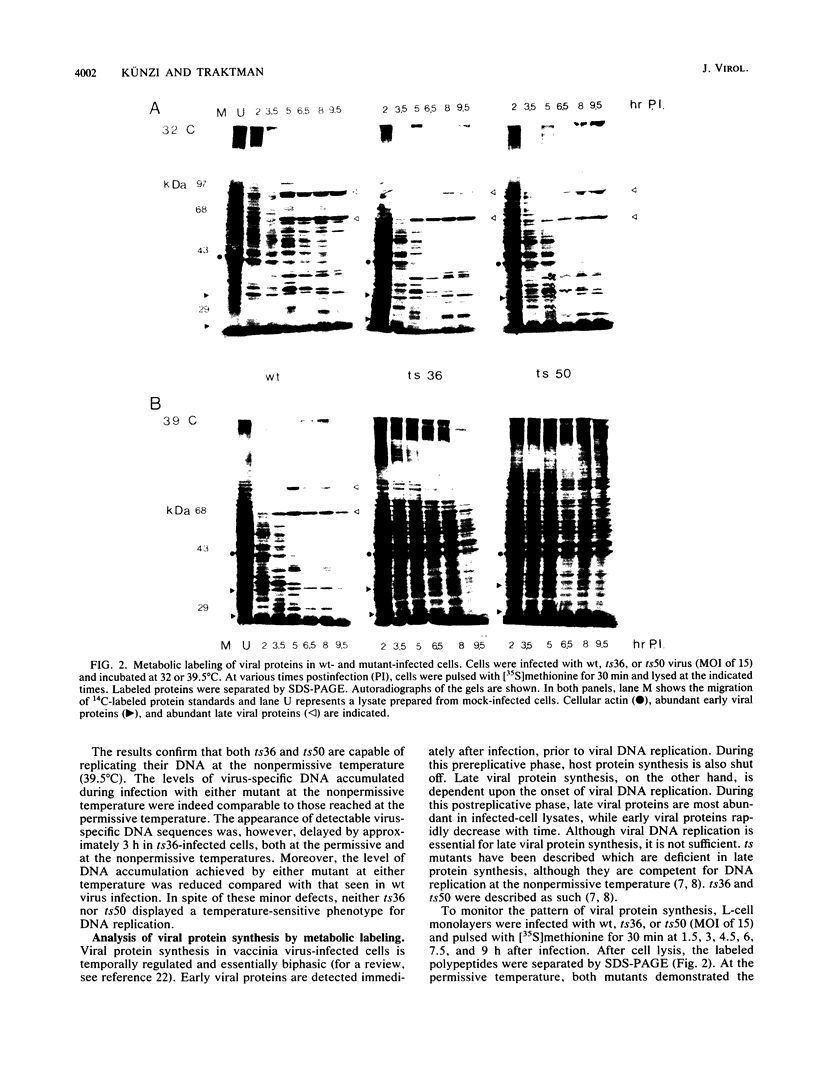
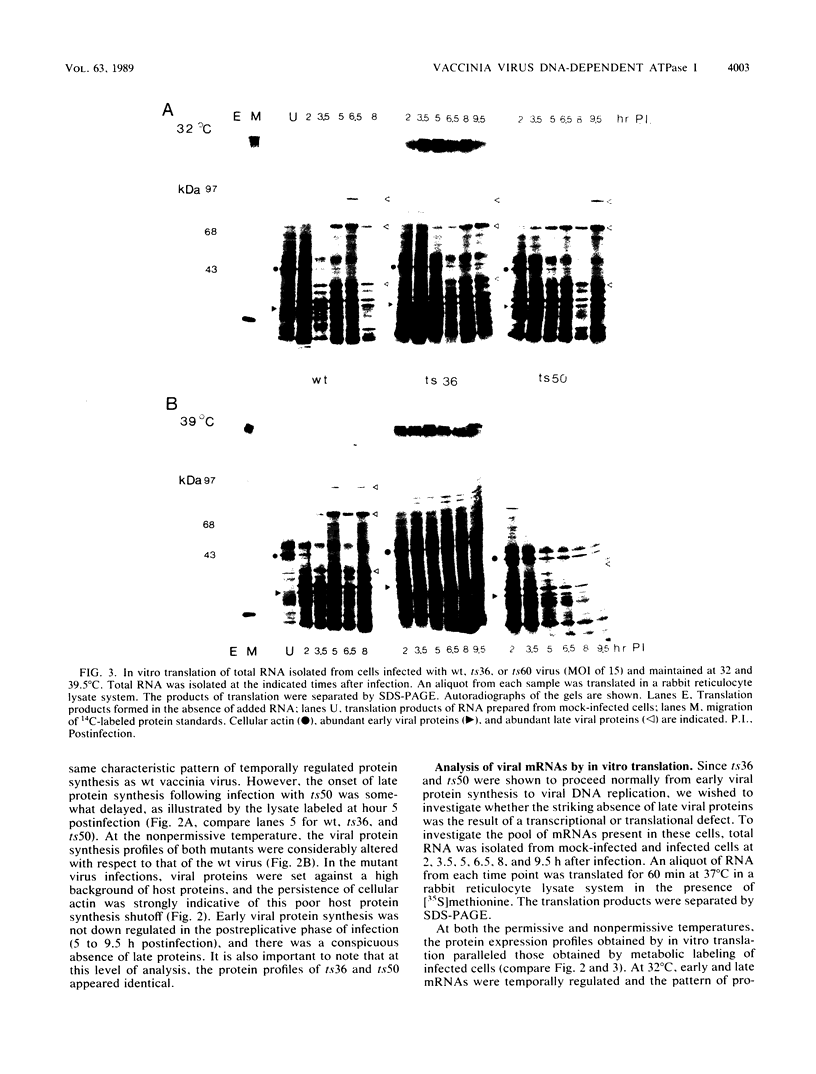
Abstract
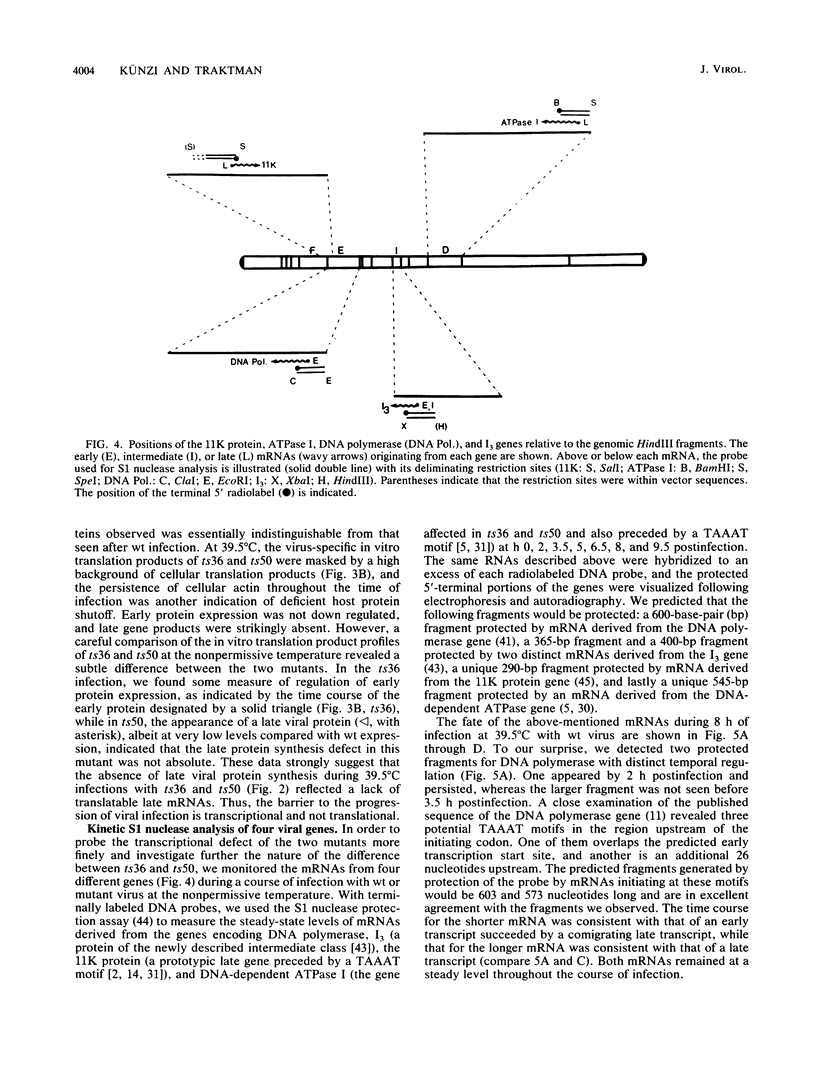
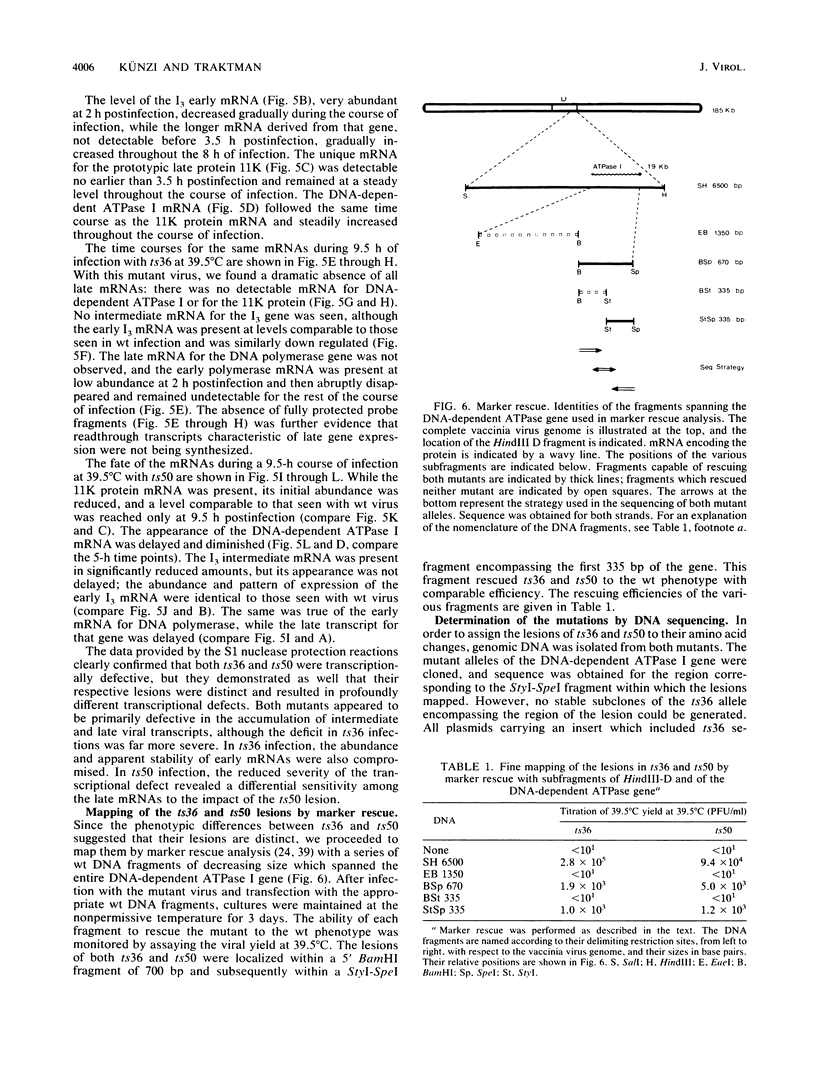
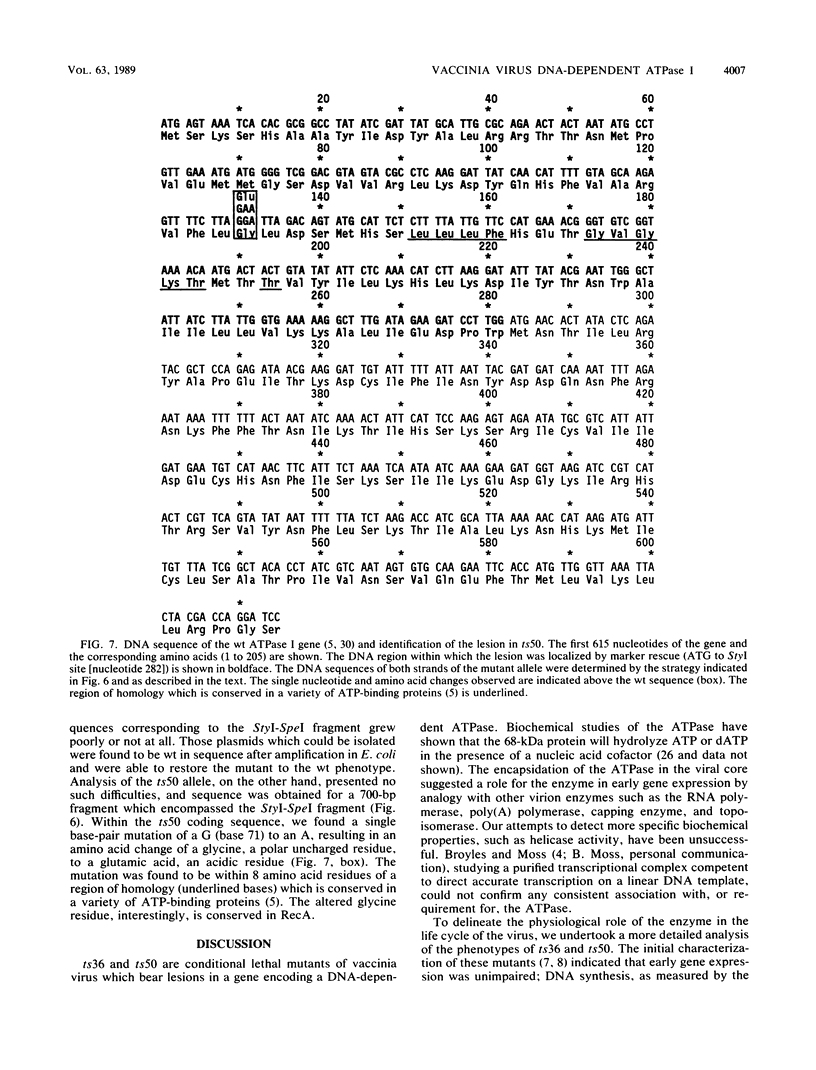
To delineate the role of the vaccinia virus-encapsidated DNA-dependent ATPase I in the life cycle of the virus, we performed a detailed study of two temperature-sensitive mutants with lesions in the gene encoding the enzyme. Profiles of viral DNA and protein accumulation during infection showed the mutants to be competent for DNA synthesis but deficient in late protein synthesis, confirming their defective late phenotype (R. C. Condit and A. Motyczka, Virology 113:224-241, 1981: R. C. Condit, A. Motyczka, and G. Spizz, Virology 128:429-443, 1983). In vitro translation of viral RNA and S1 nuclease mapping of selected mRNAs demonstrated that the deficit in late protein synthesis stemmed from a defect in the transcriptional machinery. Intermediate and late gene expression appeared to be most affected. The transcriptional defect was of unequal severity in the two mutants. However, their phenotypes were indistinguishable and their respective lesions were mapped to the same 300 nucleotides at the 5' end of the gene. DNA sequence analysis assigned a single nucleotide and amino acid change to one of the mutants.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abramson R. D., Dever T. E., Lawson T. G., Ray B. K., Thach R. E., Merrick W. C. The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J Biol Chem. 1987 Mar 15;262(8):3826–3832. [PubMed] [Google Scholar]
