Abstract
Betaine-homocysteine methyltransferase (BHMT) catalyzes the remethylation of homocysteine. BHMT2 encodes a protein 73% identical in amino acid sequence to BHMT, but the function of BHMT2 remains unclear. We set out to identify and functionally characterize common genetic variation in BHMT and BHMT2. Specifically, we sequenced exons, exon-intron splice junctions and the 5′-flanking regions (5′-FRs) of BHMT and BHMT2 using 240 DNA samples from four ethnic groups. Twenty-five single nucleotide polymorphisms (SNPs), including 4 nonsynonymous SNPs, and 39 SNPs, including 4 nonsynonymous, were observed in BHMT and BHMT2, respectively. BHMT wild type (WT) and variant allozymes were expressed in COS-1 cells. Variant allozymes showed no significant differences from WT in levels of enzyme activity or immunoreactive protein, but there were statistically significant differences in apparent Km values. Luciferase reporter gene constructs were created for the three most common BHMT 5′-FR haplotypes, and significant variation was observed in the ability of these constructs to drive transcription. Although BHMT2 mRNA has been observed in human liver and kidney, expression of the protein has not been reported. We were unable to express BHMT2 in mammalian cells, and the protein aggregated after bacterial expression. Furthermore, BHMT2 was rapidly degraded in a rabbit reticulocyte lysate, but it could be stabilized by cotransfection of COS-1 cells with BHMT and, after cotransfection, it coprecipitated with BHMT. These studies have defined common genetic variation in BHMT and BHMT2 and functionally characterized BHMT SNPs. They may also help to explain why BHMT2 has not previously been defined functionally.
Keywords: betaine-homocysteine methyltransferase, BHMT, BHMT2, homocysteine methylation, methylation, betaine, genetic polymorphisms, single nucleotide polymorphism, SNPs, functional genomics
Introduction
Betaine-homocysteine methyltransferase (BHMT, EC 2.1.1.5) catalyzes a key reaction at the convergence of the folate and the methionine cycles (Fig. 1). BHMT is a zinc-dependent cytosolic enzyme that is highly expressed in the human liver, kidney and lens of the eye [1; 2]. It catalyzes one of two major homocysteine remethylation reactions, the transfer of a methyl group from betaine to homocysteine, resulting in the formation of dimethylglycine and methionine (Fig. 1). The other homocysteine remethylation reaction is catalyzed by methyltetrahydrofolate:homocysteine methyltransferase (MTR) [1]. BHMT is thought to account for up to half of the homocysteine remethylation capacity [3]. The human BHMT gene encodes 406 amino acids, maps to chromosome 5q13.1-5q15 [4], spans approximately 20 kb, and consists of 8 exons [5]. BHMT2 encodes a protein that is 73% identical in amino acid sequence to BHMT, differing primarily in the lack of 34 amino acids at the C-terminus of BHMT2 as compared with BHMT [6]. BHMT2 is also located on chromosome 5 approximately 22.3 kb upstream of BHMT, and these two genes are thought to have originated from a tandem duplication event [6; 7]. Both BHMT and BHMT2 contain zinc-binding domains [8], and BHMT2 transcripts – like those of BHMT – are expressed in the liver and the kidney [6]. However, unlike BHMT, there have been no reports of the expression of BHMT2 protein in human tissue.
Figure 1.

The methionine and folate cycles – showing the location of BHMT.
Despite intense interest in elevated circulating homocysteine levels as a possible risk factor for cardiovascular disease, osteoporosis, dementia, and complications of pregnancy [9], surprisingly little is known with regard to common genetic variation in BHMT or BHMT2. Therefore, we set out to identify common sequence variation in these two genes – followed by functional genomic studies. Specifically, both genes were resequenced in 240 DNA samples, 60 each from individuals of four ethnic groups. We identified a total of 64 SNPs (25 in BHMT and 39 in BHMT2), including 8 nonsynonymous cSNPs (4 each in BHMT and BHMT2). We then performed functional genomic studies of BHMT variant allozymes and common 5′-FR haplotypes. However, our attempts to perform similar studies with BHMT2 were complicated by difficulty in expressing BHMT2 in mammalian cells. We also found that BHMT2 aggregates after bacterial expression and that it is rapidly degraded in a rabbit reticulocyte lysate (RRL). However, BHMT2 expression could be detected in COS-1 cells after cotransfection with BHMT, the two proteins coprecipitated during immunoprecipitation and homocysteine could “stabilize” BHMT. In summary, the present studies have defined the nature and extent of genetic variation in BHMT and BHMT2 and have defined the functional implications of BHMT polymorphisms. In addition, they have raised the possibility of a functional interaction between these two closely related proteins in vivo, and they begin to explain why little is currently known about the function of BHMT2.
Materials and methods
DNA samples
DNA samples from 60 Caucasian-American (CA), 60 African-American (AA), 60 Han Chinese-American (HCA), and 60 Mexican-American (MA) subjects were obtained from the Coriell Cell Repository (Camden, NJ). These DNA samples have been widely used for gene resequencing studies [10; 11; 12; 13]and were collected from “healthy” subjects, anonymized and deposited by the National Institute of General Medical Sciences. However, for obvious reasons related to the protection of the privacy of the donors, no additional information with regard to the subjects who donated DNA is available other than their ethnicity and gender. Written informed consent had been obtained from all subjects for the use of their DNA for research purposes. Our studies were reviewed and approved by the Mayo Clinic Institutional Review Board.
Gene resequencing
The human BHMT and BHMT2 genes were resequenced in 240 Coriell Cell Repository DNA samples. To amplify BHMT, 9 reactions were performed with primers that flanked exons approximately 200 bp on either side of each exon, as well as approximately 1 kb of the 5′-FR (see Supplemental Table 1 for primer sequences). For BHMT2, 8 amplification reactions were performed. Amplicons were sequenced on both strands in the Mayo Clinic Molecular Biology Core Facility. The GenBank accession number for the BHMT and BHMT2 reference sequences was NT_006713.14. The reference sequences for BHMT and BHMT2 mRNA were NM_001713.1 and NM_017614.3, respectively. cDNA clones for BHMT and BHMT2 were purchased from the American Type Culture Collection (Manassas, VA) (accession numbers BC012616 and BC020665, respectively). The sequences of these clones were confirmed by DNA sequencing.
BHMT and BHMT2 expression in COS-1, HepG2 and HEK293T cells
The open reading frames (ORFs) of the BHMT and BHMT2 cDNAs were cloned into the mammalian expression vector pcDNA3.1/V5-His© TOPO® TA (Invitrogen, Calsbad, CA). In addition, a C-terminal hemagglutinin (HA) tagged BHMT2 cDNA was cloned into the pcDNA3.1 directional TOPO mammalian expression vector (Invitrogen, Calsbad, CA). Site-directed mutagenesis performed with “circular PCR” was used to create expression constructs for each of the BHMT nonsynonymous SNPs. Each insert was sequenced in both directions to verify construct sequences. These constructs, as well as “empty” vector lacking an insert, were then transfected into COS-1, HepG2 and HEK293T cells, using the TransFast™ transfection reagent (Promega, Madison, WI). The cells were also cotransfected with pSV-β-galactosidase (Promega, Madison, WI) to correct for possible variation in transfection efficiency. The transfected cells were cultured for 48 h at 37°C; washed with phosphate-buffered saline; resuspended in homogenization buffer, and were lysed with a Polytron homogenizer (Brinkman Instruments). Following centrifugation at 100,000 × g for 1 h at 4°C, the supernatant (cytosol) was stored at −80°C prior to assay.
