Abstract
Archaea contain histones that have primary sequences in common with eukaryal nucleosome core histones and a three-dimensional structure that is essentially only the histone fold. Here we report the results of experiments that document that archaeal histones compact DNA in vivo into structures similar to the structure formed by the histone (H3+H4)2 tetramer at the center of the eukaryal nucleosome. After formaldehyde cross-linking in vivo, these archaeal nucleosomes have been isolated from Methanobacterium thermoautotrophicum and Methanothermus fervidus, visualized by electron microscopy on plasmid and genomic DNAs, and shown by immunogold labeling, SDS/PAGE, and immunoblotting to contain archaeal histones, cross-linked into tetramers. Archaeal nucleosomes protect ≈60 bp of DNA and multiples of ≈60 bp from micrococcal nuclease digestion, and immunoprecipitation has demonstrated that most, but not all, M. fervidus genomic DNA sequences are associated in vivo with archaeal histones.
The presence of a nuclear membrane defines the Eukarya, although the regular packaging of nuclear DNA by histones into nucleosomes and chromatin is equally definitive. Accessing genetic information within chromatin poses obvious problems, and transcription-initiating complexes are far more complex in Eukarya than in Bacteria (1). Archaea lack a nuclear membrane and therefore are, by definition, prokaryotes, but they do contain histones (2), and archaeal transcription initiation conforms to the eukaryal paradigm. Archaeal RNA polymerases have primary sequences and subunit complexities in common with eukaryal RNA polymerases, archaeal promoters contain TATA box elements, and archaeal transcription initiation requires the participation of structural and functional homologs of eukaryal TATA box binding and transcription factor TFIIB proteins (3, 4). Archaeal histones form the histone fold (5, 6) that facilitates DNA wrapping into nucleosomes by eukaryal histones (7), and archaeal genomes therefore also might be similarly constrained into nucleosome- and chromatin-related structures. An electron microscopy (EM) investigation of genomic DNA released by osmotic shock from halophilic archaeal cells revealed the presence of structures that visibly resembled nucleosomes, but their protein and nucleic contents were not determined (8). To investigate this issue, we have isolated and characterized archaeal histone–DNA complexes from two methanogens, Methanobacterium thermoautotrophicum strain Marburg and Methanothermus fervidus, and document here that these structures, designated archaeal nucleosomes, do have features in common with the eukaryal nucleosome. Specifically, they resemble the structure formed by the histone (H3+H4)2 tetramer at the center of the eukaryal nucleosome (9–11); they exhibit localized assembly, contain histone tetramers, and protect ≈60 bp of DNA from nuclease digestion.
MATERIALS AND METHODS
Growth of Methanogens, RNA Isolation, and Transcript Analyses.
Cultures of M. thermoautotrophicum strain Marburg and M. fervidus were grown at 65°C and 83°C, respectively, in a 20-liter fermentor supplied with H2 plus CO2. RNA was isolated, and the presence of specific transcripts was determined by Northern blotting as described (12, 13).
Purification of HMt and HMf and Preparation and Purification of Anti-Archaeal Histone IgG.
HMt and HMf were purified, as described (13), from M. thermoautotrophicum strain Marburg and from M. fervidus, respectively, and were used to immunize New Zealand white rabbits to obtain anti-archaeal histone antisera. Polyclonal IgG were affinity-purified from these antisera (14) using protein A affinity Pak columns (Pierce).
Isolation and Immunoblotting of Archaeal Histone–DNA Complexes.
Protoplasts were prepared from M. thermoautotrophicum strain Marburg cells, as described (15), using a pseudomurein-digesting endopeptidase isolated from Methanobacterium wolfei. Protein–DNA complexes were cross-linked in vivo by exposing the protoplasts to 1% formaldehyde (16, 17), in protoplast buffer, for 1 h at room temperature. The formaldehyde was quenched by addition of 0.4 M NH4-acetate, and the protoplasts were washed twice with protoplast buffer and lysed by resuspension in 0.1% SDS, 1 mM Na–EDTA, 0.5 mM Na–EGTA, and 10 mM Na-Hepes (pH 7.5). Lysates were digested with 100 μg of pronase per milliliter for 3 h at 37°C and centrifuged at 8000 × g for 10 min, and the resulting supernatants were loaded onto 15–50% (wt/vol) sucrose gradients dissolved in lysis buffer that contained 100 μM phenylmethylsulfonyl fluoride. After centrifugation at 4°C in a SW41 rotor for 16.5 h at 25,000 rpm, fractions were collected, and the complexes present in each fraction were separated by electrophoresis through 0.8% (wt/vol) agarose gels, ethidium bromide-stained, and probed for the presence of archaeal histones by immunoblotting using anti-archaeal histone antiserum.
