Abstract
The telomeric P elements TP5 and TP6 are associated with the P cytotype, a maternally inherited condition that represses P-element-induced hybrid dysgenesis in the Drosophila germ line. To see if cytotype repression by TP5 and TP6 might be mediated by the polypeptides they could encode, hobo transgenes carrying these elements were tested for expression of mRNA in the female germ line and for repression of hybrid dysgenesis. The TP5 and TP6 transgenes expressed more germ-line mRNA than the native telomeric P elements, but they were decidedly inferior to the native elements in their ability to repress hybrid dysgenesis. These paradoxical results are inconsistent with the repressor polypeptide model of cytotype. An alternative model based on the destruction of P transposase mRNA by Piwi-interacting (pi) RNAs was supported by finding reduced P mRNA levels in flies that carried the native telomeric P elements, which are inserted in a known major piRNA locus.
TRANSPOSABLE elements are significant components of the genomes of many organisms. These elements can cause gene mutations and chromosome breakage, and over evolutionary time, they can alter the composition and structure of genomes. There is, therefore, considerable interest in elucidating the mechanisms that foster or repress their activity. For example, many researchers have studied the regulation of P transposable elements in Drosophila melanogaster—a model family of elements in a model genetic organism (Engels 1989; Rio 1990).
P elements are cut-and-paste transposons whose activity is catalyzed by an 87-kDa polypeptide, the P transposase, which is encoded by complete members of the P-element family (Karess and Rubin 1984; Rio et al. 1986). This polypeptide is restricted to the germ line because the last of the element's three introns is removed from P RNA only in that tissue (Laski et al. 1986). In somatic cells, retention of this intron results in the production of a 66-kDa polypeptide instead of the transposase. Tissue-specific splicing is therefore an important mechanism for controlling P-element activity. However even within the germ line, where the P transposase is made, P-element activity is regulated.
Genetic evidence for this regulation was obtained from early studies that defined a maternally transmitted state called cytotype (Engels 1979; Kidwell 1981). The M cytotype permits P-element activity whereas the P cytotype represses it. Thus, when P elements are combined with the M cytotype by crossing P-bearing males to M-cytotype females, the P elements are mobilized in the offspring, where they cause a syndrome of germ-line abnormalities called hybrid dysgenesis (Kidwell et al. 1977; Engels 1989). This syndrome includes gonadal dysgenesis (GD) in both of the sexes, chromosome breakage, and elevated mutation rates. By contrast, crosses between P-bearing males and P-cytotype females produce offspring that seldom show dysgenic traits. These early studies also demonstrated that the P cytotype depends on the presence of P elements themselves (Engels 1979; Sved 1987); thus, the P-element family is autoregulated.
For many years the P cytotype has been attributed to the 66-kDa polypeptide encoded by complete P elements through alternate splicing of P transcripts (Rio 1990; Misra and Rio 1990; Roche et al. 1995). This polypeptide acts as a repressor of transposase activity and appears to be made in the germ line as well as the soma (Simmons et al. 2002a). However, recent studies have shown that incomplete P elements incapable of encoding this polypeptide are able to evoke the P cytotype (Marin et al. 2000; Stuart et al. 2002; Simmons et al. 2004). These elements are located in the telomere-associated sequences (TAS) at the left telomere of the X chromosome. When transmitted maternally, each of these elements represses germ-line P-element activity.