BHMT radiochemical enzyme assay
Methyl-14C-betaine hydrate (specific activity 29.3 mCi/mmol) was synthesized by Perkin-Elmer (Boston, MA) for use in the BHMT enzyme activity assay. The assay procedure was a modification of the method described by Garrow et al [14]. The 100 μL reaction mixture contained 400 μM L-homocysteine, 64 μM (0.16 μCi/μmol) betaine and 50 mM KH2PO4 (pH 8.0). L-homocyteine was generated from L-homocysteine thiolactone. Briefly, L-homocysteine thiolactone was treated with 5.0 N NaOH for 5 min at room temperature, followed by neutralization with 1.0 M KH2PO4. The enzyme assay was started by adding radioactive betaine to the reaction mixture in borosilicate glass tubes at 37°C. Following incubation for 2 h, the reaction was stopped by adding 1 ml of ice-cold water to each tube, and the tubes were immediately placed on ice. After vortexing, 1 ml of each reaction mixture was transferred to a Dowex 1-X4 ion exchange column that contained 2 ml of resin (Sigma-Aldrich Corp., St. Louis, MO). The commercially available Cl− form of the resin was converted to the OH− form by pre-treatment with 1 M NaOH, followed by rinsing with 4 volumes of deionized H2O. Five ml of deionized H2O was used to elute unreacted betaine from the column. The reaction product was then eluted by adding 3 ml of 1.5 N HCl, and radioactivity was measured by liquid scintillation counting. For substrate kinetic studies, eight concentrations of each substrate, prepared by serial 2-fold dilutions, were tested. Specifically, L-homocysteine concentrations varied from 3.1 to 400 μM and concentrations of betaine varied from 2 to 256 μM. Basal activities of BHMT allozymes were compared using 64 μM betaine and 400 μM L-homocysteine to perform the assays. “Empty” vector controls were used to correct for possible endogenous BHMT activity in COS-1 cells. Under these conditions, the COS-1 cells expressed less than 3% of the activity observed after transfection with the WT construct.
Western blot analysis
Peptides consisting of BHMT amino acids 384–406, an area not represented in the amino acid sequence of BHMT2, were synthesized in the Mayo Proteomics Facility. Rabbit polyclonal antisera directed against this peptide were generated by Cocalico Biologicals, Inc. (Reamstown, PA). The specificity of antibodies was tested by Western blot analysis performed with both human liver cytosol and recombinant WT human BHMT expressed in COS-1 cells. Bound antibody was detected using the ECL Western Blotting system (Amersham Pharmacia, Piscataway, NJ). Results were expressed as percentages of the WT human BHMT on that gel, corrected for small variations in transfection efficiency.
Reporter gene assays
Firefly luciferase reporter gene constructs were created in pGL3-Basic (Promega) for the three most common BHMT 5′-FR haplotypes (frequencies > 1% in at least one ethnic group). Approximately 1 kb of the BHMT 5′-FR was amplified from human genomic DNA samples containing the desired haplotype and was used to create reporter gene constructs. Inserts in these constructs were sequenced on both strands, and the constructs were used to transfect HepG2 and HEK293T cells. During transfection, 2 μg of construct DNA was cotransfected with 0.2 μg of pRL-TK (Promega) DNA encoding Renilla luciferase to make it possible to correct for variation in transfection efficiency. Cells were harvested and lysed 48 h after transfection, and reporter gene activity in cell lysates was measured using the Promega Dual Luciferase Reporter Assay system. Results are reported as the ratio of firefly luciferase light units to those for Renilla luciferase.
Rabbit reticulocyte lysate (RRL) translation and degradation studies
Transcription and translation of BHMT and BHMT2 were performed with the TNT® coupled RRL System (Promega) as described by Wang et al [15]. Briefly, 1 μg of expression construct DNA was added to 25 μL of RRL that had been treated to inhibit protein degradation, together with 2 μL T7 buffer, 1 μL T7 polymerase, 1 μL of a mixture of amino acids that lacked methionine, 1 μL RNasin and 2 μL 35S-methionine (1000 Ci/mM, 10 mCi/mL, 0.4 μM final concentration). With the exception of the RNasin (Promega) and 35S-methionine (Amersham Pharmacia Biotech), all reagents were included in the Promega kit. The reaction volume was increased to 50 μL with nuclease-free water (Promega), and the mixture was incubated at 30°C for 90 min. A 5 μL aliquot was then used to perform SDS-PAGE, followed by autoradiography. For the protein degradation experiments, 10 μL of in vitro-translated 35S-methionine-labeled protein was added to 50 μL of an adenosine 5′-triphosphate (ATP) generating system and 50 μL of “untreated” RRL. The ATP generating system consisted of 100 μL containing 1 M Tris-HCl (pH 7.8), 160 mM MgCl2, 120 mM KCl, 100 mM dithiothreitol, 100 mM ATP, 200 mM creatine phosphate and 2 mg/mL creatine kinase (all from Sigma), plus 300 μL nuclease-free water (Promega). This mixture was incubated at 37°C, and aliquots were removed at 0, 4, 8 and 24 h to perform SDS-PAGE, followed by autoradiography.
Immunoprecipitation (IP)
IP was performed using the anti-HA immunoprecipitation kit (Sigma) as suggested by the manufacturer. Specifically, cells transfected with constructs for BHMT, HA-tagged BHMT2 or both were lysed, and cell lysates were incubated for 1 h with anti-HA-agarose. The beads were then washed and bound proteins were dissolved in SDS sample buffer. These mixtures were then subjected to Western blot analysis.
Bacterial recombinant BHMT and BHMT2
Human BHMT and BHMT2 constructs in the bacterial expression vector pGEX6P1 were transformed into BL21 E. coli to express BHMT-GST and BHMT2-GST fusion proteins. The fusion proteins were purified with a GSTrap FF affinity column (Amersham). Cleavage of the GST tag was performed in 4°C with PreScission Protease (Amersham). BHMT and BHMT2 activities were measured as described above. These proteins were also subjected to FPLC gel permeation chromatography using a Superdex 200 10/300 GL (Amersham) column with 50 mM Tris-HCl, pH 8.0, as the mobile phase.
Statistical analysis
With 60 DNA samples (120 alleles) for each of the 4 ethnic groups studied, we had 90% power to detect a variant allele with a true population frequency of ≥ 2% [16]. All polymorphisms detected during resequencing were tested for Hardy-Weinberg equilibrium. Haplotypes were inferred using a program based on the E-M algorithm [17; 18; 19], and values for π, θ and Tajima’s D were calculated as described by Tajima [20]. Linkage disequilibrium analysis was performed by calculating D′ values [21; 22]. Haplotype analysis was performed as described by Schaid et al. [19] using the E-M algorithm. Apparent Km values were determined using the GraphPad Prism 4.0 software program (GraphPad, San Diego, CA). Student’s t-test was used to assess differences between mean values.
Results
Human BHMT and BHMT2 resequencing
BHMT and BHMT2 were resequenced using anonymized DNA samples from 60 AA, 60 CA, 60 HCA, and 60 MA subjects. Nine and 8 PCR amplifications were performed to resequence BHMT and BHMT2, respectively, and both strands were sequenced for all samples. A total of approximately 1.9 × 106 and 1.7 × 106 bp of DNA were sequenced and analyzed for BHMT and BHMT2, respectively. Twenty-five SNPs were observed in BHMT — 17 in AA, 8 in CA, 9 in HCA, and 10 in MA subjects (Table 1 and Fig. 2 upper panel). Fifteen of these SNPs were “common”, with a minor allele frequency (MAF) of >1% in at least one ethnic group. There were 11 polymorphisms within exons, including 4 nonsynonymous SNPs that resulted in the following changes in encoded amino acids: Arg(16)Cys, Pro(197)Ser, Gly(199)Ser, and Arg(239)Gln. Arg(16)Cys was present in only one AA subject; Pro(197)Ser only in AA and Gly(199)Ser only in CA subjects; and Arg(239)Gln was observed in all 4 ethnic groups with a MAF >10%.
Table 1.
