Abstract
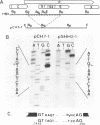
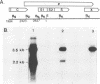
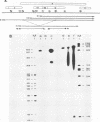
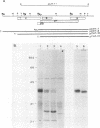
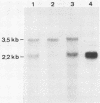
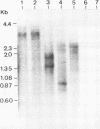
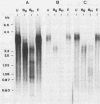
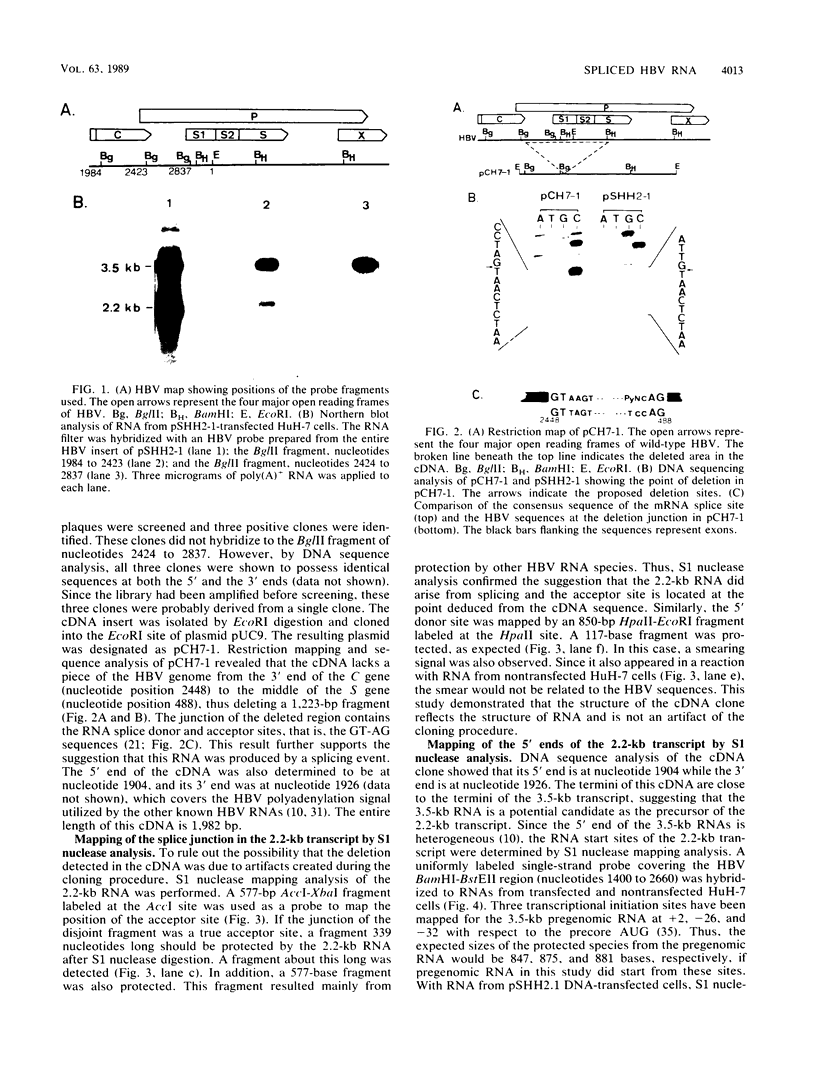
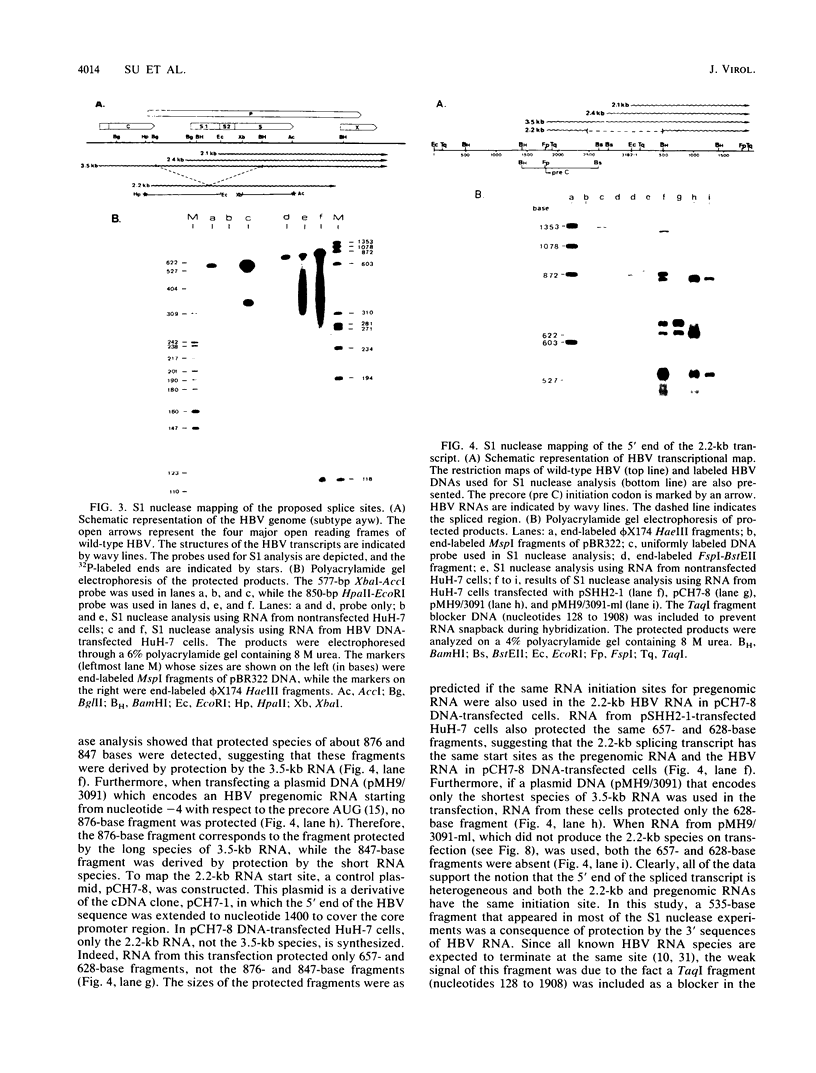
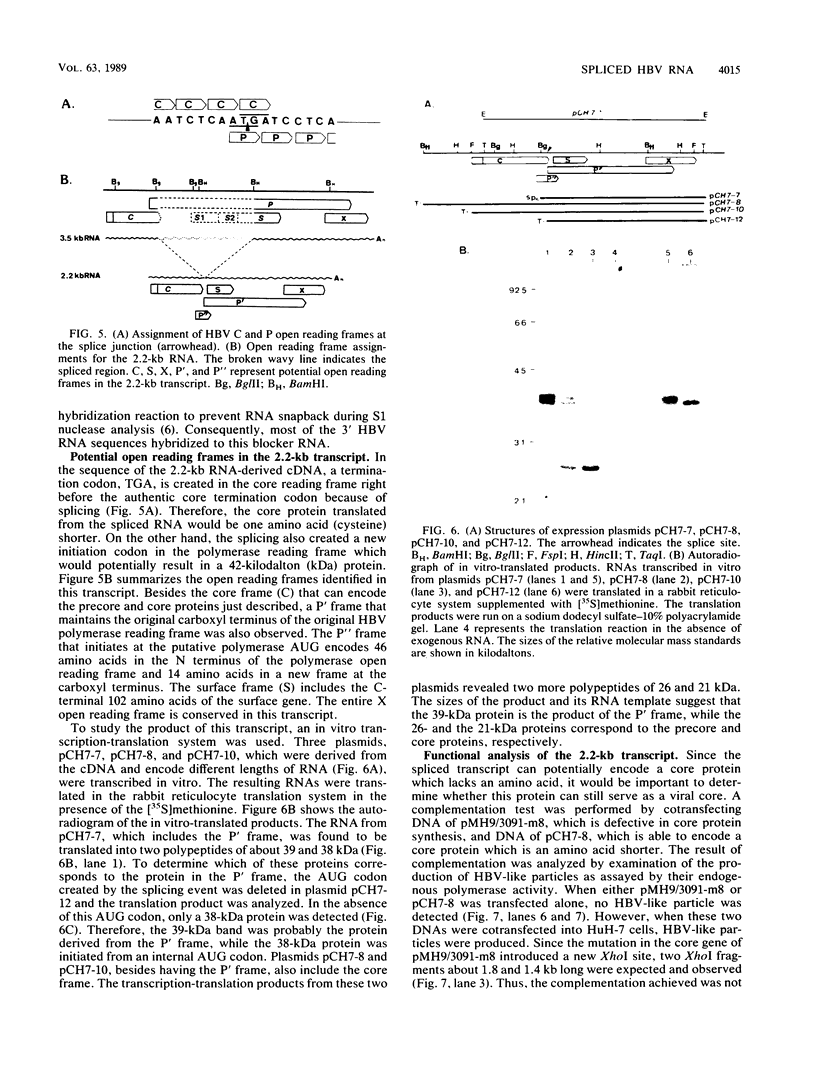
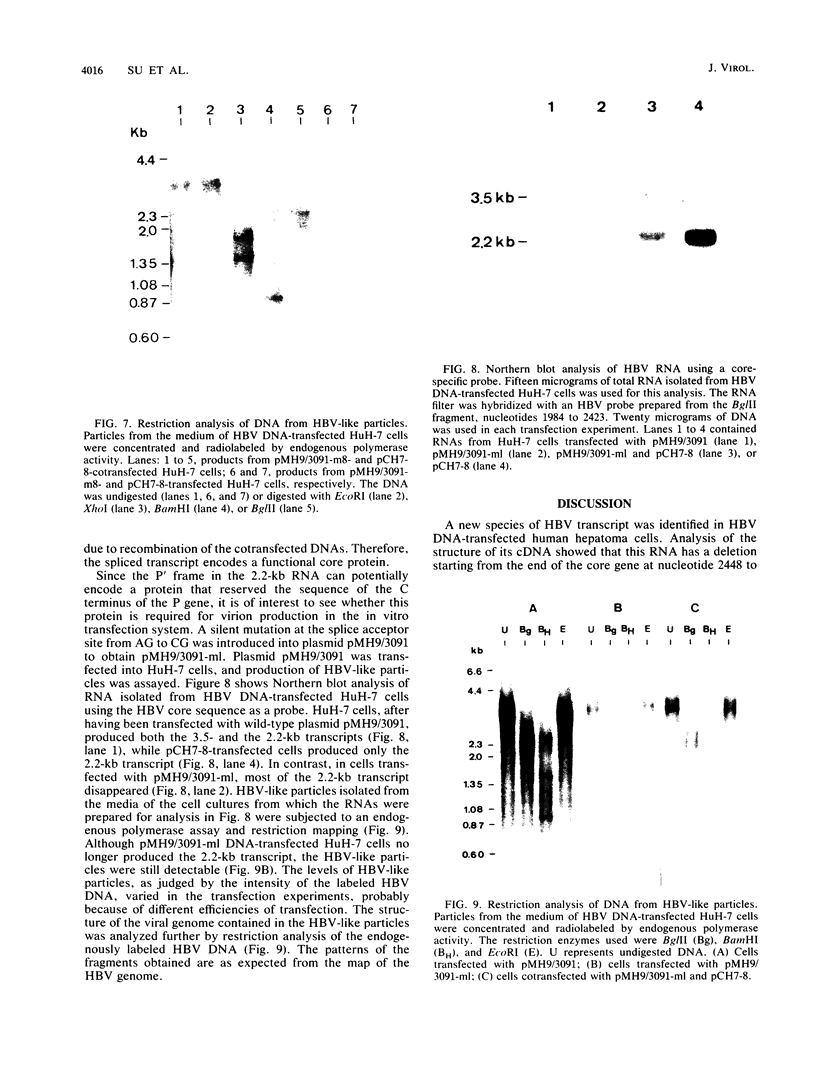
A new hepatitis B virus (HBV) transcript of about 2.2 kilobases was identified in HBV DNA-transfected human hepatoma cells. The 5' terminus of this viral RNA appears to map at one or more of the precore initiation sites, contains a deletion of 1,223 bases corresponding to the last codon of the core gene to the middle of the surface antigen gene, and terminates at the 3' polyadenylation site used by the other known HBV RNAs. The junction region of the deleted sequences showed the conserved splice donor and acceptor GT-AG sequences. Moreover, when a mutant HBV DNA in which the splice acceptor site was changed from AG to CG was transfected into human hepatoma cells, no 2.2-kilobase RNA was detected, further suggesting that this RNA represents a spliced transcript. The core gene, although an amino acid shorter, still encoded a functional viral core protein in complementation experiments. Sequence analysis of the cDNA of the 2.2-kilobase RNA suggests that this transcript can potentially encode a new protein that comprises the reverse transcriptase domain of HBV. However, genetic analysis using a transient DNA transfection system suggests that the gene product(s) of this transcript is not essential for viral replication. The function of this transcript remains to be studied.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büscher M., Reiser W., Will H., Schaller H. Transcripts and the putative RNA pregenome of duck hepatitis B virus: implications for reverse transcription. Cell. 1985 Mar;40(3):717–724. doi: 10.1016/0092-8674(85)90220-x. [DOI] [PubMed] [Google Scholar]
