Abstract
Nanoscale pores have potential to be used as biosensors and are an established tool for analysing the structure and composition of single DNA or RNA molecules1–3. Recently, nanopores have been used to measure the binding of enzymes to their DNA substrates4,5. In this technique, a polynucleotide bound to an enzyme is drawn into the nanopore by an applied voltage. The force exerted on the charged backbone of the polynucleotide by the electric field is used to examine the enzyme–polynucleotide interactions. Here we show that a nanopore sensor can accurately identify DNA templates bound in the catalytic site of individual DNA polymerase molecules. Discrimination among unbound DNA, binary DNA/polymerase complexes, and ternary DNA/polymerase/deoxynucleotide triphosphate complexes was achieved in real time using finite state machine logic. This technique is applicable to numerous enzymes that bind or modify DNA or RNA including exonucleases, kinases and other polymerases.
We describe a nanopore device that monitors ionic current through a single protein pore inserted in a lipid bilayer (Fig. 1a). The limiting aperture of the pore is just sufficient to accommodate single-stranded DNA (ssDNA)6,7, and the adjacent pore vestibule can accommodate double-stranded (duplex) DNA (dsDNA)7–9. In the absence of DNA, the open channel current (Io) through the α-haemolysin pore is 60 pA at 180 mV applied potential in 0.3 M KCl. DNA capture in the nanopore results in a decrease in the current (I). The DNA resides in the pore for a time (tD) until it leaves, moving to the trans compartment (Fig. 1b). These two parameters, I and tD, together with current noise, are typically used to report results from nanopore experiments6,10–19.
Figure 1. Detection of DNA translocation events with the nanopore device.
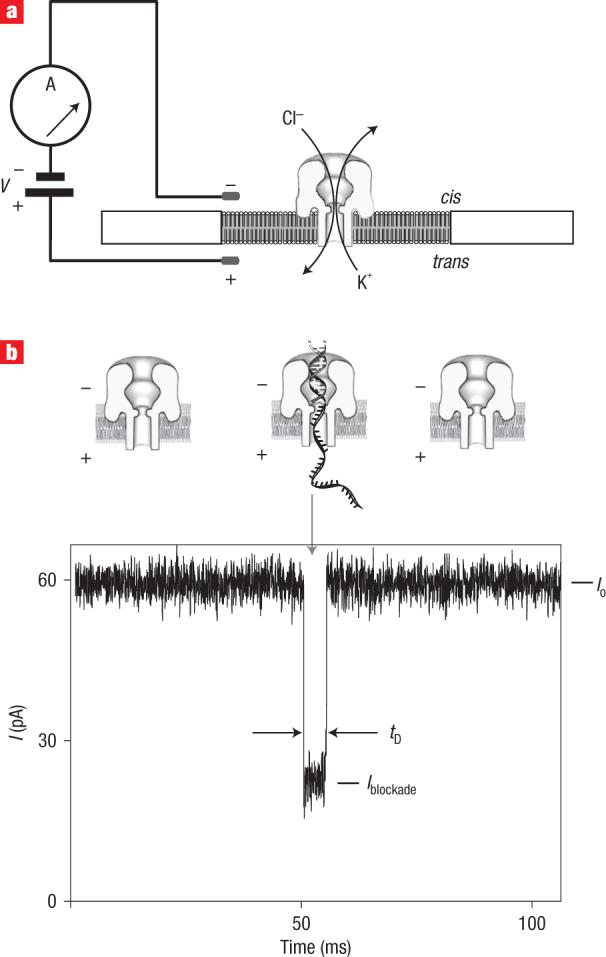
a, Schematic of the nanopore device. Voltage is applied across a single α-haemolysin channel inserted in a ∼25-μm-diameter lipid bilayer (trans-side positive). Current through the nanopore is carried by K+ and Cl− ions (0.3 M bulk phase concentration). b, Current trace for the primer/template DNA being captured, and then translocated through the nanopore under an applied voltage (bottom). An illustration of the events that yield the three sections of the current trace is shown over the trace. Io is the open channel current, I is the average current level for the blockade event, and tD is the translocation dwell time.
We used a nanopore instrument to probe the interaction of the Klenow fragment (KF) of Escherichia coli DNA polymerase I with its DNA substrate. This substrate is a duplex DNA formed by base-pairing of a short ssDNA primer with a longer template DNA. The KF catalyses DNA replication by the sequential addition of nucleotides to the primer strand, dictated by Watson–Crick complementarity to the template strand20. In contrast with earlier studies examining Exonuclease I/DNA complexes4 and EcoRI/DNA complexes5, our nanopore analysis of the KF binary (DNA/polymerase) and ternary (DNA/polymerase/dNTP) complexes was performed in the presence of Mg2+. This cofactor is essential for KF catalytic function. It is present in extensive biochemical and kinetic studies characterizing the KF reaction cycle20,21, and in X-ray crystal structures for binary complexes (DNA/KF)22,23, and ternary complexes23,24.