Human BHMT and BHMT2 genetic polymorphisms. Polymorphism locations, alterations in nucleotide and amino acid sequences, as well as minor allele frequencies for all four ethnic groups studied are listed. Polymorphisms within exons are shaded. SNPs in exons and 5′-FRs have been numbered on the basis of their locations in the cDNA, with the ‘A’ in the translation initiation codon assigned (+1). Negative numbers are located 5′, and positive numbers 3′ to this position. SNPs in introns have been numbered based on their distance to the nearest 5′ or 3′ exon-intron splice junctions, using positive and negative numbers, respectively. The final column indicates whether the polymorphism was already present in dbSNP.
| BHMT Polymorphisms | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| # | Gene Location | Nucleotide Location | Nucleotide Sequence Alteration | Amino Acid Sequence Alteration | Minor allele frequencies | dbSNP designation | |||
| AA | CA | HCA | MA | ||||||
| 1 | 5′FR | −809 | A to C | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 2 | 5′FR | −773 | T to C | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 3 | 5′FR | −448 | C to T | 0.117 | 0.117 | 0.000 | 0.042 | rs16876512 | |
| 4 | 5′FR | −412 | C to A | 0.017 | 0.000 | 0.000 | 0.000 | ||
| 5 | 5′FR | −142 | G to T | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 6 | Intron 1 | −164 | T to C | 0.067 | 0.083 | 0.100 | 0.042 | rs16876527 | |
| 7 | Intron 1 | −93 | C to T | 0.042 | 0.000 | 0.000 | 0.000 | ||
| 8 | Exon 2 | 46 | C to T | Arg(16)Cys | 0.008 | 0.000 | 0.000 | 0.000 | |
| 9 | Intron 2 | 41 | G to A | 0.008 | 0.000 | 0.000 | 0.008 | ||
| 10 | Exon 3 | 222 | C to T | 0.033 | 0.000 | 0.000 | 0.000 | ||
| 11 | Intron 3 | 132 | G to A | 0.000 | 0.000 | 0.017 | 0.000 | ||
| 12 | Exon 4 | 438 | G to A | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 13 | Intron 4 | 52 | C to T | 0.092 | 0.375 | 0.492 | 0.408 | rs567754 | |
| 14 | Exon 5 | 589 | C to T | Pro(197)Ser | 0.033 | 0.000 | 0.000 | 0.000 | |
| 15 | Exon 5 | 595 | G to A | Gly(199)Ser | 0.000 | 0.017 | 0.000 | 0.000 | |
| 16 | Intron 5 | −132 | G to C | 0.000 | 0.000 | 0.000 | 0.008 | ||
| 17 | Intron 5 | −89 | A to G | 0.392 | 0.292 | 0.150 | 0.183 | rs694290 | |
| 18 | Exon 6 | 716 | G to A | Arg(239)Gln | 0.192 | 0.292 | 0.292 | 0.383 | rs3733890 |
| 19 | Exon 6 | 792 | C to T | 0.000 | 0.000 | 0.000 | 0.200 | rs4703772 | |
| 20 | Intron 6 | 68 | T to C | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 21 | Exon 7 | 852 | C to T | 0.017 | 0.000 | 0.133 | 0.042 | ||
| 22 | Exon 7 | 858 | A to G | 0.000 | 0.008 | 0.000 | 0.000 | ||
| 23 | Exon 7 | 966 | A to G | 0.008 | 0.025 | 0.000 | 0.008 | ||
| 24 | Intron 7 | −51 | A to G | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 25 | 3′UTR | 1251 | C to G | 0.000 | 0.000 | 0.025 | 0.000 | ||
| BHMT2 Polymorphisms | |||||||||
| Gene Location | Nucleotide Location | Nucleotide Sequence Alteration | Amino Acid Sequence Alteration | Minor allele frequencies | dbSNP designation | ||||
| # | AA | CA | HCA | MA | |||||
| 1 | 5′FR | −799 | T to G | 0.033 | 0.000 | 0.000 | 0.000 | rs6877474 | |
| 2 | 5′FR | −762 | A to C | 0.442 | 0.192 | 0.175 | 0.208 | rs642934 | |
| 3 | 5′FR | −671 | T to C | 0.008 | 0.000 | 0.000 | 0.033 | ||
| 4 | 5′FR | −622 | T to C | 0.217 | 0.575 | 0.642 | 0.575 | rs4998017 | |
| 5 | 5′FR | −616 to −617 | deletion of GC | 0.150 | 0.575 | 0.642 | 0.575 | rs5868949 | |
| 6 | 5′FR | −519 | A to G | 0.175 | 0.158 | 0.050 | 0.175 | rs644191 | |
| 7 | 5′FR | −431 | A to G | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 8 | 5′FR | −406 | T to A | 0.025 | 0.000 | 0.000 | 0.008 | ||
| 9 | 5′FR | −334 | C to G | 0.000 | 0.000 | 0.000 | 0.008 | ||
| 10 | 5′FR | −265 | A to G | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 11 | 5′FR | −151 | G to C | 0.033 | 0.000 | 0.000 | 0.000 | ||
| 12 | Intron 1 | 72 | C to T | 0.033 | 0.000 | 0.000 | 0.000 | ||
| 13 | Intron 1 | 136 | A to G | 0.033 | 0.000 | 0.000 | 0.000 | ||
| 14 | Intron 1 | −262 | G to A | 0.042 | 0.000 | 0.000 | 0.000 | ||
| 15 | Intron 1 | −158 | T to C | 0.075 | 0.108 | 0.008 | 0.100 | ||
| 16 | Intron 1 | −142 | T to C | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 17 | Exon 2 | 162 | C to T | 0.333 | 0.692 | 0.625 | 0.617 | rs682985 | |
| 18 | Intron 2 | 56 | A to G | 0.025 | 0.000 | 0.000 | 0.008 | ||
| 19 | Intron 2 | −52 | T to A | 0.025 | 0.000 | 0.000 | 0.000 | ||
| 20 | Exon 3 | 197 | C to T | Ala(66)Val | 0.033 | 0.000 | 0.000 | 0.008 | |
| 21 | Intron 3 | −95 | C to T | 0.000 | 0.008 | 0.000 | 0.000 | ||
| 22 | Intron 3 | −92 | A to G | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 23 | Intron 4 | 59 | C to T | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 24 | Intron 4 | 99 | A to G | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 25 | Exon 5 | 463 | G to T | Val(155)Phe | 0.000 | 0.008 | 0.000 | 0.000 | |
| 26 | Intron 5 | 1 | G to A | 0.017 | 0.000 | 0.000 | 0.000 | ||
| 27 | Intron 5 | 45 | C to T | 0.042 | 0.117 | 0.000 | 0.033 | ||
| 28 | Intron 5 | 46 | G to A | 0.000 | 0.000 | 0.000 | 0.067 | ||
| 29 | Intron 5 | −109 | A to G | 0.000 | 0.000 | 0.000 | 0.033 | ||
| 30 | Intron 5 | −69 | C to A | 0.000 | 0.000 | 0.000 | 0.008 | ||
| 31 | Intron 5 | −48 | Deletion of A | 0.025 | 0.008 | 0.000 | 0.000 | ||
| 32 | Intron 5 | −1 | G to A | 0.008 | 0.008 | 0.000 | 0.000 | ||
| 33 | Exon 6 | 648 | G to A | 0.033 | 0.000 | 0.000 | 0.000 | ||
| 34 | Exon 6 | 653 | C to T | Thr(218)Met | 0.008 | 0.000 | 0.000 | 0.000 | |
| 35 | Exon 6 | 699 | G to T | 0.000 | 0.017 | 0.000 | 0.008 | ||
| 36 | Exon 6 | 709 | G to A | Val(237)Met | 0.000 | 0.008 | 0.000 | 0.000 | |
| 37 | Intron 6 | 17 | A to G | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 38 | Intron 7 | −90 | T to A | 0.025 | 0.000 | 0.000 | 0.008 | ||
| 39 | 3′UTR | 1126 | C to G | 0.008 | 0.000 | 0.000 | 0.000 | ||
Figure 2.