- Cattaneo R., Will H., Darai G., Pfaff E., Schaller H. Detection of an element of the SV40 late promoter in vectors used for expression studies in COS cells. EMBO J. 1983;2(4):511–514. doi: 10.1002/j.1460-2075.1983.tb01455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. M., Jeng K. S., Hu C. P., Lo S. J., Su T. S., Ting L. P., Chou C. K., Han S. H., Pfaff E., Salfeld J. Production of hepatitis B virus in vitro by transient expression of cloned HBV DNA in a hepatoma cell line. EMBO J. 1987 Mar;6(3):675–680. doi: 10.1002/j.1460-2075.1987.tb04807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L. J., Pryciak P., Ganem D., Varmus H. E. Biosynthesis of the reverse transcriptase of hepatitis B viruses involves de novo translational initiation not ribosomal frameshifting. Nature. 1989 Jan 26;337(6205):364–368. doi: 10.1038/337364a0. [DOI] [PubMed] [Google Scholar]
- Enders G. H., Ganem D., Varmus H. E. 5'-terminal sequences influence the segregation of ground squirrel hepatitis virus RNAs into polyribosomes and viral core particles. J Virol. 1987 Jan;61(1):35–41. doi: 10.1128/jvi.61.1.35-41.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders G. H., Ganem D., Varmus H. Mapping the major transcripts of ground squirrel hepatitis virus: the presumptive template for reverse transcriptase is terminally redundant. Cell. 1985 Aug;42(1):297–308. doi: 10.1016/s0092-8674(85)80125-2. [DOI] [PubMed] [Google Scholar]
- Ganem D. Persistent infection of humans with hepatitis B virus: mechanisms and consequences. Rev Infect Dis. 1982 Sep-Oct;4(5):1026–1047. doi: 10.1093/clinids/4.5.1026. [DOI] [PubMed] [Google Scholar]