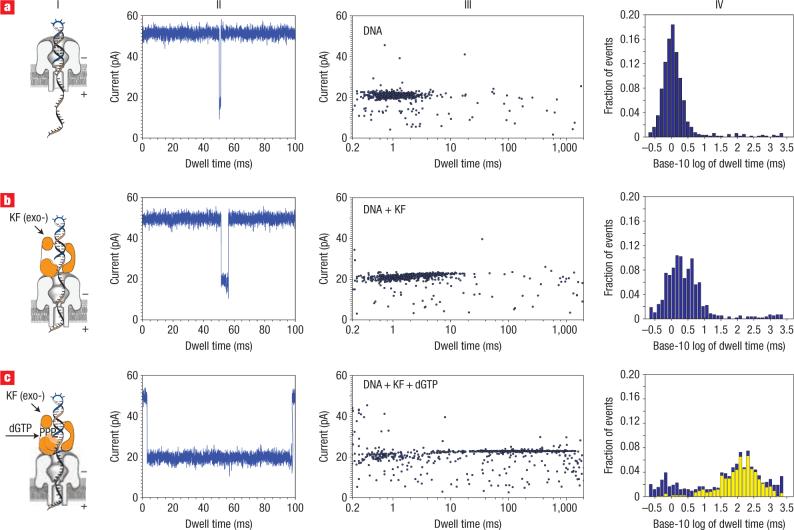
Capture and translocation of a model DNA template (14 bp hairpin with a 36-nucleotide 5′ overhang and 2′–3′ dideoxycytidine terminus) resulted in a cluster of events with a median duration of 1 ms and an average blockade amplitude I = 20 pA (Fig. 2a). When the KF (2 μM) was subsequently added to the cis compartment under conditions where catalytic activity had been demonstrated in separate experiments (see Supplementary Information, Fig. S1), a second population of events emerged with a 3-ms median dwell time and a higher blockade current (I = 23 pA, Fig. 2b). This class of events is enzyme-concentration-dependent (see Supplementary Information, Fig. S2 and Table S1), consistent with nanopore capture of a DNA/KF binary complex.
Figure 2. Distinguishing DNA, DNA/KF complexes or DNA/KF/dNTP complexes in the nanopore device.
a, Translocation, through the nanopore, of DNA alone (14-bp hairpin with a 36-nucleotide 5′ overhang and 2′–3′ dideoxycytidine terminus; template base at n = 0 is C). b,c, Translocation of the 14-bphp from complexes with KF (b) or from complexes with KF and dGTP (c). For each of rows a–c are presented a diagram of the nanopore with the associated complex (column I), a current trace (column II) and a dwell time event plot (column III). In column IV, probability histograms of the base-10 logarithm of dwell time data are shown in blue. Close examination of the event plot in c, column III, reveals that most long dwell time events are within 22 to 24 pA. A yellow subset histogram for the events within 22−24 pA is overlaid on probability histogram c, revealing that the chosen range is dominated by long dwell time events. The number of events, mean and IQR values for probability histograms a and b, and subset histogram c, are reported in Table 1.
Addition of a deoxynucleotide triphosphate (dNTP) substrate complementary to the template base in the KF catalytic site results in the formation of a higher stability ternary complex with primer/template substrates bearing a 2′–3′ dideoxy terminus25. Thus, correct addition of dNTP would be predicted to cause an increase in the median event dwell time relative to the putative binary complex. The modified primer terminus prevents enzymatic extension, exploiting the classic strategy of Sanger sequencing26. We found that the addition of deoxyguanosine triphosphate (dGTP)— the complement to deoxycytidine (dC), which is the first unpaired base of the template strand (defined as n = 0)— resulted in a large increase in the dwell time of the DNA capture events (median dwell time = 133 ms, Fig. 2c). The emergence of a population of events with a 133-ms median dwell time and the concomitant decrease in the population of events characterized by the 3-ms median dwell time both depended upon dNTP concentration (see Supplementary Information, Fig. S2 and Table S1).
For template-directed polymerases including KF, dNTP substrate discrimination is dictated by detection and amplification of the Watson–Crick complementarity between the template and the incoming dNTP (ref. 21). We therefore predicted that only the addition of complementary dNTP, and not the three non-complementary dNTPs, would yield the cluster of longer dwell time events at ∼24 pA amplitude (Fig. 2c).
