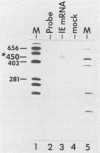
Abstract
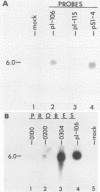
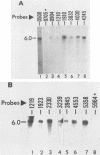
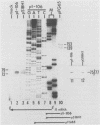
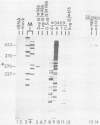
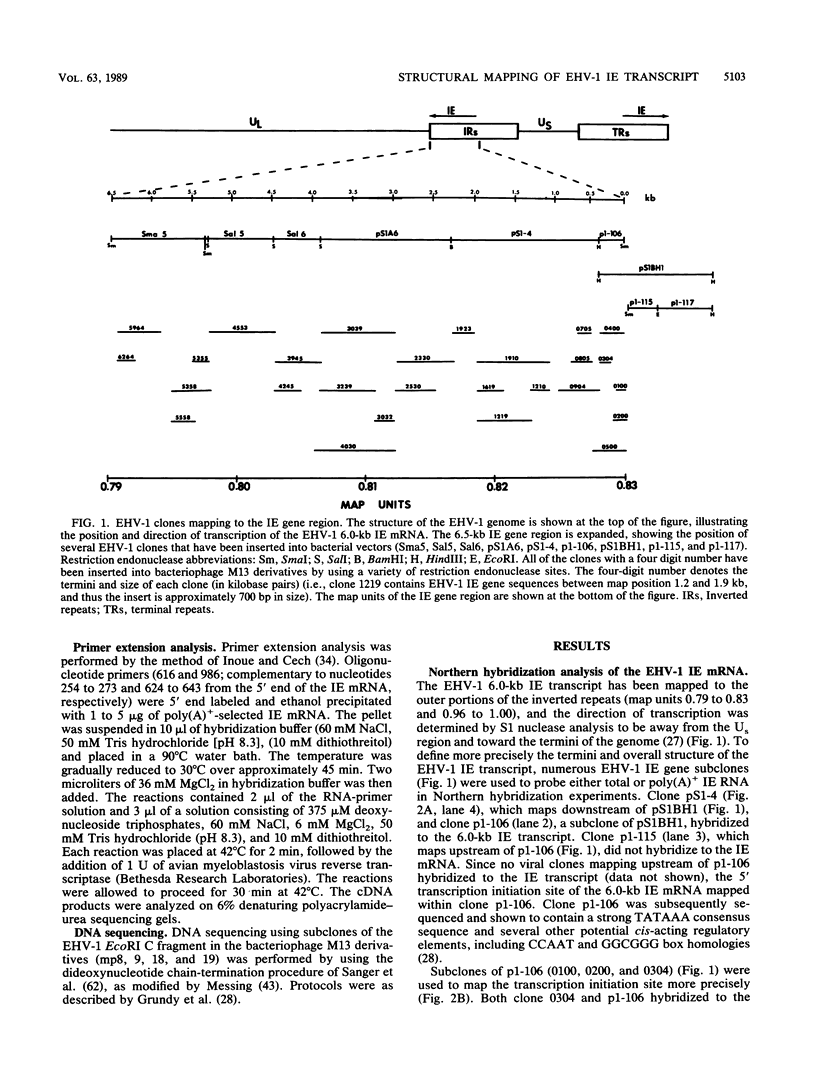
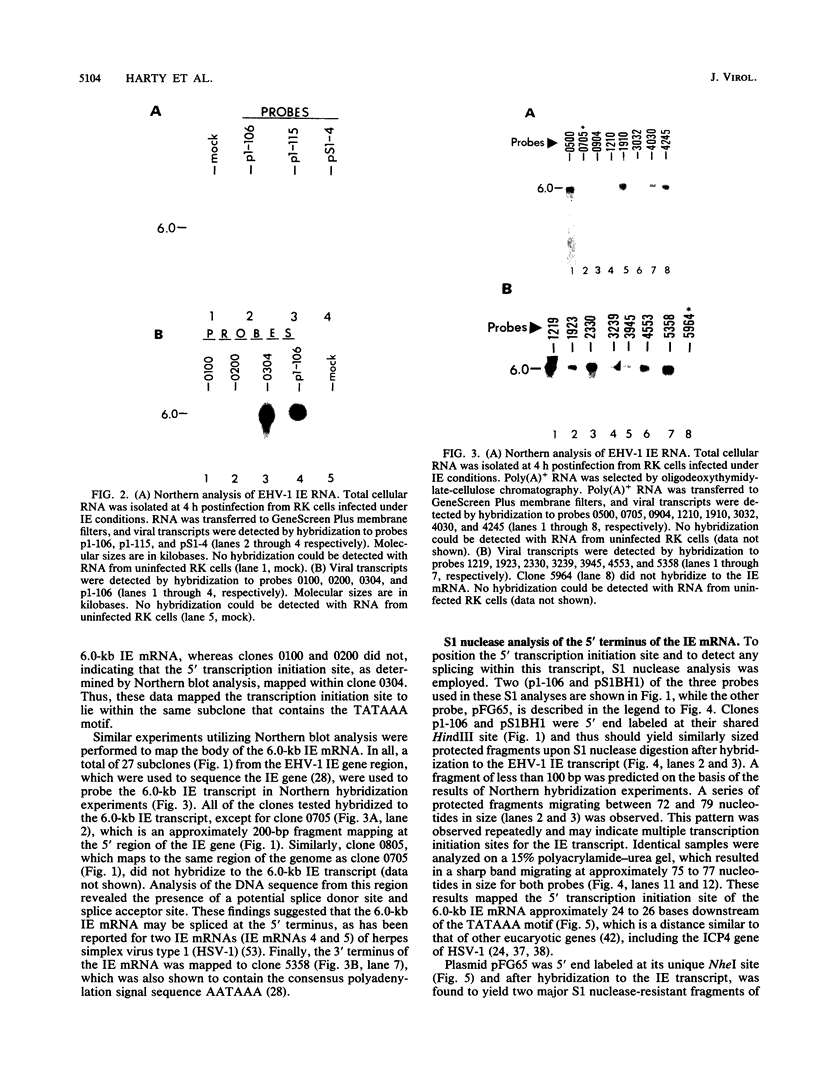
Equine herpesvirus 1 (EHV-1) has been shown to synthesize a 6.0-kilobase (kb) species of immediate-early (IE) mRNA in productively infected cells. This IE gene region maps within the outer portion (map units 0.79 to 0.83 and 0.96 to 1.00) of the two inverted repeat segments of the short genomic region, and elucidation of its DNA sequence has revealed multiple potential open reading frames (ORFs), including a major ORF of 4,461 nucleotides (F. J. Grundy, R. P. Baumann, and D. J. O'Callaghan, Virology 172:223-236, 1989). Analyses of IE polypeptides synthesized in EHV-1-infected cells (in vivo) and in vitro translation of hybrid-selected IE mRNA indicated that multiple species of IE proteins are encoded by this IE mRNA species. To address the nature of the 6.0-kb IE RNA species, Northern (RNA) blot hybridization, S1 nuclease mapping, and primer extension analyses have been employed. These data revealed that no major introns were detected within the body of the IE transcript. However, the IE mRNA was shown to be spliced at the 5' terminus, such that a 372-base intron containing two small ORFs (19 and 51 amino acids) was removed from the leader region of the transcript. This splicing event reduced the leader region from 625 to 253 bases. S1 and primer extension analyses of the 5' terminus of this transcript revealed that the transcription initiation site is located 24 to 26 bases downstream of the consensus TATAAA motif. The 3' transcription termination site was mapped by S1 nuclease analysis to approximately 10 to 20 bases downstream of the polyadenylation signal, AATAAA. The distance from the stop codon of the major ORF to the polyadenylation site is approximately 300 bases. Results from S1 nuclease experiments indicated that splicing does not occur at the 3' terminus. These studies indicated that the EHV-1 6.0-kb IE mRNA is spliced at the 5' terminus and that alternative splicing of this transcript may function in regulating translation of the IE mRNA species.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ace C. I., McKee T. A., Ryan J. M., Cameron J. M., Preston C. M. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J Virol. 1989 May;63(5):2260–2269. doi: 10.1128/jvi.63.5.2260-2269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K. P., Costa R. H., Holland L. E., Wagner E. K. Characterization of herpes simplex virus type 1 RNA present in the absence of de novo protein synthesis. J Virol. 1980 Apr;34(1):9–27. doi: 10.1128/jvi.34.1.9-27.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann R. P., Dauenhauer S. A., Caughman G. B., Staczek J., O'Callaghan D. J. Structure and genetic complexity of the genomes of herpesvirus defective-interfering particles associated with oncogenic transformation and persistent infection. J Virol. 1984 Apr;50(1):13–21. doi: 10.1128/jvi.50.1.13-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann R. P., Staczek J., O'Callaghan D. J. Cloning and fine mapping the DNA of equine herpesvirus type one defective interfering particles. Virology. 1986 Sep;153(2):188–200. doi: 10.1016/0042-6822(86)90022-x. [DOI] [PubMed] [Google Scholar]