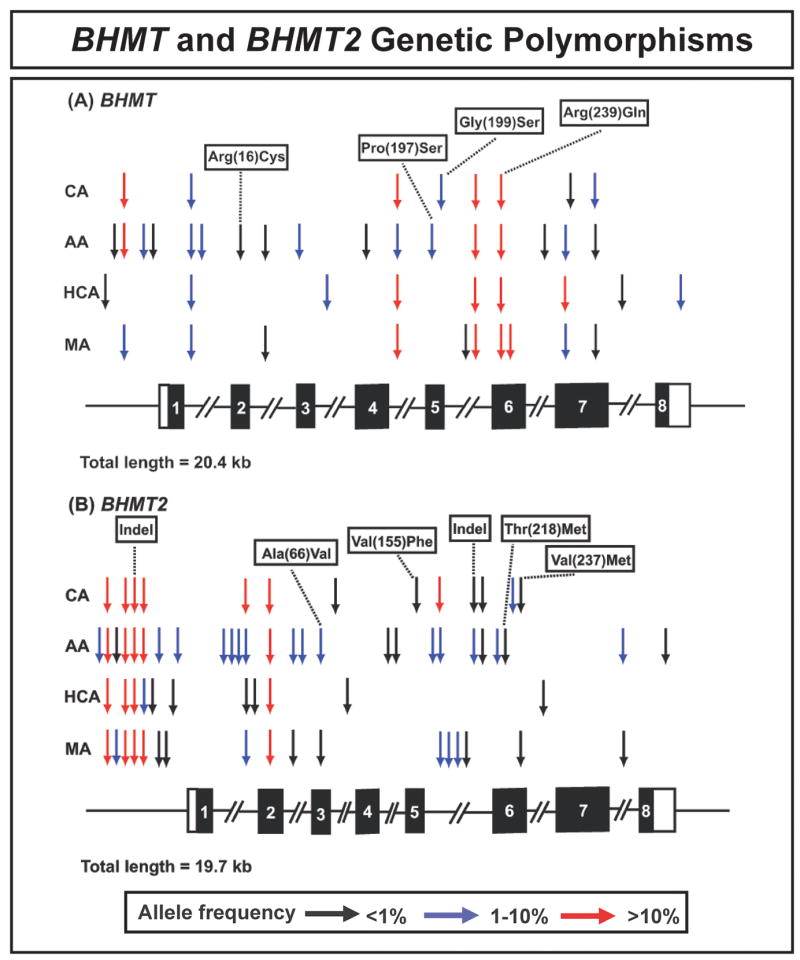
Human BHMT and BHMT2 genetic polymorphisms. The figure shows a schematic representation of the structures of (A) BHMT and (B) BHMT2, with arrow indicating the locations of polymorphisms. Exons encoding the open-reading frame (ORF) are represented as dark rectangles and portions of exons that encode untranslated region sequences are indicated by open rectangles. “Indel” represents an insertion-deletion. Colors of arrows indicate minor allele frequencies for polymorphisms.
For BHMT2, 39 SNPs were observed — 26 in AA, 13 in CA, 11 in HCA, and 17 in MA subjects (Table 1 and Fig. 2B). Twenty-four of the BHMT2 polymorphisms were common (MAF >1% in at least one ethnic group), with striking ethnic differences, i.e., 20 for AA, 8 for CA, 5 for HCA and 10 for MA subjects. There were two insertion/deletion events and 4 non-synonymous SNPs in BHMT2. The indels consisted of a GC deletion in the 5′-FR (−616 to −617) and an A deletion at intron 5 (−48). The four BHMT2 nonsynonymous SNPs resulted in the following changes in encoded amino acids: Ala(66)Val, Val(155)Phe, Thr(218)Met, and Val(237)Met.
Among the 64 polymorphisms observed in BHMT and BHMT2, six for each gene were present in dbSNP (www.ncbi.nlm.nih.gov/SNP) (Table 1). All BHMT and BHMT2 polymorphisms were in Hardy-Weinberg equilibrium (p > 0.05), with the following exceptions: nucleotide (−164) in BHMT intron 1 for CA (p = 3 × 10−5), HCA (p = 0.00027), and MA subjects (p = 0.001); the SNP at position 1251 in the BHMT 3′-UTR for HCA (p = 0.025); and SNP (−52) in BHMT2 intron 2 for AA subjects (p = 0.025).
We also calculated nucleotide diversity, a measure of genetic variation adjusted for the number of alleles studied, using 2 standard measures; π, average heterozygosity per site and θ, a population mutation measure that is theoretically equal to the neutral mutation parameter [23; 24]. π was similar across the four ethnic groups, while values for θ were largest for the AA samples (Supplemental Table 2). Tajima’s D, a test of the ‘neutral’ mutation hypothesis, did not differ significantly from zero for any of the four ethnic groups (Supplemental Table 2).
Linkage disequilibrium and haplotype analysis
Linkage disequilibrium analysis was performed for each ethnic group. Specifically, all possible pairwise SNP comparisons were used to compute R2 and D′ values [25]. Data for all pairwise SNP comparisons with R2 values ≥ 0.7 are listed in Supplemental Table 3. D′ is 1 when there is maximal association between 2 SNPs and 0 when there is random association [21; 23]. When these data were displayed graphically, no clearly defined haplotype blocks were seen over an area that included both BHMT and BHMT2 (data not shown).
Since haplotype is of increasing importance in genetic association studies [25; 26], we also performed haplotype analyses for BHMT and BHMT2, identifying both observed and inferred haplotypes. BHMT and BHMT2 haplotypes with frequencies ≥ 1% in at least one ethnic group are listed in Tables 2 and 3. Haplotype designations were based on the amino acid sequence of the encoded allozyme, with the most common amino acid sequence in AA subjects designated as *1. Based on descending haplotype frequencies, and beginning with data for AA subjects, letter designations were then added, e.g., the rank order of haplotype frequencies was *1A > *1B > *1C. For example, for BHMT, *2, *3, *4, *5 denote variants that encode Cys16, Ser197, Ser199, and Gln239 variant amino acid sequences, respectively, while for BHMT2, *2, *3, *4, *5 denote variants that encode Val66, Phe155, Met218, and Met237, respectively. These data were used in the subsequent functional genomic studies, especially the studies of common variant haplotypes in the BHMT 5′-FRs, i.e., the area presumed to contain the core promoter.
Table 2.
Human BHMT haplotypes with frequencies ≥ 1%. If a haplotype encoded a variant amino acid sequences, it was included in the table even if its frequency was less than 1%. Nucleotide positions are numbered as described in the legend for Table 1. Variant nucleotides compared with the “reference sequence” (i.e., the most common allele in AA subjects) are highlighted as white type against a black background. The initial column lists observed (o) and inferred (i) haplotypes.

|
Table 3.
Human BHMT2 haplotypes with frequencies ≥ 1%. The table legend is described in Table 2 except that “D” = deletion and “I” = insertion.

|
BHMT allozyme activity and protein levels
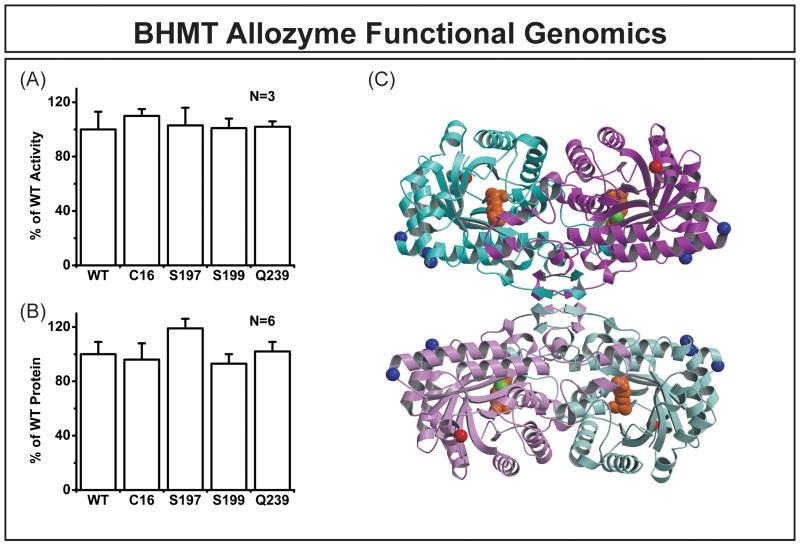
To study the possible functional effects of BHMT nonsynonymous SNPs, expression constructs were created for the WT and four variant allozymes. A mammalian expression system was used to perform these studies to ensure that mammalian post-translation modification and protein degradation systems would be present. Levels of BHMT enzyme activity and immunoreactive protein were measured for all five allozymes. Fig. 3A compares basal enzyme activity levels among BHMT allozymes, expressed as a percentage of the WT activity. No statistically significant differences were detected. Results for immunoreactive protein levels determined by quantitative Western blot analysis also failed to show significant differences among WT and variant allozymes (Fig. 3B). Finally, substrate kinetic studies were performed for all five allozymes and apparent Km values for both betaine and homocysteine are listed in Table 4. With the exception of Ser197, variant allozymes generally had lower apparent Km values than did the WT, but these differences were of a magnitude that might not be of physiologic significance.
Figure 3.