Electron Microscopy.
The DNA spreading and EM procedures have been described (18). Protoplasts were lysed in 100 mM NH4–acetate, 10 mM triethylamine⋅HCl, and 5 mM MgCl2 (pH 8), and the resulting lysates were passaged through a Sepharose 4B column, glutaraldehyde-fixed, and the genomic DNA present was spread by adsorption onto mica. For immunogold labeling, DNA–protein complexes from sucrose gradient fractions were subjected to Sepharose 4B column chromatography before incubation with anti-HMt IgG for 1 h at room temperature. The resulting complexes were repassaged through the Sepharose 4B column, incubated with protein A 5-nm gold bead conjugates (Sigma) for 1 h at room temperature, passaged again through the Sepharose 4B column, adsorbed to mica, and visualized by EM.
Micrococcal Nuclease (MN) Digestion and Analysis of Nucleosome-Protected DNAs.
After formaldehyde fixation, cells were ruptured by passage through a French pressure cell (20,000 psi), and the lysates obtained were incubated with 1.5 units of MN (Sigma) per gram wet weight of cell paste for 0, 1, 2, 4, 6, 8, 10, 15, 30, and 45 min. Reactions were quenched with 0.2% (wt/vol) SDS and 20 mM EDTA, and the reaction mixtures then were incubated with 300 μg of proteinase K per milliliter for 3 h at 37°C. Protein–DNA cross-links were reversed by incubating the resulting complexes for 6 h at 65°C (16), and the DNA molecules present were isolated by phenol-chloroform extraction, subjected to electrophoresis through 4% Nusieve-GTG low melting temperature agarose gels (FMC), ethidium bromide-stained, and photographed.
Isolation and Analysis of Cross-linked Proteins from Archaeal Nucleosomes.
Lysates from formaldehyde-fixed cells were digested for 10 min with MN and then quenched, as described above. The complexes present were purified by Sepharose 4B column chromatography followed by electrophoresis through Nusieve-GTG agarose gels. The region of the gel that contained complexes was excised, and the agarose was melted at 70°C and then digested by incubation with β-agarase overnight at 45°C. The proteins that remained after an incubation with 100 μg of DNase I/ml for 30 min at 37°C were separated by tricine–SDS/PAGE (19), visualized by silver staining, and identified by immunoblotting (14).
Immunoprecipitation.
DNA–protein complexes formaldehyde-cross-linked in vivo were separated from proteins and protein-free DNA by sedimentation through preformed CsCl step gradients (20). Fractions that contained histone–DNA complexes, identified by immunoblotting, were pooled and digested for 8 h with restriction enzymes. Formalin-fixed Staphylococcus A cells were added [20% (vol/vol) pansorbin; (Calbiochem–Novabiochem) in 0.2% sarkosyl/10 mM Tris⋅HCl, pH 7.5/1 mM EDTA], and nonspecific associations that formed in the absence of antibodies were removed by centrifugation. Anti-archaeal histone IgG was added, and the mixtures were incubated overnight, with shaking, at 4°C. Control mixtures were incubated overnight with nonspecific polyclonal IgG. Immune complexes that bound to the pansorbin were collected by centrifugation, washed 10 times as described (20), and eluted by four 20-min washes with 200 μl of 50 mM Tris⋅HCl (pH 8.5), 1% SDS, and 2 mM EDTA that contained 1.5 μg of sonicated carrier DNA/ml. The eluted material was incubated with 100 μg RNase A per milliliter for 30 min at 37°C and then with 100 μg of proteinase K per milliliter for 6 h at 65°C. The presence of specific sequences in the DNA fragments that remained was determined by Southern blotting using [32P]-end-labeled probes (21).