- Baumann R. P., Staczek J., O'Callaghan D. J. Equine herpesvirus type 1 defective-interfering (DI) particle DNA structure: the central region of the inverted repeat is deleted from DI DNA. Virology. 1987 Jul;159(1):137–146. doi: 10.1016/0042-6822(87)90356-4. [DOI] [PubMed] [Google Scholar]
- Baumann R. P., Sullivan D. C., Staczek J., O'Callaghan D. J. Genetic relatedness and colinearity of genomes of equine herpesvirus types 1 and 3. J Virol. 1986 Mar;57(3):816–825. doi: 10.1128/jvi.57.3.816-825.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann R. P., Yalamanchili V. R., O'Callaghan D. J. Functional mapping and DNA sequence of an equine herpesvirus 1 origin of replication. J Virol. 1989 Mar;63(3):1275–1283. doi: 10.1128/jvi.63.3.1275-1283.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985 Dec;43(3 Pt 2):729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Caughman G. B., Robertson A. T., Gray W. L., Sullivan D. C., O'Callaghan D. J. Characterization of equine herpesvirus type 1 immediate early proteins. Virology. 1988 Apr;163(2):563–571. doi: 10.1016/0042-6822(88)90297-8. [DOI] [PubMed] [Google Scholar]
- Caughman G. B., Staczek J., O'Callaghan D. J. Equine herpesvirus type 1 infected cell polypeptides: evidence for immediate early/early/late regulation of viral gene expression. Virology. 1985 Aug;145(1):49–61. doi: 10.1016/0042-6822(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Clements J. B., Watson R. J., Wilkie N. M. Temporal regulation of herpes simplex virus type 1 transcription: location of transcripts on the viral genome. Cell. 1977 Sep;12(1):275–285. doi: 10.1016/0092-8674(77)90205-7. [DOI] [PubMed] [Google Scholar]
- Cohen J. C., Perdue M. L., Randall C. C., O'Callaghan D. J. Herpesvirus transcription: altered regulation induced by FUdR. Virology. 1977 Feb;76(2):621–633. doi: 10.1016/0042-6822(77)90244-6. [DOI] [PubMed] [Google Scholar]
- Cohen J. C., Randall C. C., O'Callaghan D. J. Transcription of equine herpesvirus type 1: evidence for classes of transcripts differing in abundance. Virology. 1975 Dec;68(2):561–565. doi: 10.1016/0042-6822(75)90299-8. [DOI] [PubMed] [Google Scholar]
- Dauenhauer S. A., Robinson R. A., O'Callaghan D. J. Chronic production of defective-interfering particles by hamster embryo cultures of herpesvirus persistently infected and oncogenically transformed cells. J Gen Virol. 1982 May;60(Pt 1):1–14. doi: 10.1099/0022-1317-60-1-1. [DOI] [PubMed] [Google Scholar]
- Davison A. J., McGeoch D. J. Evolutionary comparisons of the S segments in the genomes of herpes simplex virus type 1 and varicella-zoster virus. J Gen Virol. 1986 Apr;67(Pt 4):597–611. doi: 10.1099/0022-1317-67-4-597. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Scott J. E. DNA sequence of the major inverted repeat in the varicella-zoster virus genome. J Gen Virol. 1985 Feb;66(Pt 2):207–220. doi: 10.1099/0022-1317-66-2-207. [DOI] [PubMed] [Google Scholar]