BHMT allozyme functional genomics. (A) BHMT allozyme activity levels assayed with 400 μM L-homocysteine and 64 μM betaine as substrates. Activities are expressed as percentages of that of the WT. (B) BHMT allozyme immunoreactive protein levels. Protein levels are expressed as percentages of that of the WT. Values for (A) and (B) represent mean ± SEM. (C) Locations in the human BHMT crystal structure of residues altered by nonsynonymous SNPs. Human BHMT crystallizes as a dimer of dimers [27]. The monomers in the upper dimer are depicted as light blue and purple ribbon structures, and monomers in the lower dimer have lighter shades of the same colors. Bound in the active site of each monomer are a zinc ion (green) and the bi-substrate analog S(δ-carboxybutyl)-L-homocysteine (orange), both shown as space-filling structures. The Cα atoms of the four residues altered by SNPs (Arg16 red; Pro197, Gly199, and Arg239 blue) are drawn as spheres.
Table 4.
Recombinant human BHMT allozyme apparent Km values. Values are mean ± SEM. One way anova and protected Student’s t-test were used to assess differences between the wild type and variant allozymes.
| Allozyme | Betaine Km, μM | P value | L-homocysteine Km, μM | P value |
|---|---|---|---|---|
| Wild Type | 22.7 ± 1.9 | ------ | 29.1 ± 3.4 | ------ |
| Cys16 | 13.9 ± 1.8 | <0.005 | 7.6 ± 1.2 | <0.001 |
| Ser197 | 21.3 ± 2.0 | 0.5 | 21.6 ± 2.2 | <0.001 |
| Ser199 | 13.9 ± 3.5 | <0.005 | 20.6 ± 0.9 | <0.001 |
| Gln239 | 12.0 ± 2.7 | <0.005 | 15.8 ± 1.3 | <0.001 |
We also examined the x-ray crystal structure of human BHMT [27] in an attempt to understand the possible structural consequences of the nonsynonymous polymorphisms in BHMT. Variant amino acid residues altered by nonsynonymous cSNPs were mapped onto the tetrameric structure of BHMT (Fig. 3C). Variant residues 197, 199 and 239 were surface exposed, so their substitutions as a result of nonsynonymous SNPs could be easily accommodated sterically. The side chain of Arg16 was mostly buried and formed a salt bridge with Asp115. The Arg16-Asp115 interaction tethered the N-terminal helix of BHMT to the central beta-sheet. However, even though removing interaction with Asp115 as a result of the Arg(16)Cys polymorphism might affect local or global structure, the Arg16Cys substitution could be tolerated sterically in terms of side chain size. Finally, none of the amino acid substitutions were located near the active site or at an intermolecular protein-protein contact. These results were consistent with our observation that none of the BHMT nonsynonymous cSNPs had a dramatic effect on either enzyme activity or protein level (Fig. 3A and Fig. 3B).
BHMT reporter gene assay
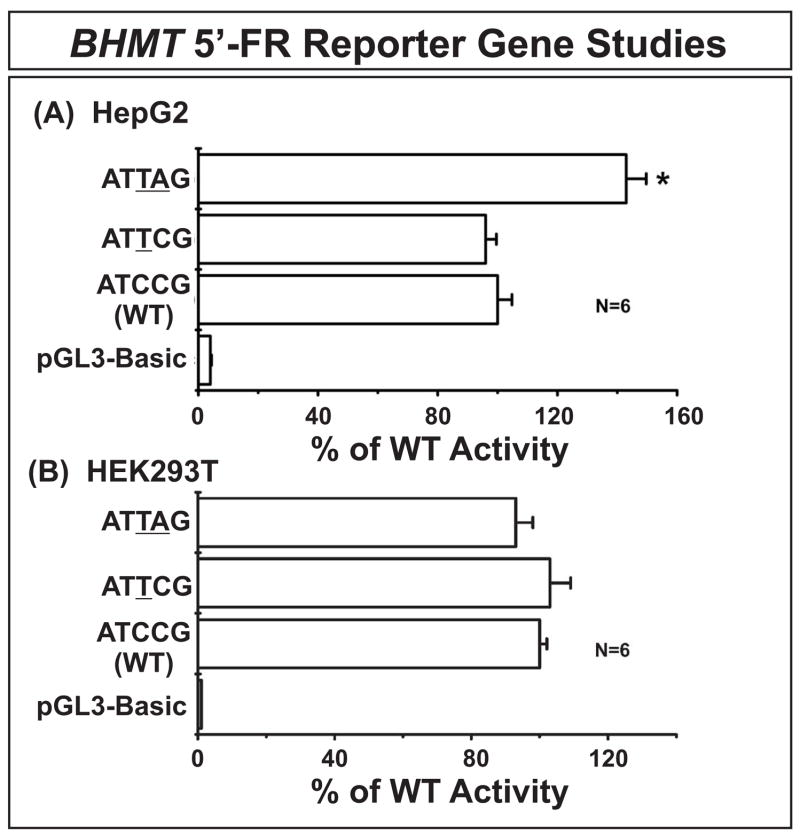
Our resequencing studies had identified 5 SNPs within the initial 1 kb of the BHMT 5′-FR (Table 1 and Fig. 2). To determine whether those SNPs might influence transcription, haplotypes within this region of BHMT were either determined or inferred (Table 5), and luciferase reporter gene constructs were created for all BHMT 5′-FR haplotypes with frequencies greater than 1% in at least one of the populations studied. The ATCCG haplotype was most common in all populations and was designated as the WT. ATTCG was present at a frequency of over 10% in both the AA and CA populations, and ATTAG was found only in the AA population, with a frequency of 2%. These three constructs were used to transfect HepG2 and HEK293T cells, and dual-luciferase assays were performed (Fig. 4A). There was cell line-dependent variation in transcription. In HepG2 cells, but not the HEK293T cells, the ATTAG haplotype showed an increase of approximately 40% in reporter gene activity when compared with the WT (p < 0.01) (Fig. 4A and Fig. 4B). When we compared the nucleotides at positions (−412) and (−448) with those in the mouse and rat, we found that the WT nucleotide in humans at (−412) was conserved in both rodent species, while the variant was present in both at nucleotide (−448).
Table 5.
Human BHMT 5′-FR haplotypes. The table lists haplotypes for 5′-FR sequences. Nucleotide positions are numbered as described in the legend for Table 1.
| Frequency, % | 5′-FR Nucleotides | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Reporter Gene Constructs | AA | CA | HCA | MA | (−809) | (−773) | (−448) | (−412) | (−142) |
| ATCCG (WT) | 87 | 88 | 99 | 96 | A | T | C | C | G |
| ATTCG | 10 | 12 | --- | 4 | A | T | T | C | G |
| ATTAG | 2 | --- | --- | --- | A | T | T | A | G |
Figure 4.
Human BHMT 5′-FR reporter gene studies. Luciferase activity levels for reporter gene constructs containing BHMT 5′-FR haplotypes transfected into (A) HepG2 or (B) HEK293T cells are shown as percentages of values for the WT haplotype. See Table 4 for the locations of SNPs. Each bar represents the average of six independent experiments (mean ± SEM); * p < 0.01.
BHMT2 expression and degradation
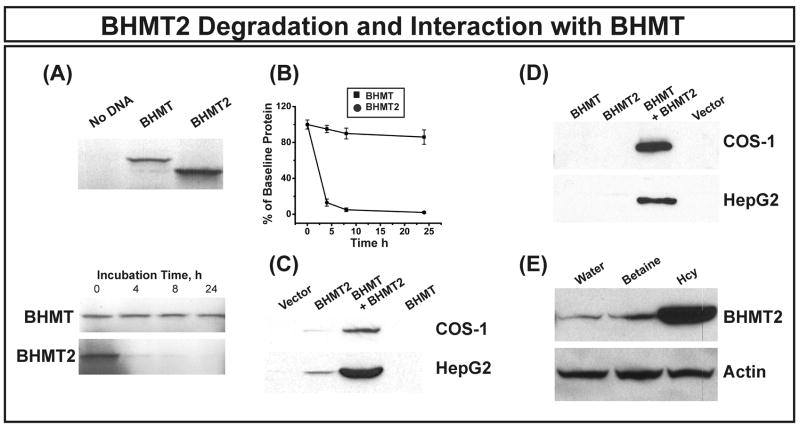
The human BHMT2 gene is predicted to encode a 363 amino acid protein with 73% amino acid sequence identity to BHMT. The encoded protein differs primarily in that it lacks 34 amino acids at the C-terminus as compared with BHMT. As mentioned previously, these two genes probably originated as a result of a tandem duplication event. Human BHMT2 mRNA is expressed in the liver and kidney [6], but there are no published reports regarding the properties – or even the presence – of this protein in human tissues. When we attempted functional genomic studies for BHMT2 which paralleled those described in preceding paragraphs for BHMT, we found one possible explanation for this lack of functional information. All of our attempts to express BHMT2 in mammalian cell lines were unsuccessful, raising the possibility that this protein might be unstable and/or rapidly degraded. Therefore, we performed BHMT2 protein degradation studies using the rabbit reticulocyte lysate (RRL), an experimental system that has been widely used to study protein degradation [15; 28; 29]. As a first step, BHMT and BHMT2 were translated in vitro using a “treated” RRL. Both proteins could be synthesized (Fig. 5A). However, when these [35S]methionine radioactively labeled proteins were added to an untreated RRL that included an ATP generating system, BHMT2 was degraded much more rapidly than was BHMT, with a loss of nearly 90% of BHMT2 in 4 h (Fig. 5A and 5B).