It is possible that these incomplete, telomeric P elements encode small polypeptides that, like the 66-kDa repressor, regulate P-element activity. Some incomplete P elements are known to encode such polypeptides (Black et al. 1987; Rasmusson et al. 1993; Andrews and Gloor 1995; Lee et al. 1998; Simmons et al. 2002b) and these types of elements are prevalent in natural populations, presumably because natural selection has favored their spread; however, unlike telomeric P elements, repressor-producing P elements need not be transmitted maternally in crosses to exert their effects (Black et al. 1987; Simmons et al. 2002b). Furthermore, although many—perhaps most—Drosophila in natural populations have P elements inserted at the left end of the X chromosome (Ajioka and Eanes 1989), the particular kinds of incomplete P elements found there are not widespread (Stuart et al. 2002). These observations suggest that cytotype regulation by telomeric P elements has little to do with the ability of these elements to produce repressor polypeptides; rather, it may involve a different class of regulatory molecules. In this regard, Brennecke et al. (2007) have shown that the TAS at the left end of the X chromosome generate small RNAs that associate with some of the proteins involved in RNA interference—the Piwi-type proteins. These Piwi-interacting (pi) RNAs may therefore play a role in regulating transposon families whose members have inserted within or near the TAS.
To test these alternate models of cytotype regulation, we have examined the expression and biological functions of two incomplete telomeric P elements, TP5 and TP6, in their native positions within the TAS of the X chromosome and in transgenes inserted elsewhere in the genome. The polypeptide repressor model predicts that the native telomeric P elements, which are powerful repressors of hybrid dysgenesis (Stuart et al. 2002), should produce coding mRNAs in the germ line; furthermore, it predicts that if the transgenic counterparts of these elements also express coding mRNAs, they too should repress hybrid dysgenesis. By contrast, the piRNA model predicts that the native telomeric P elements should be underexpressed relative to the transgenic elements because of a shift from mRNA production to piRNA production and that only the native telomeric P elements should be effective regulators of the P family because they are inserted in a piRNA locus. Our data indicate that the native telomeric P elements are underexpressed compared to the transgenic P elements and that only the native elements are effective repressors of hybrid dysgenesis—findings that clearly favor the piRNA model of cytotype regulation.
MATERIALS AND METHODS
Drosophila stocks and husbandry:
The genetic markers and special chromosomes that were used in this work are explained in the FlyBase web site, in Lindsley and Zimm (1992), or in references cited in the text. Experimental cultures were reared on a standard cornmeal–molasses–dried yeast medium at 25° unless stated otherwise.
RNA isolation and RT–PCR:
RNA was isolated from groups of 30 virgin females using TRIZOL reagent (Invitrogen) according to the supplier's instructions. RNA pellets were rehydrated in 20 μl diethylpyrocarbonate (DEPC)-treated water and two 4-μl aliquots from each sample were removed for analysis. One aliquot was subjected to reverse transcription (RT) by the ThermoScript reverse transcriptase (Invitrogen) using an oligo-dT primer in a total volume of 20 μl according to the supplier's instructions. The other aliquot was added to 16 μl of DEPC-treated water to serve as a non-RT control. After RT was completed, both RT and control samples were treated with 1 μl RNase A (10 mg/ml) to clear them of RNA. The DNA in 2-μl aliquots from these samples was then amplified by polymerase chain reaction (PCR) in a total volume of 25 μl using appropriate primers and temperature profiles (supplemental Table S1). Except where noted, all amplifications were over 30 cycles. The PCR products were analyzed on 1% agarose gels run at 70 V; 2 μl 6× tracking dye was mixed into each reaction tube and 20 μl of the mix were inserted into the gel for analysis. Band intensities on the gels were quantified using ImageJ software (Abramoff et al. 2004).
Construction of transgenes and creation of transgenic stocks:
Transgenes designed to express the TP5 and TP6 elements were constructed using (1) the hobo transformation vector pHawN (B. Calvi, personal communication), (2) a cloned fragment containing the hsp70 promoter from Drosophila melanogaster, and (3) PCR-amplified fragments containing the coding regions of the TP5 and TP6 elements. The resulting transgenes (supplemental Figure S1), denoted H(hsp/TP5) and H(hsp/TP6), contain the native promoter of each P element as well as the hsp70 promoter, which is positioned immediately upstream of the P sequence. They also contain a marker gene, mini-white, situated between the 5′ and 3′ segments of the hobo element in pHawN. This gene confers the ability to make eye pigment in stocks that are mutant for the native white gene. Because the P elements in each of these constructs are terminally truncated, they cannot be excised or transposed by the action of the P transposase.