- Bertholet C., Drillien R., Wittek R. One hundred base pairs of 5' flanking sequence of a vaccinia virus late gene are sufficient to temporally regulate late transcription. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2096–2100. doi: 10.1073/pnas.82.7.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. A., Dombroski A. J., Platt T. Transcription termination factor rho is an RNA-DNA helicase. Cell. 1987 Mar 27;48(6):945–952. doi: 10.1016/0092-8674(87)90703-3. [DOI] [PubMed] [Google Scholar]
- Broyles S. S., Moss B. DNA-dependent ATPase activity associated with vaccinia virus early transcription factor. J Biol Chem. 1988 Aug 5;263(22):10761–10765. [PubMed] [Google Scholar]
- Broyles S. S., Moss B. Identification of the vaccinia virus gene encoding nucleoside triphosphate phosphohydrolase I, a DNA-dependent ATPase. J Virol. 1987 May;61(5):1738–1742. doi: 10.1128/jvi.61.5.1738-1742.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles S. S., Moss B. Sedimentation of an RNA polymerase complex from vaccinia virus that specifically initiates and terminates transcription. Mol Cell Biol. 1987 Jan;7(1):7–14. doi: 10.1128/mcb.7.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit R. C., Motyczka A. Isolation and preliminary characterization of temperature-sensitive mutants of vaccinia virus. Virology. 1981 Aug;113(1):224–241. doi: 10.1016/0042-6822(81)90150-1. [DOI] [PubMed] [Google Scholar]
- Condit R. C., Motyczka A., Spizz G. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology. 1983 Jul 30;128(2):429–443. doi: 10.1016/0042-6822(83)90268-4. [DOI] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. Enzymes of general recombination. Annu Rev Biochem. 1987;56:229–262. doi: 10.1146/annurev.bi.56.070187.001305. [DOI] [PubMed] [Google Scholar]