- DeLuca N. A., Schaffer P. A. Activation of immediate-early, early, and late promoters by temperature-sensitive and wild-type forms of herpes simplex virus type 1 protein ICP4. Mol Cell Biol. 1985 Aug;5(8):1997–2008. doi: 10.1128/mcb.5.8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca N. A., Schaffer P. A. Physical and functional domains of the herpes simplex virus transcriptional regulatory protein ICP4. J Virol. 1988 Mar;62(3):732–743. doi: 10.1128/jvi.62.3.732-743.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenwick M. L., McMenamin M. Synthesis of alpha (immediate-early) proteins in Vero cells infected with pseudorabies virus. J Gen Virol. 1984 Sep;65(Pt 9):1449–1456. doi: 10.1099/0022-1317-65-9-1449. [DOI] [PubMed] [Google Scholar]
- Gelman I. H., Silverstein S. Dissection of immediate-early gene promoters from herpes simplex virus: sequences that respond to the virus transcriptional activators. J Virol. 1987 Oct;61(10):3167–3172. doi: 10.1128/jvi.61.10.3167-3172.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godowski P. J., Knipe D. M. Transcriptional control of herpesvirus gene expression: gene functions required for positive and negative regulation. Proc Natl Acad Sci U S A. 1986 Jan;83(2):256–260. doi: 10.1073/pnas.83.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W. L., Baumann R. P., Robertson A. T., Caughman G. B., O'Callaghan D. J., Staczek J. Regulation of equine herpesvirus type 1 gene expression: characterization of immediate early, early, and late transcription. Virology. 1987 May;158(1):79–87. doi: 10.1016/0042-6822(87)90240-6. [DOI] [PubMed] [Google Scholar]
- Gray W. L., Baumann R. P., Robertson A. T., O'Callaghan D. J., Staczek J. Characterization and mapping of equine herpesvirus type 1 immediate early, early, and late transcripts. Virus Res. 1987 Sep;8(3):233–244. doi: 10.1016/0168-1702(87)90018-9. [DOI] [PubMed] [Google Scholar]
- Gray W. L., Yalamanchili R., Raengsakulrach B., Baumann R. P., Staczek J., O'Callaghan D. J. Viral transcripts in cells infected with defective interfering particles of equine herpesvirus type 1. Virology. 1989 Sep;172(1):1–10. doi: 10.1016/0042-6822(89)90101-3. [DOI] [PubMed] [Google Scholar]
- Grundy F. J., Baumann R. P., O'Callaghan D. J. DNA sequence and comparative analyses of the equine herpesvirus type 1 immediate early gene. Virology. 1989 Sep;172(1):223–236. doi: 10.1016/0042-6822(89)90124-4. [DOI] [PubMed] [Google Scholar]
- Henry B. E., Newcomb W. W., O'Callaghan D. J. Alterations in virus protein synthesis and capsid production in infection with DI particles of herpesvirus. J Gen Virol. 1980 Apr;47(2):343–353. doi: 10.1099/0022-1317-47-2-343. [DOI] [PubMed] [Google Scholar]
- Henry B. E., Newcomb W. W., O'Callaghan D. J. Biological and biochemical properties of defective interfering particles of equine herpesvirus type 1. Virology. 1979 Jan 30;92(2):495–506. doi: 10.1016/0042-6822(79)90152-1. [DOI] [PubMed] [Google Scholar]
- Henry B. E., Robinson R. A., Dauenhauer S. A., Atherton S. S., Hayward G. S., O'Callaghan D. J. Structure of the genome of equine herpesvirus type 1. Virology. 1981 Nov;115(1):97–114. doi: 10.1016/0042-6822(81)90092-1. [DOI] [PubMed] [Google Scholar]