- Ganem D., Varmus H. E. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Imazeki F., Yaginuma K., Omata M., Okuda K., Kobayashi M., Koike K. RNA transcripts of hepatitis B virus in hepatocellular carcinoma. Hepatology. 1987 Jul-Aug;7(4):753–757. doi: 10.1002/hep.1840070423. [DOI] [PubMed] [Google Scholar]
- Junker M., Galle P., Schaller H. Expression and replication of the hepatitis B virus genome under foreign promoter control. Nucleic Acids Res. 1987 Dec 23;15(24):10117–10132. doi: 10.1093/nar/15.24.10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möröy T., Etiemble J., Trépo C., Tiollais P., Buendia M. A. Transcription of woodchuck hepatitis virus in the chronically infected liver. EMBO J. 1985 Jun;4(6):1507–1514. doi: 10.1002/j.1460-2075.1985.tb03810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi H., Taketa K., Miyano K., Yamane T., Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982 Sep;42(9):3858–3863. [PubMed] [Google Scholar]
- O'hare K., Levis R., Rubin G. M. Transcription of the white locus in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6917–6921. doi: 10.1073/pnas.80.22.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito I., Oya Y., Shimojo H. Novel RNA family structure of hepatitis B virus expressed in human cells, using a helper-free adenovirus vector. J Virol. 1986 May;58(2):554–560. doi: 10.1128/jvi.58.2.554-560.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicht H. J., Radziwill G., Schaller H. Synthesis and encapsidation of duck hepatitis B virus reverse transcriptase do not require formation of core-polymerase fusion proteins. Cell. 1989 Jan 13;56(1):85–92. doi: 10.1016/0092-8674(89)90986-0. [DOI] [PubMed] [Google Scholar]