- Crute J. J., Mocarski E. S., Lehman I. R. A DNA helicase induced by herpes simplex virus type 1. Nucleic Acids Res. 1988 Jul 25;16(14A):6585–6596. doi: 10.1093/nar/16.14.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl P. L., Jones E. V., Moss B. Homology between DNA polymerases of poxviruses, herpesviruses, and adenoviruses: nucleotide sequence of the vaccinia virus DNA polymerase gene. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3659–3663. doi: 10.1073/pnas.83.11.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensinger M. J. Phenotypic characterization of temperature-sensitive mutants of vaccinia virus with mutations in a 135,000-Mr subunit of the virion-associated DNA-dependent RNA polymerase. J Virol. 1987 Jun;61(6):1842–1850. doi: 10.1128/jvi.61.6.1842-1850.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geider K., Hoffmann-Berling H. Proteins controlling the helical structure of DNA. Annu Rev Biochem. 1981;50:233–260. doi: 10.1146/annurev.bi.50.070181.001313. [DOI] [PubMed] [Google Scholar]
- Hooda-Dhingra U., Thompson C. L., Condit R. C. Detailed phenotypic characterization of five temperature-sensitive mutants in the 22- and 147-kilodalton subunits of vaccinia virus DNA-dependent RNA polymerase. J Virol. 1989 Feb;63(2):714–729. doi: 10.1128/jvi.63.2.714-729.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänggi M., Bannwarth W., Stunnenberg H. G. Conserved TAAAT motif in vaccinia virus late promoters: overlapping TATA box and site of transcription initiation. EMBO J. 1986 May;5(5):1071–1076. doi: 10.1002/j.1460-2075.1986.tb04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübscher U., Stalder H. P. Mammalian DNA helicase. Nucleic Acids Res. 1985 Aug 12;13(15):5471–5483. doi: 10.1093/nar/13.15.