- Huang H. L., Szabocsik J. M., Randall C. C., Gentry G. A. Equine abortion (herpes) virus-specific RNA. Virology. 1971 Aug;45(2):381–389. doi: 10.1016/0042-6822(71)90339-4. [DOI] [PubMed] [Google Scholar]
- Ihara S., Feldman L., Watanabe S., Ben-Porat T. Characterization of the immediate-early functions of pseudorabies virus. Virology. 1983 Dec;131(2):437–454. doi: 10.1016/0042-6822(83)90510-x. [DOI] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kristie T. M., Roizman B. Alpha 4, the major regulatory protein of herpes simplex virus type 1, is stably and specifically associated with promoter-regulatory domains of alpha genes and of selected other viral genes. Proc Natl Acad Sci U S A. 1986 May;83(10):3218–3222. doi: 10.1073/pnas.83.10.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackem S., Roizman B. Differentiation between alpha promoter and regulator regions of herpes simplex virus 1: the functional domains and sequence of a movable alpha regulator. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4917–4921. doi: 10.1073/pnas.79.16.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackem S., Roizman B. Regulation of herpesvirus macromolecular synthesis: transcription-initiation sites and domains of alpha genes. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7122–7126. doi: 10.1073/pnas.77.12.7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dolan A., Donald S., Brauer D. H. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 1986 Feb 25;14(4):1727–1745. doi: 10.1093/nar/14.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Muller M. T. Binding of the herpes simplex virus immediate-early gene product ICP4 to its own transcription start site. J Virol. 1987 Mar;61(3):858–865. doi: 10.1128/jvi.61.3.858-865.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan D. J., Hyde J. M., Gentry G. A., Randall C. C. Kinetics of viral deoxyribonucleic acid, protein, and infectious particle production and alterations in host macromolecular syntheses in equine abortion (herpes) virus-infected cells. J Virol. 1968 Aug;2(8):793–804. doi: 10.1128/jvi.2.8.793-804.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare P., Hayward G. S. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985 Mar;53(3):751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdue M. L., Kemp M. C., Randall C. C., O'Callaghan D. J. Studies of the molecular anatomy of the L-M cell strain of equine herpes virus type 1: proteins of the nucleocapsid and intact virion. Virology. 1974 May;59(1):201–216. doi: 10.1016/0042-6822(74)90216-5. [DOI] [PubMed] [Google Scholar]
- Preston C. M., Cordingley M. G., Stow N. D. Analysis of DNA sequences which regulate the transcription of a herpes simplex virus immediate early gene. J Virol. 1984 Jun;50(3):708–716. doi: 10.1128/jvi.50.3.708-716.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rixon F. J., Clements J. B. Detailed structural analysis of two spliced HSV-1 immediate-early mRNAs. Nucleic Acids Res. 1982 Apr 10;10(7):2241–2256. doi: 10.1093/nar/10.7.