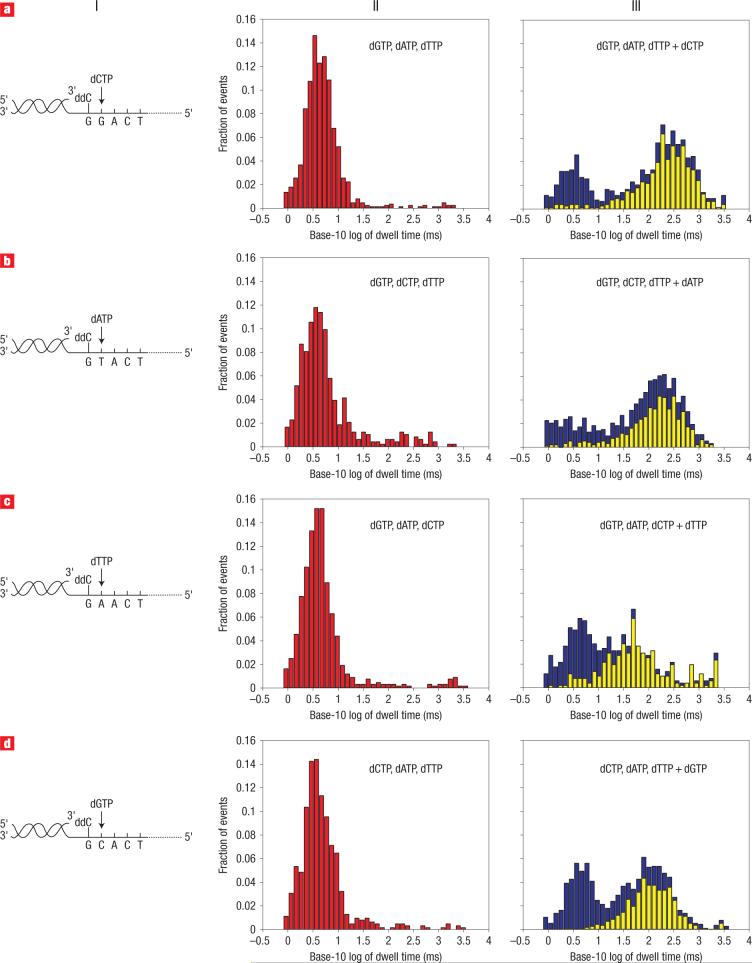
To test this prediction, we examined a series of DNA duplexes comprised of 100 nt templates annealed to 21 nt primers with 2′–3′ dideoxycytidine termini (Fig. 3, column I). The four DNA constructs were identical except for the template nucleotide at the ssDNA–dsDNA junction (that is, dG, dC, dA, dT at n = 0 in the KF catalytic site). In all four cases, addition of non-complementary dNTPs to the cis compartment containing primer/template, Mg2+ and KF resulted in a cluster of events with an average blockade current I = 20 pA and a median dwell time of ∼4 ms (Fig. 3, column II; Table 1). When the correct dNTP was subsequently added to each appropriate mixture, a subpopulation of events emerged that had an average amplitude of I = 23−24 pA, with median dwell times of 242, 159, 51 and 111 ms for dCTP, dATP, dTTP and dGTP respectively (Fig. 3, column III; Table 1). These event clusters were, in fact, very similar to those measured for the ternary complex assembled with dGTP using the 14 bphp (14 bp hairpin) template (Fig. 2c). In the absence of Mg2+, the longer dwell time cluster was not observed despite the presence of all four dNTPs (median dwell time = 2.6 ms). This is consistent with the critical role of two Mg2+ ions in the formation of the high-affinity KF catalytic domain20,23,27. We conclude that the nanopore assay allows the identification of a ternary complex between the KF, the primer/template and the dNTP substrate in a sequence-dependent manner.
Figure 3. Detection of correct dNTP binding to the KF/primer-template complex.
Column I depicts four primer/template hybrids, which differ only in the template base (position n = 0). The primer has a 3′ ddC terminus, and the correct incoming dNTP, complementary to n = 0, is indicated. a–d, Translocation event dwell time histograms when n = 0 is G (a), T (b), A (c) and C (d), respectively. Column II shows probability histograms of event dwell times for primer/template pairs in the presence of KF and all three incorrect dNTPs (in red). Column III shows probability histograms after the fourth complementary dNTP was added (in blue). Most long dwell time events are within the range of −35 to −32 pA (a,b,d) and −41 to −37 pA (c). Subset histograms for the events within these ranges (in yellow) are overlaid on the probability histograms. The number of events, mean and IQR values for probability histograms (II) and subset histograms (III) are reported in Table 1. Because the histograms in this figure are based on data from multiple experiments, blockade events are reported relative to a 0 pA open channel current. Consequently, the reported blockade amplitudes fall within the −50 to −20 pA range.