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodadek T., Alberts B. M. Stimulation of protein-directed strand exchange by a DNA helicase. Nature. 1987 Mar 19;326(6110):312–314. doi: 10.1038/326312a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lasken R. S., Kornberg A. The primosomal protein n' of Escherichia coli is a DNA helicase. J Biol Chem. 1988 Apr 25;263(12):5512–5518. [PubMed] [Google Scholar]
- Matson S. W., George J. W. DNA helicase II of Escherichia coli. Characterization of the single-stranded DNA-dependent NTPase and helicase activities. J Biol Chem. 1987 Feb 15;262(5):2066–2076. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nakai H., Richardson C. C. Leading and lagging strand synthesis at the replication fork of bacteriophage T7. Distinct properties of T7 gene 4 protein as a helicase and primase. J Biol Chem. 1988 Jul 15;263(20):9818–9830. [PubMed] [Google Scholar]
- Nakano E., Panicali D., Paoletti E. Molecular genetics of vaccinia virus: demonstration of marker rescue. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1593–1596. doi: 10.1073/pnas.79.5.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
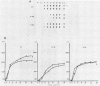
- Niles E. G., Condit R. C., Caro P., Davidson K., Matusick L., Seto J. Nucleotide sequence and genetic map of the 16-kb vaccinia virus HindIII D fragment. Virology. 1986 Aug;153(1):96–112. doi: 10.1016/0042-6822(86)90011-5. [DOI] [PubMed] [Google Scholar]
- Paolette E., Rosemond-Hornbeak H., Moss B. Two nucleid acid-dependent nucleoside triphosphate phosphohydrolases from vaccinia virus. Purification and characterization. J Biol Chem. 1974 May 25;249(10):3273–3280. [PubMed] [Google Scholar]