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. S., Boundy A., O'Hare P., Pizzorno M. C., Ciufo D. M., Hayward G. S. Direct correlation between a negative autoregulatory response element at the cap site of the herpes simplex virus type 1 IE175 (alpha 4) promoter and a specific binding site for the IE175 (ICP4) protein. J Virol. 1988 Nov;62(11):4307–4320. doi: 10.1128/jvi.62.11.4307-4320.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A. T., Caughman G. B., Gray W. L., Baumann R. P., Staczek J., O'Callaghan D. J. Analysis of the in vitro translation products of the equine herpesvirus type 1 immediate early mRNA. Virology. 1988 Oct;166(2):451–462. doi: 10.1016/0042-6822(88)90516-8. [DOI] [PubMed] [Google Scholar]
- Robinson R. A., O'Callaghan D. J. A specific viral DNA sequence is stably integrated in herpesvirus oncogenically transformed cells. Cell. 1983 Feb;32(2):569–578. doi: 10.1016/0092-8674(83)90476-2. [DOI] [PubMed] [Google Scholar]
- Robinson R. A., Tucker P. W., Dauenhauer S. A., O'Callaghan D. J. Molecular cloning of equine herpesvirus type 1 DNA: analysis of standard and defective viral genomes and viral sequences in oncogenically transformed cells. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6684–6688. doi: 10.1073/pnas.78.11.6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson R. A., Vance R. B., O'Callaghan D. J. Oncogenic transformation by by equine herpesviruses. II. Coestablishment of persistent infection and oncogenic transformation of hamster embryo cells by equine herpesvirus type 1 preparations enriched for defective interfering particles. J Virol. 1980 Oct;36(1):204–219. doi: 10.1128/jvi.36.1.204-219.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyechan W. T., Dauenhauer S. A., O'Callaghan D. J. Electron microscopic study of equine herpesvirus type 1 DNA. J Virol. 1982 Apr;42(1):297–300. doi: 10.1128/jvi.42.1.297-300.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988 Oct 6;335(6190):563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Struhl K. The DNA-binding domains of the jun oncoprotein and the yeast GCN4 transcriptional activator protein are functionally homologous. Cell. 1987 Sep 11;50(6):841–846. doi: 10.1016/0092-8674(87)90511-3. [DOI] [PubMed] [Google Scholar]
- Triezenberg S. J., Kingsbury R. C., McKnight S. L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988 Jun;2(6):718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- Watson R. J., Preston C. M., Clements J. B. Separation and characterization of herpes simplex virus type 1 immediate-early mRNA's. J Virol. 1979 Jul;31(1):42–52. doi: 10.1128/jvi.31.1.42-52.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalley J. M., Robertson G. R., Davison A. J. Analysis of the genome of equine herpesvirus type 1: arrangement of cleavage sites for restriction endonucleases EcoRI, BglII and BamHI. J Gen Virol. 1981 Dec;57(Pt 2):307–323. doi: 10.1099/0022-1317-57-2-307. [DOI] [PubMed] [Google Scholar]
- Zarkower D., Wickens M. Formation of mRNA 3' termini: stability and dissociation of a complex involving the AAUAAA sequence. EMBO J. 1987 Jan;6(1):177–186. doi: 10.1002/j.1460-2075.1987.tb04736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]