- Simonsen C. C., Levinson A. D. Analysis of processing and polyadenylation signals of the hepatitis B virus surface antigen gene by using simian virus 40-hepatitis B virus chimeric plasmids. Mol Cell Biol. 1983 Dec;3(12):2250–2258. doi: 10.1128/mcb.3.12.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T. S., Lui W. Y., Lin L. H., Han S. H., P'eng F. K. Analysis of hepatitis B virus transcripts in infected human livers. Hepatology. 1989 Feb;9(2):180–185. doi: 10.1002/hep.1840090203. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiollais P., Pourcel C., Dejean A. The hepatitis B virus. Nature. 1985 Oct 10;317(6037):489–495. doi: 10.1038/317489a0. [DOI] [PubMed] [Google Scholar]
- Toh H., Hayashida H., Miyata T. Sequence homology between retroviral reverse transcriptase and putative polymerases of hepatitis B virus and cauliflower mosaic virus. 1983 Oct 27-Nov 2Nature. 305(5937):827–829. doi: 10.1038/305827a0. [DOI] [PubMed] [Google Scholar]
- Treinin M., Laub O. Identification of a promoter element located upstream from the hepatitis B virus X gene. Mol Cell Biol. 1987 Jan;7(1):545–548. doi: 10.1128/mcb.7.1.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Padgett R. A., Stark G. R. Gene amplification causes overproduction of the first three enzymes of UMP synthesis in N-(phosphonacetyl)-L-aspartate-resistant hamster cells. J Biol Chem. 1979 Sep 10;254(17):8679–8689. [PubMed] [Google Scholar]
- Yaginuma K., Shirakata Y., Kobayashi M., Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci U S A. 1987 May;84(9):2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosuka O., Omata M., Imazeki F., Ito Y., Okuda K. Hepatitis B virus RNA transcripts and DNA in chronic liver disease. N Engl J Med. 1986 Nov 6;315(19):1187–1192. doi: 10.1056/NEJM198611063151903. [DOI] [PubMed] [Google Scholar]