Determination of the Histone-to-DNA Ratio in Vivo.
Cell lysates resulting from four sequential passages through the French pressure cell in a high salt buffer (3 M KCl/50 mM Tris⋅HCl, pH 8/0.5 mM phenylmethylsulfonyl fluoride/0.1 mM EDTA) were centrifuged at 2000 × g for 10 min to remove unbroken cells and then dialyzed against 200 mM NaCl and 50 mM Tris⋅HCl/0.1 mM EDTA (pH 8). The protein and DNA concentrations of these cell lysates were measured using a commercial BCA assay (Sigma) with BSA as the standard and using the Burton variation of the diphenylamine assay (22) with calf thymus DNA as the standard, respectively. Aliquots of the lysate were subjected to SDS/PAGE in parallel with increasing amounts of purified archaeal histones (13), and immunoblots were generated by incubation overnight at 4°C with rabbit anti-histone antisera followed by incubation with 125I-labeled protein A (DuPont/NEN). Radioactively labeled regions of the blot were excised, and their 125I content was measured by γ radiation counting. The histone content of the lysates was calculated by comparing the 125I bound to experimental samples with 125I bound to the standard curve.
RESULTS
Archaeal Histones Are Present in Archaeal Nucleosomes in Vivo.
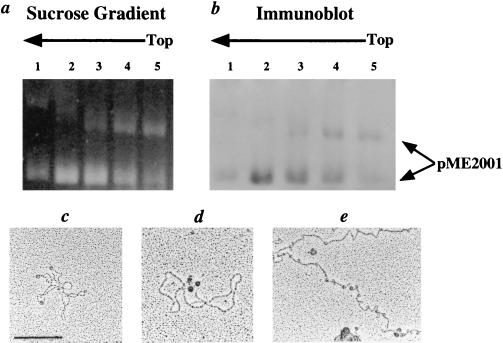
Previous in vitro studies have focused primarily on the HMf histones from the hyperthermophile, M. fervidus, although archaeal histones with sequences >80% identical to the HMf histones are present in other Archaea (2, 4). This includes the HMt histones in M. thermoautotrophicum (23), which was included in this investigation because an endopeptidase is available that generates protoplasts from M. thermoautotrophicum cells (15) that could be lysed gently by detergent addition, thus eliminating the need for a physically disruptive cell breakage step. The Marburg strain of M. thermoautotrophicum was chosen for study because this methanogen contains a plasmid, pME2001 (24), which added the opportunity to isolate and characterize discrete protein–DNA complexes. M. thermoautotrophicum strain Marburg protoplasts were exposed to formaldehyde and lysed by SDS addition, and the complexes present were separated by sedimentation through sucrose gradients. Complexes in each fraction from the sucrose gradients were further separated by agarose gel electrophoresis, stained, and then probed by immunoblotting using anti-HMt antiserum. Two pME2001-containing complexes, with mobilities consistent with the presence of supercoiled and relaxed plasmid DNAs, were separated by the electrophoresis, and both contained HMt (Figs. 1 a and b). EM visualization of these plasmid-containing complexes (Fig. 1c) and of genomic DNA spread directly from ruptured protoplasts (Fig. 1e), demonstrated the presence of structures that visibly resembled nucleosomes, and immunogold-labeling localized HMt to these structures (Fig. 1d). Genomic DNA also contained many small DNA loops, consistent with the remains of archaeal nucleosomes that had lost their protein components after their adsorption to the mica support used for EM visualization (18).
Figure 1.
Isolation and EM of archaeal nucleosomes from M. thermoautotrophicum strain Marburg. (a) Electrophoretic separation of pME2001-containing complexes in sucrose gradient fractions from a lysate of formaldehyde-fixed protoplasts. (b) Immunoblot of the complexes in a, generated with anti-HMt antiserum. (c) EM of a pME2001–HMt complex (Bar = 200 nm). (d) pME2001–HMt–anti-HMt–IgG complex immunogold-labeled (black dots) with a protein A–gold bead conjugate. (e) EM of genomic DNA from a formaldehyde-fixed protoplast.