Transgenic stocks were obtained by injecting the H(hsp/TP) constructs into embryos from the y67c23 w stock following the procedure of Simmons et al. (2002a). Each of the transgene insertions was made homozygous by inbreeding and then mapped to a specific chromosome by segregation against dominant markers. All the insertions used in the analysis were located on chromosomes 2 or 3. Stocks carrying autosomal insertions of hobo transgenes containing other types of P elements—CP (complete P), KP, or SP—were also used in this work; all these stocks have been described previously (Simmons et al. 2002a,b).
Gonadal dysgenesis assay for P-element activity:
GD was induced by crossing females from the stocks under test to males from Harwich, a P strain that is homozygous for a null mutation of the white gene. The tests were carried out at 29° according to standard procedures (Simmons et al. 2007a).
Mutability assay for P-element activity:
The snw allele is due to the insertion of two incomplete P elements in the 5′ untranslated region of the singed gene (Roiha et al. 1988). In the presence of the P transposase, either of these elements can be excised at high frequency to create singed alleles that have different phenotypes: extreme singed (sne) or pseudo-wild type (sn+). Because these P-excision events occur in the germ line, the different bristle phenotypes are manifested in the next generation, and their combined frequency can be used to measure the amount of germ-line P-element activity. To assess the effect of the H(hsp/P) transgenes on this activity, each of the transgenes was crossed into a w snw stock. Then homozygous w snw; H(hsp/P) females were crossed at 21° to males homozygous for the stable P transposase source P(ry+, Δ2-3)99B (Robertson et al. 1988) to produce w snw; H(hsp/P)/+ sons that were heterozygous for P(ry+, Δ2-3)99B. These sons were individually mated at 25° to C(1)DX, y f females from a P strain, and their male offspring were scored for the three singed bristle phenotypes—weak, pseudo-wild type, and extreme—as described (Stuart et al. 2002). The frequency of the last two phenotypes among all the flies counted was used to estimate the mutability of snw induced by P(ry+, Δ2-3)99B in the presence of the H(hsp/P) transgene. Control flies without the H(hsp/P) transgenes were produced by crossing w snw, TP5 w snw, or TP6 w snw females (see Stuart et al. 2002) to P(ry+, Δ2-3)99B males at 21°. The (TP) w snw; P(ry+, Δ2-3)99B/+ sons were then individually crossed to C(1)DX, y f females from a P strain at 25° to assess snw mutability in the germ line.
Statistical analyses:
Variances (and standard errors) were computed empirically among data collected from replicate cultures. Differences between the means of experimental groups were assessed by performing z-tests.
RESULTS
Expression of telomeric P elements:
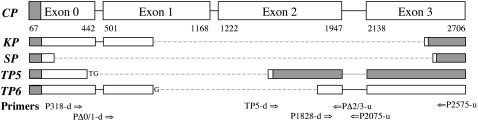
The transcriptional units in the telomeric P elements TP5 and TP6 and in the other P elements CP, KP, and SP are diagrammed in Figure 1. Each of these units was inserted into a transgenic construct downstream of a heat-shock-inducible promoter (hsp70); however, because each element also has its own promoter, expression of these transgenic elements does not require heat shock, and none was employed in any of the experiments. The positions of the primers that were used in the RT–PCR analysis of P-element expression are also shown in Figure 1. One of the primers is specific for TP5 because it spans the deletion breakpoints in this element, and two of them (PΔ0/1-d and PΔ2/3-u) are specific for cDNA derived from spliced P RNA because they span introns. The PΔ2/3-u primer is additionally specific for germ-line cDNA because it spans the last P intron, which is removed only in germ-line cells.
Figure 1.—
Transcribed portions of the CP, KP, SP, TP5, and TP6 elements and binding sites of PCR primers. The first and last nucleotides of each exon are given below the complete element (CP). Deletions in the incomplete elements are indicated by dashed lines; their breakpoints have been reported previously—KP (Black et al. 1987), SP (Rasmusson et al. 1993), TP5 and TP6 (Stuart et al. 2002). Shaded boxes indicate regions that are untranslated or that are translated out of frame compared to the complete element. Arrows show positions of the primer binding sites (sequences can be found in supplemental Table S1).