- Paoletti E., Moss B. Two nucleic acid-dependent nucleoside triphosphate phosphohydrolases from vaccinia virus. Nucleotide substrate and polynucleotide cofactor specificities. J Biol Chem. 1974 May 25;249(10):3281–3286. [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rodriguez J. F., Kahn J. S., Esteban M. Molecular cloning, encoding sequence, and expression of vaccinia virus nucleic acid-dependent nucleoside triphosphatase gene. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9566–9570. doi: 10.1073/pnas.83.24.9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosel J. L., Earl P. L., Weir J. P., Moss B. Conserved TAAATG sequence at the transcriptional and translational initiation sites of vaccinia virus late genes deduced by structural and functional analysis of the HindIII H genome fragment. J Virol. 1986 Nov;60(2):436–449. doi: 10.1128/jvi.60.2.436-449.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J. F., Stunnenberg H. G. Sequence and transcriptional analysis of the vaccinia virus HindIII I fragment. J Virol. 1988 Jun;62(6):1889–1897. doi: 10.1128/jvi.62.6.1889-1897.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Enomoto T., Yanagisawa J., Hanaoka F., Ui M. Further characterization of DNA helicase activity of mouse DNA-dependent adenosinetriphosphatase B (DNA helicase B). Biochemistry. 1988 Mar 8;27(5):1766–1771. doi: 10.1021/bi00405a057. [DOI] [PubMed] [Google Scholar]
- Stahl H., Dröge P., Knippers R. DNA helicase activity of SV40 large tumor antigen. EMBO J. 1986 Aug;5(8):1939–1944. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt B. L. Escherichia coli transcription termination protein rho has three hydrolytic sites for ATP. J Biol Chem. 1988 Aug 15;263(23):11130–11137. [PubMed] [Google Scholar]