MN Digestion Generates ≈60-bp Ladders from Archaeal Chromatin.
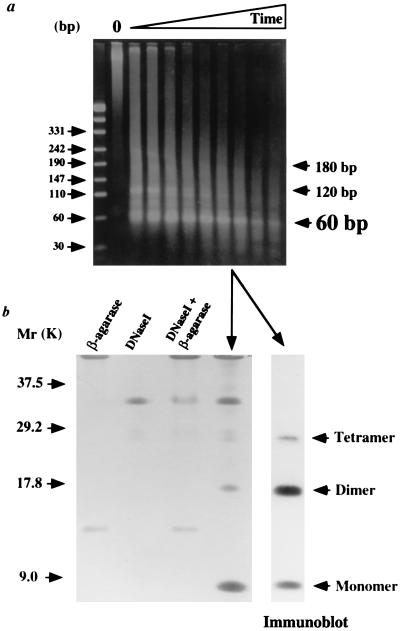
Nuclease digestion of eukaryal chromatin, formaldehyde cross-linked in vivo, generates a ladder of DNA molecules with lengths that are ≈146 bp, and multiples of ≈146 bp, that result from nuclease protection by one or more adjacent nucleosomes (17, 18, 25). Exposure of nucleoprotein complexes isolated from formaldehyde-treated M. thermoautotrophicum or M. fervidus to MN generated nuclease-protected fragments that were ≈60 bp, and multiples of ≈60 bp, in length, and with increasing exposure to MN only ≈60-bp fragments remained (Fig. 2a).
Figure 2.
DNA and protein content of archaeal nucleosomes. (a) Electrophoretic separation of DNA fragments protected from MN digestion in nucleoprotein complexes isolated from formaldehyde-fixed M. fervidus. MN digestion was for 1, 2, 4, 6, 8, 10, 15, 30, and 45 min at 37°C. Control tracks contained size standards and undigested complexes (O). (b) Electrophoretic separation and silver staining of the proteins isolated from the complexes indicated in a that protected ≈60 bp fragments of DNA from MN digestion. Purification of these proteins involved incubations with β-agarase and DNase I, and the control tracks demonstrate the polypeptides present in these reagents. Small amounts of these polypeptides remained in the experimental material. An immunoblot of the purified proteins generated with anti-HMf antibodies is shown adjacent to the stained gel.
Archaeal Histones Are Cross-Linked in Vivo into Tetramers.
Complexes shown to protect ≈60-bp DNA fragments from MN digestion were purified, after exposure to MN, by gel filtration chromatography and agarose gel electrophoresis. After DNase I digestion, the proteins that remained were separated by tricine–SDS/PAGE, silver-stained, and immunoblotted. As shown in Fig. 2b, the only polypeptides present other than DNase I detectable by silver staining were identified by immunoblotting as histone monomers, dimers, and tetramers. There were no cross-linked trimers, nor was there evidence for cross-linking of the archaeal histones into oligomers larger than tetramers.
Archaeal Histone to DNA Ratio in Vivo and Genome Localization.
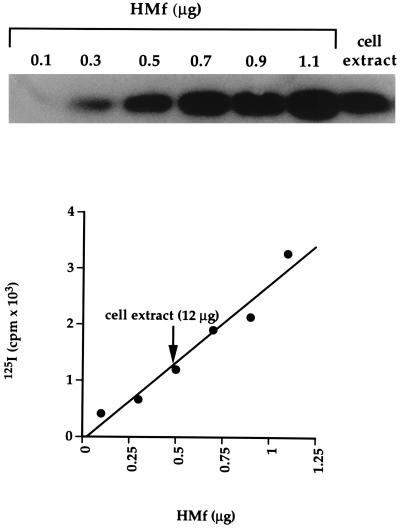
Eukaryal histones are abundant proteins that package almost all of the nuclear DNA into chromatin, although some nucleosomes are positioned, and the localized assembly and disassembly of nucleosomes play an important role in regulating eukaryal gene expression (25). To determine if these features might also exist in Archaea, the histone-to-DNA ratio was established, and the presence or absence of specific regions of the M. fervidus genome within histone–DNA complexes in vivo was determined. Quantitation of 125I-labeled immunoblots revealed that HMf constitutes ≈4% of the total soluble protein in M. fervidus (Fig. 3). Based on the total protein-to-DNA ratio and a genome size of 1.7 ± 0.2 Mbp, the size range of genomes of closely related methanogens (26), this amounts to one histone tetramer per ≈67 bp of genomic DNA. It appears, therefore, that almost all of the M. fervidus genome could be packaged by HMf tetramers into structures that contain ≈60 bp of DNA.