Table 1.
| Components | Template base | No. of events | Median (ms) | IQR (ms) | Figure no. |
|---|---|---|---|---|---|
| DNA 1a | C | 686 | 1.1 | 1.0 | 2a, IV |
| DNA 1, KF | C | 589 | 2.0 | 3.3 | 2b, IV |
| DNA 1, KF, dGTPc | C | 525 | 133.3 | 215.5 | 2c, IV |
| DNA 2b, KF, dCTP, dATP, dTTP | C | 618 | 4.0 | 4.1 | 3d, II |
| DNA 2, KF, dCTP, dATP, dTTP + dGTPd | C | 490 | 110.8 | 167.7 | 3d, III |
| DNA 2, KF, dGTP, dATP, dCTP | A | 686 | 3.9 | 3.5 | 3c, II |
| DNA 2, KF, dGTP, dATP, dCTP + dTTPe | A | 276 | 51.0 | 102.8 | 3c, III |
| DNA 2, KF, dGTP, dCTP, dTTP | T | 484 | 4.1 | 5.1 | 3b, II |
| DNA 2, KF, dGTP, dCTP, dTTP + dATPd | T | 460 | 158.8 | 248.1 | 3b, III |
| DNA 2, KF, dGTP, dATP, dTTP | G | 906 | 4.3 | 3.9 | 3a, II |
| DNA 2, KF, dGTP, dATP, dTTP + dCTPd | G | 472 | 242.0 | 359.2 | 3a, III |
DNA 1 denotes the 14-bphp template;
DNA 2 denotes the primer/template hybrids depicted in Fig. 3a;
Values based on a data subset (yellow bins Fig. 2c, column IV) with amplitude range of 22−24 pA;
Values based on data subsets (yellow bins Fig. 3, column III) with amplitude range of −35 to −32 pA;
Values based on a data subset (yellow bins Fig. 3c, column III) with amplitude range of −41 to −37 pA. Correct incoming dNTPs are indicated in bold italic text.
A possible mechanistic explanation for the higher blockade currents associated with KF complexes emerged from close inspection of the raw traces. For example, a representative trace for a ternary complex (Fig. 4a) was characterized by a long segment (∼90 ms) at 24 pA, followed by a shorter terminal segment (∼5 ms) at 20 pA amplitude. This two-step process was observed in more than 97% of the ternary complex events in Fig. 3. We reasoned that the first portion of this current pattern represents the initial capture of the ternary complex, with the duplex DNA segment perched outside the pore vestibule in association with the polymerase (Fig. 4a, i). Upon dissociation of the ternary complex, the duplex segment slips down into the vestibule and causes a current decrease (Fig. 4a, ii). This series of events must occur prior to translocation of the template and primer as single-stranded molecules (Fig. 4a, iii).
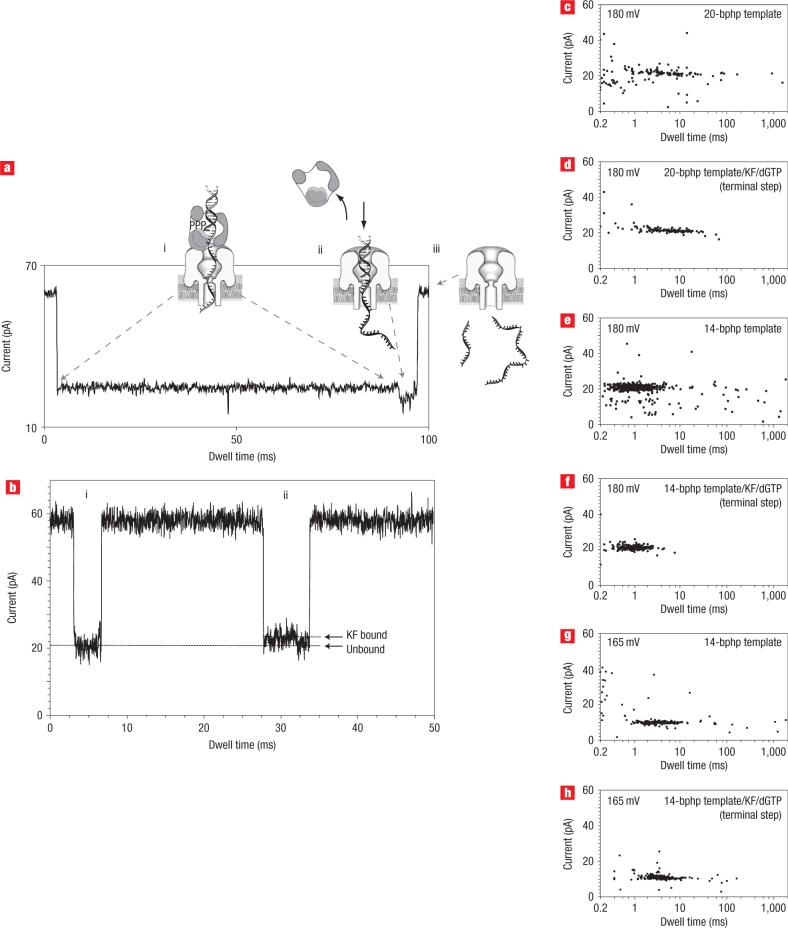
Figure 4. Proposed mechanism for voltage-facilitated dissociation of DNA from KF/DNA or KF/DNA/dNTP complexes.