Figure 5.
BHMT2 translation, degradation and interaction with BHMT. (A) Human BHMT and BHMT2 in vitro translation in a rabbit reticulocyte lysate (RRL) is shown in the top panel. Representative autoradiographs for 35S-methionine radioactively-labeled BHMT and BHMT2 at different time points in degradation experiments are shown in the bottom panel. The data shown are representative of three independent experiments. (B) BHMT and BHMT2 protein remaining at each time point in the RRL degradation study, expressed as percentages of the basal value. Each point is the mean ± SEM for 3 independent experiments. BHMT2 differed significantly from BHMT at each point (p < 0.0001). (C) Immunoblot analysis of HA-tagged BHMT2 expressed in COS-1 and HepG2 cells either together with BHMT or alone. Western blotting was performed with anti-HA antibody. Samples were loaded on the SDS-PAGE gels based on cotransfected β-galactosidase activity to correct for possible variation in transfection efficiency. (D) BHMT2 and BHMT interaction. HA-tagged BHMT2 was transfected or cotransfected with BHMT into COS-1 and HepG2 cells. IP was performed with equal quantities of total protein and anti-HA antibody, followed by immunoblot analysis with anti-BHMT antibody. Results shown are representative of 3 independent experiments. (E) HA-tagged BHMT2 expressed in HEK293T cells was stabilized in the presence of 1 mM homocysteine. Cells expressing HA-tagged BHMT2 were cultured with 1 mM homocysteine (Hcy), 1 mM betaine or water for 24 h. Western blot analysis was performed with anti-HA antibody, and actin was used as a control for loading.
SDS-PAGE and gel filtration chromatography of partially purified BHMT suggested that it is a hexamer with identical subunits [14; 30; 31; 32]. However, subsequent crystallization and sedimentation velocity data demonstrated that a tetramer, as depicted in Figure 3C, is the most likely molecular structure [27; 33; 34]. Since we had observed that BHMT2 was rapidly degraded, we asked whether BHMT2 might interact with BHMT to form hetero-oligomers, and whether this interaction might stabilize BHMT2 in the cellular environment. To test that hypothesis, we coexpressed HA-tagged BHMT2 with BHMT in COS-1 and HepG2 cells, probed those preparations with anti-HA antibody, and found that HA-tagged BHMT2 was stabilized in the presence of BHMT (Fig. 5C). IP was also performed to determine whether BHMT2 might interact with BHMT. Specifically, when anti-HA antibody was used to pull down HA-tagged BHMT2, it also precipitated BHMT (Fig. 5D). Finally, we tested the hypothesis that one of the cosubstrates for the BHMT-catalyzed reaction, betaine or homocysteine, might stabilize BHMT2. We found that BHMT2 was stabilized when HEK293T cells expressing this protein were cultured in the presence of 1 mM homocysteine (Fig. 5E), but there was no effect when a similar experiment was performed in the presence of betaine. We also expressed BHMT2 in bacteria.
Bacterial expression and BHMT2 activity
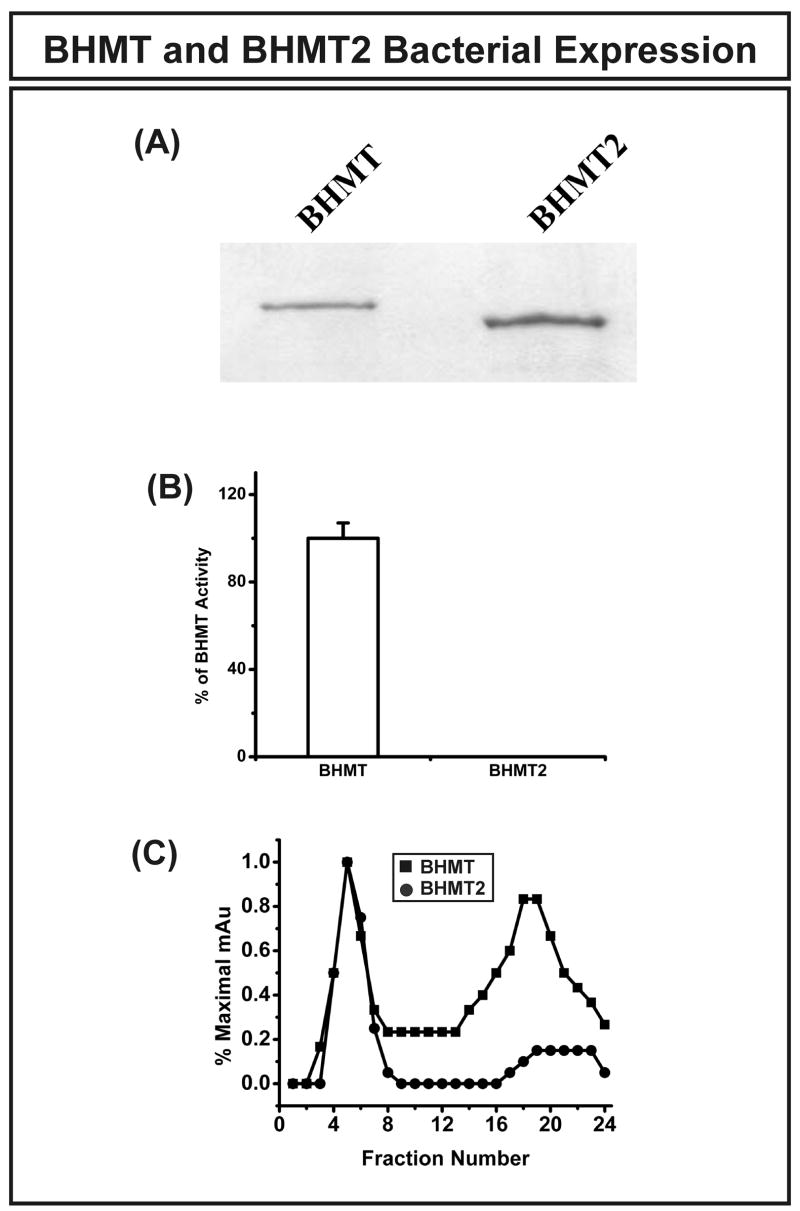
BHMT and BHMT2 were expressed in E. coli as GST fusion proteins. The GST-tags were removed after purification, and similar quantities of BHMT and BHMT2 (Fig. 6A) were used to test their ability to transfer a methyl group from betaine to homocysteine. Bacterially expressed BHMT showed very high activity with this assay, but BHMT2 was unable to catalyze the reaction (Fig. 6B). That observation could indicate either that BHMT2 was unable to catalyze the reaction or that the protein was inactive. To determine whether BHMT2 might aggregate after E. coli expression, we performed size-exclusion chromatography with recombinant GST-BHMT and GST-BHMT2. BHMT2 eluted from the column almost entirely with the void volume, but only approximately 50% of BHMT did so (Fig. 6C). Therefore, the size-exclusion chromatography data were consistent with the results of our enzyme assays, i.e. this experiment indicated that BHMT2 tends to aggregate and, as a result, might be expected to be catalytically inactive.
Figure 6.