Figure 3.
Quantitation of HMf. The immunoblot of a 125I-labeled standard curve, adjacent to an aliquot of a M. fervidus lysate, is shown above a graphical quantitation of this result.
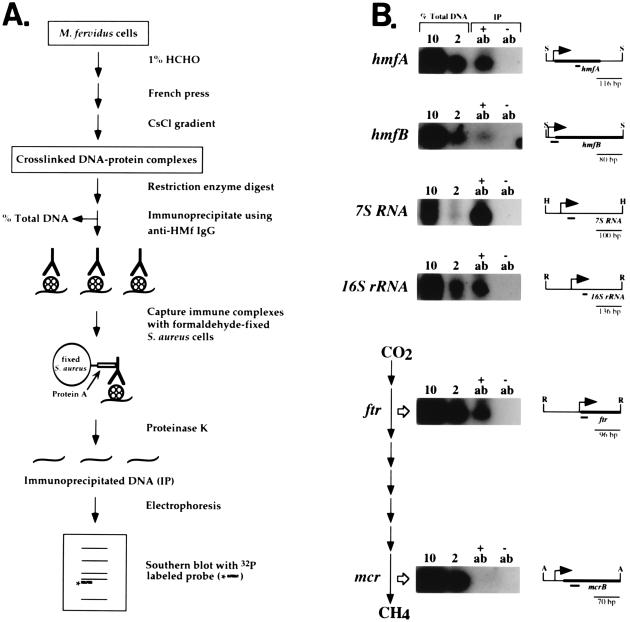
To determine if specific regions of the M. fervidus genome were associated with HMf in vivo, cross-linked complexes were separated from proteins and protein-free DNA by CsCl buoyant density centrifugation, subjected to restriction enzyme digestion, and immunoprecipitated by anti-HMf IgG (Fig. 4A and ref. 20). After deproteinization, the DNA fragments that remained were probed by Southern blotting for the presence of upstream intergenic regions and coding regions of the 7S and 16S stable RNA genes, the histone-encoding hmfA and hmfB genes, and several genes that encode enzymes involved in methanogenesis (2, 12, 18, 27). Hybridization was observed in every case, to a restriction fragment of the correct size, except when probes specific for the mcr operon were used (Fig. 4B). Consistent differences in signal intensities were observed. The 7S and 16S sequences were always present at relatively high levels in immunoprecipitated complexes, hmfA sequences were more abundant than hmfB sequences and, despite the use of different probes and restriction enzymes, signals for the mcr operon were undetectable. This was not a general case for all methane genes. The ftr and mcr genes encode the enzymes that catalyze the second and seventh steps in methanogenesis from CO2 and H2, respectively (12, 27), and ftr sequences were present in immunoprecipitated complexes (Fig. 4B).
Figure 4.
(A) Outline of the procedure used to isolate and immunoprecipitate histone–DNA complexes cross-linked in vivo (20). (B) Southern blots generated with probes specific for the M. fervidus genes listed (27, 28). The probes (short heavy lines) were designed to hybridize, as indicated, to similar-sized restriction fragments, ApoI (A), HincII (H), RsaI (R), and SspI (S), that spanned the sites of transcription (bent arrow) and translation initiation (thick bar). As illustrated, the ftr and mcr genes encode the enzymes that catalyze the second and seventh steps in CO2 reduction to CH4 (27–29). At the time of formaldehyde fixation, the cells were actively synthesizing methane, and Northern blot analyses demonstrated that the ftr, mcr, hmfA, hmfB, 7S, and 16S transcripts were abundant. Aliquots (10% and 2%) of the total DNA present before the immunoprecipitation (see A) were electrophoresed in the tracks adjacent to the tracks that contained the DNA isolated from complexes immunoprecipitated (IP) by anti-HMf IgG (+ab) or by the control antiserum (−ab).