a, Representative trace at 180 mV showing the current levels associated with nanopore capture of the ternary complex, and illustration of the molecular events proposed to occur at each step. i, The long 24 pA blockade arises from the initial capture of the ternary complex, with the duplex DNA held on top the pore vestibule because of association with the KF. ii, The shorter 20 pA terminal step occurs upon KF dissociation, when duplex DNA is pulled into the nanopore vestibule. iii, Finally, primer and template strands are dissociated, translocating through the nanopore as single-stranded molecules and resulting in a return to the open channel current. b, Current trace from an experiment with DNA and KF in the nanopore chamber, showing two translocation events. i, Trace shows the 20 pA signature characteristic of events in the presence of DNA only. ii, Trace is characteristic of events that only occur in the presence of both KF and DNA (without the correct dNTP). c–h, Two-dimensional plots show event clusters for 20-bp DNA hairpin translocation at 180 mV (c); 20-bp DNA hairpin/KF/dGTP complex terminal step at 180 mV (d); 14-bp DNA hairpin translocation at 180 mV (e); 14-bp DNA hairpin/KF/dGTP complex terminal step at 180 mV (f); 14-bp DNA hairpin translocation at 165 mV (g); 14-bp DNA hairpin/KF/dGTP complex terminal step at 165 mV (h).
This model makes three testable predictions. (1) The dwell time and amplitude of the terminal step extracted from ternary complex events in Fig. 3 must correspond to values for capture of the unbound primer/template duplex. (2) The two-step mechanism should be observed for the binary complex as well as the ternary complex. (3) The duration of the terminal step (Fig. 4a, ii) must scale with the length of the duplex region of the captured DNA molecules.
To test the first prediction, we measured the duration of the terminal step in the current traces for the ternary complex events summarized in Fig. 3a (column III). The median duration for these terminal steps was 4 ms, compared with 4 ms for the 21-bp primer/template events summarized in Fig. 3a (column II). Average blockade currents I were 20 pA in both cases. To test the second prediction, we re-examined the binary complex data from Fig. 2b. The current traces in the presence of KF and DNA revealed that more than 90% of events were characterized by two sequential current states (Fig. 4b, ii), the first centred at 23 pA, followed by a brief 20 pA step. In the presence of DNA alone, less than 1% of events fit this pattern. To test the third prediction, we compared blockade events for a 20-bp DNA hairpin to events for a 14-bp DNA hairpin. The median dwell times for DNA alone, captured in the pore at 180 mV, decreased from 4 ms (20 bphp) to 1 ms (14 bphp) (Fig. 4c,e), as did the median duration for terminal steps of their respective ternary complexes (Fig. 4d,f). The blockade current amplitudes averaged ∼20 pA in each case.
It remained a formal possibility that the terminal current step was not due to KF dissociation just before DNA translocation across the nanopore, as modeled in Fig. 4a, but instead was due to the exit of the complex from the nanopore into the cis compartment. To discriminate between these two possibilities, we repeated the 14-bp hairpin experiments shown in Fig. 4e,f, but at a reduced voltage (165 mV in Fig. 4g,h). At 165 mV, the force acting on the DNA in the nanopore is reduced relative to the force at 180 mV. The model shown in Fig. 4a predicts that the terminal step duration would increase when the force pulling DNA to the trans compartment is reduced. In contrast, the terminal step duration should decrease as force is reduced if complex release into the cis compartment accounts for the terminal current step. When voltage was reduced from 180 mV to 165 mV, the median dwell time for the terminal step increased from 1 ms (Fig. 4f ) to 3.8 ms (Fig. 4 h). Similarly, the median dwell time for the 14-bp hairpin DNA alone increased, from 1 ms at 180 mV (Fig. 4e) to 3.2 ms at 165 mV (Fig. 4 g). This increase in the duration of the terminal step as voltage is reduced is consistent with the model in Fig. 4a, and is inconsistent with release of the complex into the cis compartment.
Our demonstration that three different molecular states (unbound DNA, DNA/KF binary complex, DNA/KF/dNTP ternary complex) can be distinguished by current amplitude and dwell time depended upon post-experiment analysis. To extend our ability to probe mechanistic features of molecular complexes, and to exert the rapid, precise manipulation likely to be required for applications such as DNA sequencing, we sought to acquire real-time control of single molecules in the nanopore device28.