BHMT and BHMT2 bacterial expression. (A) BHMT and BHMT2 were expressed as GST fusion proteins in BL21 E. coli. The GST-tags were removed by PreScission Protease after protein purification. (B) Recombinant BHMT and BHMT2 enzyme activity. Equal quantities of bacterially expressed BHMT and BHMT2 were used to perform enzyme assays with L-homocysteine and betaine as substrates. (C) Size-exclusion chromatography of purified bacterially expressed recombinant GST-BHMT and GST-BHMT2 proteins. Protein profiles are expressed as percentages of peak milliabsorbance units (mAu). The initial peaks for both preparations eluted with the void volume.
Discussion
BHMT is a key enzyme for the remethylation of homocysteine to form methionine (Fig. 1). However, despite its abundance in the liver and kidney and its important role in homocysteine remethylation, very little is known with regard to common genetic variation in BHMT. Even less information is available with regard to BHMT2 – with virtually nothing currently known about the function of the protein encoded by this gene. The present study was conducted to systematically identify common sequence variation in the genes encoding BHMT and BHMT2 and to study the effect of that variation on the function of variant allozymes and on transcription. We were able to achieve those goals for BHMT, but – in the course of our studies – it became apparent that the current lack of information with regard to the function of BHMT2 may result, in part, from the fact that this protein is rapidly degraded. Those observations also led us to study possible interactions between BHMT and BHMT2.
The first step in our studies involved the resequencing of BHMT and BHMT2 using 240 DNA samples from 4 ethnic groups. In BHMT, 25 SNPs were observed, including 4 nonsynonymous SNPs that resulted in the following changes in encoded amino acids: Arg(16)Cys, Pro(197)Ser, Gly(199)Ser, and Arg(239)Gln. The Gly(199)Ser and Arg(239)Gln variants have been reported previously [35] and do not appear to influence circulating homocysteine levels [36] – observations compatible with our functional genomic results (Fig. 3A and Fig. 3B). Thirty-nine polymorphisms were observed in BHMT2, including 4 nonsynonymous SNPs that resulted in the following alterations in predicted amino acid sequence: Ala(66)Val, Val(155)Phe, Thr(218)Met, and Val(237)Met. As discussed subsequently, we were unable to characterize the possible functional implications of these BHMT2 polymorphisms.
Functional genomic studies performed with recombinant BHMT variant allozymes did not reveal significant changes in levels of either immunoreactive protein or enzyme activity (Fig. 3A and Fig. 3B). BHMT substrate kinetic studies for WT enzyme yielded apparent Km values for homocysteine and betaine that compared favorably with previous reports in other species [2; 7; 14; 30; 37], and there were significant, but not striking, variations among variant allozymes in apparent Km values (Table 4). However, when we attempted functional genomic studies with BHMT2, we were unable to express this protein in mammalian cell lines at levels that could provide useful data. Although BHMT2 mRNA expression has been reported in human liver and kidney [6], there have been no reports with regard to the function of this protein in humans. We found that BHMT2 was rapidly degraded in a RRL system, helping to explain why it has proven difficult to characterize functionally. We also obtained evidence that BHMT2 can interact with BHMT to form hetero-oligomers, and that interaction can at least partially stabilize BHMT2 (Fig. 5C and Fig. 5D). Finally, one of the two cosubstrates for BHMT, homocysteine, was able to stabilize BHMT2 (Fig. 5E). These observations raise the possibility of substrate-dependent regulation of BHMT2 – a hypothesis that should be explored in the course of future studies.
BHMT2 differs from BHMT primarily by the absence of residues 372 to 406 at the C-terminus. The crystal structure of human BHMT shows that the main feature of each subunit is an (α/β)8 barrel composed of residues 11–318 [27]. Residues downstream of 318 form the so-called dimerization arm which interacts with L7 of the opposite monomer. A loop composed of residues 362–365 in the dimerization arm (the hook) wraps around an equivalent loop in the other monomer (see Figure 3C). Partial elimination of the dimerization arm might destabilize BHMT2, contributing to its rapid degradation [38]. However, BHMT might partially stabilize BHMT2 – as shown in Figure 5D – by forming hetero-oligomers through elements other than the C-terminus. Obviously, that conclusion remains speculative until it is tested experimentally.
In summary, BHMT lies at a critical juncture of the methionine and folate cycles (Fig. 1). This enzyme plays an important functional role in homocysteine remethylation. BHMT2 is an intriguing protein with high sequence homologous to BHMT (73% identity in amino acid sequence), but for which functional data are currently sparse. We set out to resequence both BHMT and BHMT2 and to study the functional implication of common sequence variation in these closely related genes. We were able to accomplish those goals for BHMT. However, while performing these experiments, we discovered one reason why so little is known with regard to the function of BHMT2. This protein is rapidly degraded after mammalian expression, possibly as a result of misfolding and aggregation. Furthermore, we demonstrated that BHMT and BHMT2 can interact in the cellular environment – with at least partial stabilization of BHMT2. Whether this interaction has functional implications in vivo remains to be determined, but we can now explore the biological function of BHMT2 with these observations in mind. It will also be possible to utilize sequence variation in these two genes to perform genotype-phenotype correlation studies that may also help us to understand individual genetically-determined variation in the cyclic remethylation of homocysteine in liver and kidney – with potential functional implications for a variety of disease states.
Supplementary Material
Acknowledgments
We thank Luanne Wussow for her assistance with the preparation of this manuscript. This work was supported in part by National Institutes of Health Grants R01 GM28157, R01 GM35720 and U01 GM61388, the Pharmacogenetics Research Network as well as a PhRMA Foundation “Center of Excellence in Clinical Pharmacology Award. Dr. Lee was supported by a Detweiler Traveling Fellowship from the Royal College of Physicians and Surgeons of Canada and a Clinical Research Initiative Fellowship from the Canadian Institutes of Health Research. The gene resequencing data in this paper have been deposited in the National Institutes of Health database Pharm GKB (accession number PS206422 and PS206424).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carmel R, Jacobsen DW. Homocysteine in Health and Disease. Cambridge University Press; New York: 2001. [Google Scholar]
- 2.Millian NS, Garrow TA. Human betaine-homocysteine methyltransferase is a zinc metalloenzyme. Arch Biochem Biophys. 1998;356:93–8. doi: 10.1006/abbi.1998.0757. [DOI] [PubMed] [Google Scholar]
- 3.Finkelstein JD, Martin JJ. Methionine metabolism in mammals. Distribution of homocysteine between competing pathways. J Biol Chem. 1984;259:9508–13. [PubMed] [Google Scholar]
- 4.Sunden SL, Renduchintala MS, Park EI, Miklasz SD, Garrow TA. Betaine-homocysteine methyltransferase expression in porcine and human tissues and chromosomal localization of the human gene. Arch Biochem Biophys. 1997;345:171–4. doi: 10.1006/abbi.1997.0246. [DOI] [PubMed] [Google Scholar]
- 5.Park EI, Garrow TA. Interaction between dietary methionine and methyl donor intake on rat liver betaine-homocysteine methyltransferase gene expression and organization of the human gene. J Biol Chem. 1999;274:7816–24. doi: 10.1074/jbc.274.12.7816. [DOI] [PubMed] [Google Scholar]