DISCUSSION
The eukaryal nucleosome is a tripartite structure, in which two histone (H2A+H2B) dimers flank a central (H3+H4)2 histone tetramer, and together this histone octamer wraps ≈146 bp in a left-handed superhelix (7, 25). The (H3+H4)2 tetramer recognizes the nucleosome positioning signals, initiates nucleosome assembly, and can form stable structures that wrap ≈120 bp but protect only ≈73 bp from nuclease digestion (9–11). The results reported here document the presence of structures in Archaea that appear analogous and may be homologous to the structure formed by the eukaryal (H3+H4)2 histone tetramer. The primary sequences of the archaeal histones are most similar to H3 and H4 sequences (2, 4), and they assemble into structures that contain histone tetramers that protect ≈60 bp from nuclease digestion. The eukaryal histones are larger, with additional N- and C-terminal domains that extend beyond the nucleosome that provide sites for posttranslational regulatory modifications, but these regulatory regions are not essential (25). (H3+H4)2 tetramer-containing structures still assemble and position correctly after the removal of these regions by protease digestion (10). The handedness of the DNA superhelix constrained in the archaeal nucleosome in vivo remains to be determined. Archaeal histones wrap DNA in a right-handed superhelix in vitro, and this seemed to be a fundamental difference from the eukaryal nucleosome (30). However, it recently has been proposed that only a slight change in the dimer–dimer interface within an (H3+H4)2 tetramer would result in a structure that wrapped DNA in a right-handed superhelix (11). If such dimer:dimer interface movement can occur within a histone tetramer, then nucleosomes could constrain DNA in both left-handed and right-handed superhelices and may shift from left-handed to right-handed wrapping depending on the superhelical density of the surrounding DNA (11). If this is correct, it would explain how archaeal histone binding to closed circular DNAs, at low histone to DNA ratios, introduces negative superhelicity that spontaneously becomes positive superhelicity when the histone-to-DNA ratio is increased (2, 4, 30).
Possibly histones evolved, in an ancestor of the Archaea and Eukarya, to overcome problems similar to those still faced by M. fervidus, namely maintaining genome integrity and function while pursuing a hyperthermophilic lifestyle. Inventing a DNA wrapping solution, which coincidentally compacted DNA and introduced biochemically usable superhelical tension, may have been a major step in cellular evolution. It would have solved DNA structure and space constraint problems but mandated a transcription system that could recognize and access initiation signals and activate genes wrapped within these structures. Still having to cope with these architectural constraints may explain why the basic components of the archaeal and eukaryal transcription initiation systems remain so similar (3, 4). Regulated transcription initiation has been documented in Archaea (27), but archaeal histones do not have the sites for the posttranslational modifications that endow eukaryal histones with regulatory functions, so it seems unlikely that archaeal histones regulate specific gene expression. The lack of histone association with mcr sequences in vivo (Fig. 4B) does, however, suggest that histones might regulate the levels of gene expression. The mcr operon encodes methyl coenzyme M reductase, the most abundant enzyme in methanogens, and mcr transcription occurs at very high levels and exhibits unique regulation, or lack of regulation. Transcripts of the mcr operon continue to be synthesized when the substrate supply is reduced to a level insufficient for growth, conditions under which all other methane gene transcription is terminated (12, 27–29).
Acknowledgments
We thank our colleagues, particularly K. Sandman, for their continued interest and intellectual contributions to this work. This work was supported by National Institutes of Health Grant GM53185.
ABBREVIATIONS
- EM
electron microscopy
- MN
micrococcal nuclease
Footnotes
A commentary on this article begins on page 12251.