To accomplish this, we built on earlier nanopore computational work8,29 to develop a finite-state machine (FSM), which is a logic construct composed of a finite number of discrete states, transitions between those states, and commands executed in response to those states and transitions30. Measurements determine which state the system is in, and therefore which command is executed. We implemented FSM logic on a field-programmable gate array (FPGA), digital hardware that is reprogrammable and offers fast reaction times31.
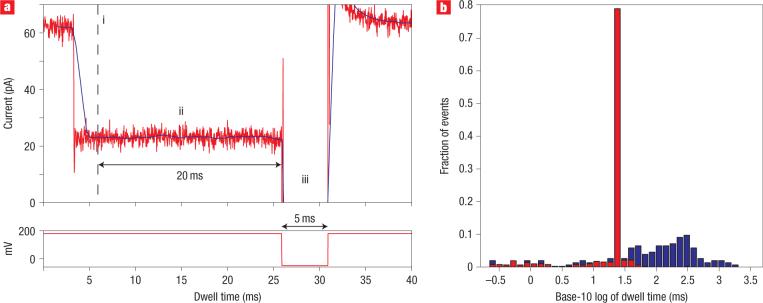
In an initial test of FSM/FPGA control (Fig. 5), our objective was to detect ternary complex events, one at a time, and reduce the dwell time of each event from 100−150 ms to 20−23 ms. For this experiment we added 14-bphp DNA, KF and dGTP to the nanopore cis compartment. We programmed FSM logic to detect the 24 pA current blockade characteristic of KF bound DNA, and then to continuously monitor the current level for 20 ms. This 20-ms period is longer than the binary complex events, and thus identifies ternary complex capture. After 20 ms, the FSM executed a programmed voltage reversal, ejecting the complex from the nanopore before translocation could occur. Dwell time probability histograms of events within the 22−25 pA amplitude range from this experiment (Fig. 5b) reveal that FSM/FPGA control condensed the ternary complex events from a median of 123 ms (235 ms interquartile range, IQR) to a median of 23 ms (0.3 ms IQR).
Figure 5. Recognition and control of DNA complexes in real time using FPGA.
a, A representative ternary complex event under FPGA control. The FPGA samples the ionic current every 5.3 μs and computes a windowed mean amplitude (solid dark line) based on the previous 1.5 ms of signal. Every 0.4 ms, it tests whether or not the mean is within 24 ± 2.8 pA. i, If the mean remains within this range for four consecutive tests, the FSM logic diagnoses the blockade as a KF binding event. The total delay for diagnosis of a KF binding event is 2.7 ms (1.5 ms for the windowed mean to enter the 24 ± 2.8 pA range, plus 1.2 ms for three consecutive subsequent tests). ii, Upon diagnosis of a KF binding event, the FPGA continues to monitor the windowed mean. If the mean remains in the 24 ± 2.8 pA range for 20 ms, the FSM logic diagnoses the blockade as resulting from a ternary complex. The 20 ms cutoff was used because 60% of events are longer than 20 ms in the presence of the correct dNTP, but only 2% of events are longer than 20 ms in the 24 ± 2.8 pA range in the absence of the correct dNTP. iii, Upon diagnosis that a ternary complex is in the pore, the FPGA reverses the voltage to −50 mV for 5 ms, ejecting the complex from the pore. The 180 mV capture voltage is then restored. For each event in which KF binding is not diagnosed, or in which KF binding is diagnosed but the mean leaves the 24 ± 2.8 pA range before 20 ms, the voltage is kept at 180 mV. b, Dwell time probability histograms for 24 ± 2.8 pA events with FPGA control (527 total events, in red) and without FPGA control (155 total events, in blue).
We have demonstrated the ability to distinguish among current signatures for unbound DNA, DNA/KF binary complexes and DNA/KF/dNTP ternary complexes using a nanopore device. The identity of DNA template bases in single KF ternary complexes was faithfully reported. Real-time discrimination and control of single molecules, achieved by integrating the nanopore device with FSM logic, can be used for probing mechanistic features of other enzymes that bind or modify DNA or RNA, including polymerases, exonucleases and kinases.
METHODS
NANOPORE EXPERIMENTS
The Klenow fragment (exo-) was obtained from New England Biolabs (100,000 U ml−1; specific activity 20,000 U mg−1). DNA oligonucleotides were synthesized by Operon Biotechnologies or Integrated DNA Technologies, and were purified by denaturing PAGE. The DNA oligonucleotides are listed in the following. In all cases, the base at the 2′–3′ terminus of the primer was dideoxycytidine (ddC).