- 6.Chadwick LH, McCandless SE, Silverman GL, Schwartz S, Westaway D, Nadeau JH. Betaine-homocysteine methyltransferase-2: cDNA cloning, gene sequence, physical mapping, and expression of the human and mouse genes. Genomics. 2000;70:66–73. doi: 10.1006/geno.2000.6319. [DOI] [PubMed] [Google Scholar]
- 7.Pajares MA, Perez-Sala D. Betaine homocysteine S-methyltransferase: just a regulator of homocysteine metabolism? Cell Mol Life Sci. 2006;63:2792–803. doi: 10.1007/s00018-006-6249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu H, Curry S, Wen S, Wicker NJ, Shaw GM, Lammer EJ, Yang W, Jafarov T, Finnell RH. Are the betaine-homocysteine methyltransferase (BHMT and BHMT2) genes risk factors for spina bifida and orofacial clefts? Am J Med Genet A. 2005;135:274–7. doi: 10.1002/ajmg.a.30739. [DOI] [PubMed] [Google Scholar]
- 9.Herrmann W. Significance of hyperhomocysteinemia. Clin Lab. 2006;52:367–74. [PubMed] [Google Scholar]
- 10.Martin YN, Olson JE, Ingle JN, Vierkant RA, Fredericksen ZS, Pankratz VS, Wu Y, Schaid DJ, Sellers TA, Weinshilboum RM. Methylenetetrahydrofolate reductase haplotype tag single-nucleotide polymorphisms and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:2322–2324. doi: 10.1158/1055-9965.EPI-06-0318. [DOI] [PubMed] [Google Scholar]
- 11.Shield AJ, Thomae BA, Eckloff BW, Wieben ED, Weinshilboum RM. Human catechol O-methyltransferase genetic variation: gene resequencing and functional characterization of variant allozymes. Mol Psychiatr. 2004;9:151–160. doi: 10.1038/sj.mp.4001386. [DOI] [PubMed] [Google Scholar]
- 12.Thomae BA, Eckloff BW, Freimuth RR, Wieben ED, Weinshilboum RM. Human sulfotransferase SULT2A1 pharmacogenetics: genotype-to-phenotype studies. The Pharmacogenomics J. 2002;2:48–56. doi: 10.1038/sj.tpj.6500089. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Thomae B, Eckloff B, Wieben E, Weinshilboum R. Human histamine N-methyltransferase pharmacogenetics: gene resequencing, promoter characterization, and functional studies of a common 5 ′-flanking region single nucleotide polymorphism (SNP) Biochem Pharmacol. 2002;64:699–710. doi: 10.1016/s0006-2952(02)01223-6. [DOI] [PubMed] [Google Scholar]
- 14.Garrow TA. Purification, kinetic properties, and cDNA cloning of mammalian betaine-homocysteine methyltransferase. J Biol Chem. 1996;271:22831–8. doi: 10.1074/jbc.271.37.22831. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Sullivan W, Toft D, Weinshilboum R. Thiopurine S-methyltransferase pharmacogenetics: chaperone protein association and allozyme degradation. Pharmacogenetics. 2003;13:555–64. doi: 10.1097/01.fpc.0000054124.14659.99. [DOI] [PubMed] [Google Scholar]
- 16.Mukherjee B, Salavaggione OE, Pelleymounter LL, Moon I, Eckloff BW, Schaid DJ, Wieben ED, Weinshilboum RM. Glutathione S-transferase omega 1 and omega 2 pharmacogenomics. Drug Met Dispos. 2006;34:1237–46. doi: 10.1124/dmd.106.009613. [DOI] [PubMed] [Google Scholar]
- 17.Long JC, Williams RC, Urbanek M. An E-M algorithm and testing strategy for multiple-locus haplotypes. Am J Hum Genet. 1995;56:799–810. [PMC free article] [PubMed] [Google Scholar]
- 18.Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12:921–7. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 19.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartl DL, Clark AG. Principles of Population Genetics. Sinauer Associates; Sunderland, MA: 1997. [Google Scholar]
- 22.Hedrick PW. Genetics of Populations. Jones and Bartlett Publishers; Sudbury, Mass: 2000. [Google Scholar]
- 23.Martin YN, Salavaggione OE, Eckloff BW, Wieben ED, Schaid DJ, Weinshilboum RM. Human methylenetetrahydrofolate reductase pharmacogenomics: gene resequencing and functional genomics. Pharmacogenet Genomics. 2006;16:265–77. doi: 10.1097/01.fpc.0000194423.20393.08. [DOI] [PubMed] [Google Scholar]
- 24.Fullerton SM, Clark AG, Weiss KM, Nickerson DA, Taylor SL, Stengard JH, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, Sing CF. Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am J Hum Genet. 2000;67:881–900. doi: 10.1086/303070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roden DM, Altman RB, Benowitz NL, Flockhart DA, Giacomini KM, Johnson JA, Krauss RM, McLeod HL, Ratain MJ, Relling MV, Ring HZ, Shuldiner AR, Weinshilboum RM, Weiss ST. Pharmacogenomics: challenges and opportunities. Ann Intern Med. 2006;145:749–57. doi: 10.7326/0003-4819-145-10-200611210-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7:781–91. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 27.Evans JC, Huddler DP, Jiracek J, Castro C, Millian NS, Garrow TA, Ludwig ML. Betaine-homocysteine methyltransferase: zinc in a distorted barrel. Structure. 2002;10:1159–71. doi: 10.1016/s0969-2126(02)00796-7. [DOI] [PubMed] [Google Scholar]
- 28.Tai HL, Fessing MY, Bonten EJ, Yanishevsky Y, d’Azzo A, Krynetski EY, Evans WE. Enhanced proteasomal degradation of mutant human thiopurine S-methyltransferase (TPMT) in mammalian cells: mechanism for TPMT protein deficiency inherited by TPMT*2, TPMT*3A, TPMT*3B or TPMT*3C. Pharmacogenetics. 1999;9:641–50. doi: 10.1097/01213011-199910000-00011. [DOI] [PubMed] [Google Scholar]
- 29.Siegel D, Anwar A, Winski SL, Kepa JK, Zolman KL, Ross D. Rapid polyubiquitination and proteasomal degradation of a mutant form of NAD(P)H:quinone oxidoreductase 1. Mol Pharmacol. 2001;59:263–8. doi: 10.1124/mol.59.2.263. [DOI] [PubMed] [Google Scholar]
- 30.Skiba WE, Taylor MP, Wells MS, Mangum JH, Awad WM., Jr Human hepatic methionine biosynthesis. Purification and characterization of betaine:homocysteine S-methyltransferase. J Biol Chem. 1982;257:14944–8. [PubMed] [Google Scholar]
- 31.Finkelstein JD, Harris BJ, Kyle WE. Methionine metabolism in mammals: kinetic study of betaine-homocysteine methyltransferase. Arch Biochem Biophys. 1972;153:320–4. doi: 10.1016/0003-9861(72)90451-1. [DOI] [PubMed] [Google Scholar]
- 32.Lee KH, Cava M, Amiri P, Ottoboni T, Lindquist RN. Betaine:homocysteine methyltransferase from rat liver: purification and inhibition by a boronic acid substrate analog. Arch Biochem Biophys. 1992;292:77–86. doi: 10.1016/0003-9861(92)90053-y. [DOI] [PubMed] [Google Scholar]
- 33.Bose N, Momany C. Crystallization and preliminary X-ray crystallographic studies of recombinant human betaine-homocysteine S-methyltransferase. Acta Crystallogr D Biol Crystallogr. 2001;57:431–3. doi: 10.1107/s0907444900020576. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez B, Pajares MA, Too HP, Garrido F, Blundell TL, Sanz-Aparicio J. Crystallization and preliminary X-ray study of recombinant betaine-homocysteine S-methyltransferase from rat liver. Acta Crystallogr D Biol Crystallogr. 2002;58:1507–10. doi: 10.1107/s0907444902011885. [DOI] [PubMed] [Google Scholar]
- 35.Morin I, Platt R, Weisberg I, Sabbaghian N, Wu Q, Garrow TA, Rozen R. Common variant in betaine-homocysteine methyltransferase (BHMT) and risk for spina bifida. Am J Med Genet A. 2003;119:172–6. doi: 10.1002/ajmg.a.20115. [DOI] [PubMed] [Google Scholar]
- 36.Heil SG, Lievers KJ, Boers GH, Verhoef P, den Heijer M, Trijbels FJ, Blom HJ. Betaine-homocysteine methyltransferase (BHMT): genomic sequencing and relevance to hyperhomocysteinemia and vascular disease in humans. Mol Genet Met. 2000;71:511–9. doi: 10.1006/mgme.2000.3078. [DOI] [PubMed] [Google Scholar]
- 37.Castro C, Gratson AA, Evans JC, Jiracek J, Collinsova M, Ludwig ML, Garrow TA. Dissecting the catalytic mechanism of betaine-homocysteine S-methyltransferase by use of intrinsic tryptophan fluorescence and site-directed mutagenesis. Biochemistry. 2004;43:5341–51. doi: 10.1021/bi049821x. [DOI] [PubMed] [Google Scholar]
- 38.Szegedi SS, Garrow TA. Oligomerization is required for betaine-homocysteine S-methyltransferase function. Arch Biochem Biophys. 2004;426:32–42. doi: 10.1016/j.abb.2004.03.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.