References
- 1.Nikolov D B, Burley S K. Proc Natl Acad Sci USA. 1997;94:15–22. doi: 10.1073/pnas.94.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grayling R A, Sandman K, Reeve J N. FEMS Microbiol Rev. 1996;16:203–213. doi: 10.1111/j.1574-6976.1996.tb00237.x. [DOI] [PubMed] [Google Scholar]
- 3.Langer D, Hain J, Thuriaux P, Zillig W. Proc Natl Acad Sci USA. 1996;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reeve J N, Sandman K, Daniels C J. Cell. 1997;89:999–1002. doi: 10.1016/s0092-8674(00)80286-x. [DOI] [PubMed] [Google Scholar]
- 5.Arents G, Moudrianakis E N. Proc Natl Acad Sci USA. 1995;92:11170–11174. doi: 10.1073/pnas.92.24.11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starich M R, Sandman K, Reeve J N, Summers M F. J Mol Biol. 1996;255:187–203. doi: 10.1006/jmbi.1996.0016. [DOI] [PubMed] [Google Scholar]
- 7.Arents G, Moudrianakis E N. Proc Natl Acad Sci USA. 1993;90:10489–10493. doi: 10.1073/pnas.90.22.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takayanagi S, Morimura S, Kusaoke H, Yokoyama Y, Kano K, Shoda M. J Bacteriol. 1992;174:7207–7216. doi: 10.1128/jb.174.22.7207-7216.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes J J, Clark D J, Wolffe A P. Proc Natl Acad Sci USA. 1991;88:6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong F, van Holde K E. Proc Natl Acad Sci USA. 1991;88:10596–10600. doi: 10.1073/pnas.88.23.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamiche A, Carot V, Alilat M, De Lucia F, O’Donohue M-F, Révet B, Prunell A. Proc Natl Acad Sci USA. 1996;93:7588–7593. doi: 10.1073/pnas.93.15.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pihl T D, Sharma S, Reeve J N. J Bacteriol. 1994;176:6384–6391. doi: 10.1128/jb.176.20.6384-6391.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandman K, Grayling R A, Dobrinski B, Lürz R, Reeve J N. Proc Natl Acad Sci USA. 1994;87:5788–5791. doi: 10.1073/pnas.87.15.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W. Current Protocols in Immunology. New York: Wiley; 1991. [Google Scholar]
- 15.Mori H, Koga Y. J Ferment Bioeng. 1992;73:6–10. [Google Scholar]
- 16.Solomon M J, Varshavasky A. Proc Natl Acad Sci USA. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solomon M J, Larsen P L, Varshavasky A. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 18.Sandman K, Krzycki J A, Dobrinski B, Lürz R, Reeve J N. Proc Natl Acad Sci USA. 1990;87:5788–5791. doi: 10.1073/pnas.87.15.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schägger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 20.Gilmore D S, Rougvie A E, Lis J Y. Methods Cell Biol. 1991;35:369–381. doi: 10.1016/s0091-679x(08)60580-4. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Habor Lab. Press; 1989. [Google Scholar]
- 22.Burton K. Biochem J. 1956;62:315–323. doi: 10.1042/bj0620315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabassum R, Sandman K M, Reeve J N. J Bacteriol. 1992;174:7890–7895. doi: 10.1128/jb.174.24.7890-7895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bokranz M, Klein A, Meile L. Nucleic Acid Res. 1990;18:363. doi: 10.1093/nar/18.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elgin S C R. Chromatin Structure and Gene Expression. Oxford: IRL Press; 1995. [Google Scholar]
- 26.Stettler R, Leisinger T. J Bacteriol. 1992;174:7227–7234. doi: 10.1128/jb.174.22.7227-7234.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reeve, J. N., Nölling, J., Morgan, R. M. & Smith, D. R. (1997) J. Bacteriol. 179, in press. [DOI] [PMC free article] [PubMed]
- 28.Weil C F, Cram D S, Sherf B A, Reeve J N. J Bacteriol. 1988;170:4718–4726. doi: 10.1128/jb.170.10.4718-4726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan R M, Pihl T D, Nölling J, Reeve J N. J Bacteriol. 1997;179:889–898. doi: 10.1128/jb.179.3.889-898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musgrave D R, Sandman K M, Reeve J N. Proc Natl Acad Sci USA. 1991;88:10397–10401. doi: 10.1073/pnas.88.23.10397. [DOI] [PMC free article] [PubMed] [Google Scholar]