14 bphp: 5′–ACT ATC ATT ATC TAC ATC CAT TAC ATC ACT ACT CCG CAT GCA GGT AGC CTT TTG GCT ACC TGC ATG ddC–3′
20 bphp: 5’–CTC ACC TAT CCT TCC ACT CAT TTT CCT TAA CCA TCG CAT TCT CAT GCA GGT AGC CTT TTG GCT ACC TGC ATG AGA ATG ddC–3′
21mer primer: 5′–GGT ATG GTA GCG TGA GGA GTddC–3′
100mer templates: 5′–CTC ACC TAT CCT TCC ACT CAT TTT CCT TAA CCA TTT CAT TCA CCC ATC TCA CTA TCA TTA TCT ACA TCC ATT ACA TCA NGA CTC CTC ACG CTA CCA TAC C–3′
Four versions of the 100mer template were used. These DNA templates were identical except for the base immediately following the primer–template junction (designated N in the sequence above), which was dC, dA, dG, or dT.
Nanopore experiments were conducted at 23 °C in 10 mM HEPES/KOH, pH 8.0 ± 0.05, 0.3 M KCl and 5 mM MgCl2, except for controls without Mg2+, where MgCl2 was replaced by 0.5 mM ethylenediamine tetraacetic acid (EDTA). We chose 0.3 M KCl in this study because it supports KF catalytic function (see Supplementary Information, Fig. S1). Unlike solid-state pores where DNA capture increases current at less than 0.4 M KCl (ref. 32), DNA capture impedes ionic current at 0.3 M KCl for the α-haemolysin pore. DNA, KF and dNTPs were added to the cis chamber as indicated in the figures and tables. Note that dNTP refers to any of four possible deoxynucleotide triphosphates where N = G, A, T or C. Except where otherwise indicated, DNA was used at a final concentration of 1 μM, KF at 2 μM and dNTPs at 200 μM. Primer/template hybrids were annealed by heating to 95 °C for 2 min and cooling slowly to 25 °C before use. Hairpin-containing DNA oligomers were annealed by heating to 95 °C for 2 min then cooling rapidly on ice to prevent interstrand hybridization.
The nanopore chamber design and methods to insert single α-haemolysin channels have been described in detail elsewhere8,10. Ionic current flux through the α-haemolysin channel was measured using an Axopatch 200B integrating patch-clamp amplifier (Molecular Devices) in voltage-clamp mode. Data were acquired with a Digidata 1440A analog-to-digital converter (Molecular Devices) at 20-ms intervals in the whole cell configuration, and filtered at 10 kHz using a low-pass Bessel filter unless otherwise noted. Current blockades were examined at 180 mV (trans-positive) for the constant-voltage experiments.
FPGA EXPERIMENTS
In the FPGA control experiments, events corresponding to the current signatures of molecular complexes in the nanopore chamber were diagnosed and responded to using the National Instruments PCI-7831R FPGA board, which was programmed using LabVIEW 8 software. Current amplitude values for state-detection logic were determined from the current signatures for DNA alone or in complexes with KF, measured at 180 mV constant voltage. The mean and standard deviation current amplitude values were calculated to quantify the most likely amplitude level, and the spread around that level.
DATA ANALYSIS
Two-dimensional event plots display blockade amplitudes in picoamperes on the vertical axis and dwell times in milliseconds on the logarithmic horizontal axis. Each point represents capture and translocation of one DNA molecule. For duplex DNA, the plotted dwell time range of 0.8−4,000 ms contains all relevant DNA and DNA–KF events. For DNA hairpins, shearing occurs in approximately 1 ms, therefore the plotted range is expanded to 0.2−4,000 ms. Events longer than 4 s indicate that DNA is stuck inside the nanopore. The plotted amplitude range is typically 0−50 pA. For histograms generated by aggregating data from multiple experiments, blockade events were reported relative to a 0 pA open channel reference current. In those cases, the histograms were based on data between −50 and −20 pA.
Histograms were generated by binning the base-10 logarithm of event dwell times using 36 bins. The dwell time probability histogram was generated by plotting each bin normalized by the total number of events, revealing the fraction of events in each bin. By normalizing, the area under the probability histogram sums to one. To examine the contribution of a subset of events that fall within a restricted current amplitude range, a subset histogram for only those events was generated and overlaid on the probability histogram. To preserve the fraction-of-events scale, each bin in the subset histogram was normalized by the total number of events in the probability histogram. To maintain alignment of the bins, 36 bins kept at the original bin centre locations were used for the subset histogram. Visually, the resulting subset histograms overlaid on the original probability histograms reveal the fraction of events that fall within the restricted current amplitude range and contribute to each respective bin.
We used median and IQR values to calculate the most likely dwell time and the spread for each data set. The IQR is the range between the 75th percentile and the 25th percentile. Thus, the IQR includes about 50% of the data and is a measure of statistical dispersion that minimizes the effect of outliers.
Acknowledgements
We wish to thank D. Branton, V. Tabard-Cossa, M. Wiggin, D. Krapf and R. Mathies for reading early versions of this manuscript. We also thank A. Kottas for guidance on statistical analysis. This work was supported by National Institutes of Health grants GM073617-01A1 (M.A.), HG003703-01 (M.A., D.D.) and HG004035-01 (W.D.).