- Sugino A., Ryu B. H., Sugino T., Naumovski L., Friedberg E. C. A new DNA-dependent ATPase which stimulates yeast DNA polymerase I and has DNA-unwinding activity. J Biol Chem. 1986 Sep 5;261(25):11744–11750. [PubMed] [Google Scholar]
- Sung P., Prakash L., Matson S. W., Prakash S. RAD3 protein of Saccharomyces cerevisiae is a DNA helicase. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8951–8955. doi: 10.1073/pnas.84.24.8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tengelsen L. A., Slabaugh M. B., Bibler J. K., Hruby D. E. Nucleotide sequence and molecular genetic analysis of the large subunit of ribonucleotide reductase encoded by vaccinia virus. Virology. 1988 May;164(1):121–131. doi: 10.1016/0042-6822(88)90627-7. [DOI] [PubMed] [Google Scholar]
- Traktman P., Kelvin M., Pacheco S. Molecular genetic analysis of vaccinia virus DNA polymerase mutants. J Virol. 1989 Feb;63(2):841–846. doi: 10.1128/jvi.63.2.841-846.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traktman P., Sridhar P., Condit R. C., Roberts B. E. Transcriptional mapping of the DNA polymerase gene of vaccinia virus. J Virol. 1984 Jan;49(1):125–131. doi: 10.1128/jvi.49.1.125-131.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos J. C., Stunnenberg H. G. Derepression of a novel class of vaccinia virus genes upon DNA replication. EMBO J. 1988 Nov;7(11):3487–3492. doi: 10.1002/j.1460-2075.1988.tb03224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittek R., Hänggi M., Hiller G. Mapping of a gene coding for a major late structural polypeptide on the vaccinia virus genome. J Virol. 1984 Feb;49(2):371–378. doi: 10.1128/jvi.49.2.371-378.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]