S, Voyiaziakis E, Markenson D F, Sokol K A, Hayek T, Breslow J L. J Clin Invest. 1995;96:2152–2161. doi: 10.1172/JCI118269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maeda N, Li H, Lee D, Oliver P, Quarfordt S H, Osada J. J Biol Chem. 1994;269:23610–23616. [PubMed] [Google Scholar]
- 60.Zhang S H, Reddick R L, Piedrahita J A, Maeda N. Science. 1992;258:468–471. doi: 10.1126/science.1411543. [DOI] [PubMed] [Google Scholar]
- 61.Weinstock P H, Bisgaier C L, Aalto-Setala K, Radner H, Ramakrishnan R, Levak-Frank S, Essenburg A D, Zechner R, Breslow J L. J Clin Invest. 1995;96:2555–2568. doi: 10.1172/JCI118319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakai N, Vaisman B L, Koch C A, Hoyt R F, Jr, Meyn S M, Talley G D, Paiz J A, Brewer H B, Jr, Santamarina-Fojo S. J Biol Chem. 1997;272:7506–7510. doi: 10.1074/jbc.272.11.7506. [DOI] [PubMed] [Google Scholar]
- 63.Nakamuta M, Hung-Junn Chang B, Zsigmond E, Kobayashi K, Lei H, Ishida B Y, Li E, Chan L. J Biol Chem. 1996;271:25981–25988. doi: 10.1074/jbc.271.42.25981. [DOI] [PubMed] [Google Scholar]
- 64.Jiang X-C, Omar L, Francone O L, Bruce C, Milne R, Mar J, Walsh A, Breslow J L, Tall A R. J Clin Invest. 1996;98:2373–2380. doi: 10.1172/JCI119050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rader D J, Ikewaki K. Curr Opin Lipidol. 1996;7:117–123. doi: 10.1097/00041433-199606000-00002. [DOI] [PubMed] [Google Scholar]
- 66.Calvo D, Vega M A. J Biol Chem. 1993;268:18929–18935. [PubMed] [Google Scholar]
- 67.Guerra R, Wang J, Grundy S M, Cohen J C. Proc Natl Acad Sci USA. 1997;94:4532–4537. doi: 10.1073/pnas.94.9.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoeg J M, Santamarina-Fojo S, Berard A M, Cornhill J F, Herderick E E, Feldman S J, Haudenschild C C, Vaisman B L, Hoyt R F, Jr, Demosky S J, Jr, Kauffman R D, Hazel C M, Marcovina S M, Brewer H B., Jr Proc Natl Acad Sci USA. 1996;93:11448–11453. doi: 10.1073/pnas.93.21.11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Temel, R., Trigatt, B. L., De Mattos, R., Azhar, S., Krieger, M. & Williams, D. (1997) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]