Footnotes
Supplementary information accompanies this paper on www.nature.com/naturenanotechnology.
Supplementary Material
References
- 1.Deamer DW, Branton D. Characterization of nucleic acids by nanopore analysis. Acc. Chem. Res. 2002;35:817–825. doi: 10.1021/ar000138m. [DOI] [PubMed] [Google Scholar]
- 2.Dekker C. Solid-state nanopores. Nature Nanotechnol. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 3.Siwy Z, et al. Protein biosensors based on biofunctionalized conical gold nanotubes. J. Am. Chem. Soc. 2005;127:5000–5001. doi: 10.1021/ja043910f. [DOI] [PubMed] [Google Scholar]
- 4.Hornblower B, et al. Single-molecule analysis of DNA–protein complexes using nanopores. Nature Methods. 2007;4:315–317. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Q, et al. Detecting SNPs using a synthetic nanopore. Nano Lett. 2007;7:1680–1685. doi: 10.1021/nl070668c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl Acad. Sci. USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song LZ, et al. Structure of Staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 8.Vercoutere W, et al. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nature Biotechnol. 2001;19:248–252. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 9.Vercoutere WA, et al. Discrimination among individual Watson–Crick base pairs at the termini of single DNA hairpin molecules. Nucleic Acids Res. 2003;31:1311–1318. doi: 10.1093/nar/gkg218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys. J. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashkenasy N, Sanchez-Quesada J, Bayley H, Ghadiri MR. Recognizing a single base in an individual DNA strand: A step toward DNA sequencing in nanopores. Angew. Chem. Int. Edn. 2005;44:1401–1404. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howorka S, Cheley S, Bayley H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nature Biotechnol. 2001;19:636–639. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 13.Butler TZ, Gundlach JH, Troll MA. Determination of RNA orientation during translocation through a biological nanopore. Biophys. J. 2006;90:190–199. doi: 10.1529/biophysj.105.068957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Unzipping kinetics of double-stranded DNA in a nanopore. Phys. Rev. Lett. 2003;90:23801. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 15.Storm AJ, et al. Fast DNA translocation through a solid-state nanopore. Nano Lett. 2005;5:1193–1197. doi: 10.1021/nl048030d. [DOI] [PubMed] [Google Scholar]
- 16.Nakane J, Akeson M, Marziali A. Evaluation of nanopores as candidates for electronic analyte detection. Electrophoresis. 2002;23:2592–2601. doi: 10.1002/1522-2683(200208)23:16<2592::AID-ELPS2592>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 17.Li J, et al. Ion-beam sculpting at nanometre length scales. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 18.Gu L-Q, Cheley S, Bayley H. Capture of a single molecule in a nanocavity. Science. 2001;291:636–640. doi: 10.1126/science.291.5504.636. [DOI] [PubMed] [Google Scholar]
- 19.Iqbal SM, Akin D, Bashir R. Solid-state nanopore channels with DNA selectivity. Nature Nanotechnol. 2007;2:243–248. doi: 10.1038/nnano.2007.78. [DOI] [PubMed] [Google Scholar]
- 20.Joyce CM, Steitz TA. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 21.Joyce CM, Benkovic SJ. DNA polymerase fidelity: Kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 22.Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: Structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl Acad. Sci. USA. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purohit V, Grindley NDF, Joyce CM. Use of 2-aminopurine fluorescence to examine conformational changes during nucleotide incorporation by DNA polymerase I (Klenow fragment). Biochemistry. 2003;42:10200–10211. doi: 10.1021/bi0341206. [DOI] [PubMed] [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steitz TA, Smerdon SJ, Jager J, Joyce CM. A unified polymerase mechanism for nonhomologous DNA and RNA polymerases. Science. 1994;266:2022–2025. doi: 10.1126/science.7528445. [DOI] [PubMed] [Google Scholar]
- 28.Bates M, Burns M, Meller A. Dynamics of DNA molecules in a membrane channel probed by active control techniques. Biophys. J. 2003;84:2366–2372. doi: 10.1016/S0006-3495(03)75042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winters-Hilt S, et al. Highly accurate classification of Watson–Crick basepairs on termini of single DNA molecules. Biophys. J. 2003;84:967–976. doi: 10.1016/S0006-3495(03)74913-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gill A. Introduction to the Theory of Finite-State Machines. McGraw-Hill; 1962. [Google Scholar]
- 31.Trimberger SM. Field-Programmable Gate Array Technology. Springer; 1994. [Google Scholar]
- 32.Smeets RMM, et al. Salt dependence of ion transport and DNA translocation through solid-state nanopores. Nano Lett. 2006;6:89–95. doi: 10.1021/nl052107w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.