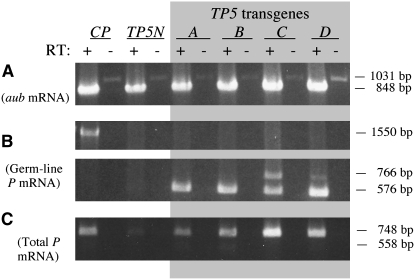
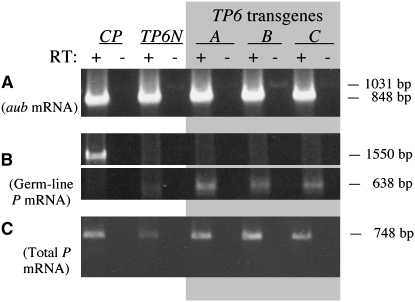
Figures 2 and 3 present the results of the RT–PCR experiments to analyze the expression of TP5 and TP6 in their native telomeric locations on the X chromosome and in hobo transgenes inserted on either of the major autosomes. An autosomal insertion of the CP transgene [H(hsp/CP)2], which produces the P transposase in the germ line (Simmons et al. 2002a), was included in these experiments as a control. RNA isolated from whole adult females carrying these P elements or transgenes was reverse transcribed using an oligo-dT primer. Success of the RT reaction was assessed by amplifying samples of the products with primers located in different exons of the aubergine (aub) gene, which is expressed in the female germ line (Brennecke et al. 2007). All the RT–PCRs yielded approximately equal amounts of the 848-bp DNA product expected from amplification of spliced aub cDNA (Figures 2A and 3A); some of the non-RT samples yielded traces of a 1031-bp product, indicating the presence of contaminating genomic DNA in these RNA isolates.
Figure 2.—
RT–PCR analysis of expression of native (N) and transgenic (A-D) TP5 elements. The CP transgene, which expresses P-transposase mRNA in the germ line, was used as a control. Samples with (+) and without (−) reverse transcription (RT) were analyzed by PCR amplification with appropriate primers. (A) Amplification using primers Aub-d and Aub-u to detect aubergine mRNA. (B) Amplification using primers P318-d and PΔ2/3-u to detect spliced germ-line mRNA from the CP transgene (top) and the TP5 elements (bottom). (C) Amplification over 23 cycles using primers P1828-d and P2575-u to detect total P-element mRNA.
Figure 3.—
RT–PCR analysis of expression of native (N) and transgenic (A-C) TP6 elements. The CP transgene, which expresses P-transposase mRNA in the germ line, was used as a control. Samples with (+) and without (−) reverse transcription (RT) were analyzed by PCR amplification with appropriate primers. (A) Amplification using primers Aub-d and Aub-u to detect aubergine mRNA. (B) Amplification using primers P318-d and PΔ2/3-u to detect germ-line mRNA from the CP transgene (top) and the TP6 elements (bottom). (C) Amplification over 23 cycles using primers P1828-d and P2575-u to detect total P-element mRNA.
The RT and non-RT samples were then amplified with the P-element primers P318-d and PΔ2/3-u, the latter being specific for germ-line P cDNA (Figures 2B and 3B). RT–PCR with the H(hsp/CP) transgene yielded a 1550-bp product showing that fully spliced germ-line CP cDNA was present. The RT–PCRs with the samples from the H(hsp/TP5) and H(hsp/TP6) transgenes also yielded germ-line cDNA products (576 bp for TP5 and 638 bp for TP6); some of these reactions also yielded products ∼190 bp longer, probably because the PΔ2/3-u primer binds downstream of the 2/3 intron in unspliced cDNA and primes DNA synthesis through it. Within each set of results, the amounts of the germ-line cDNA product from different samples of the TP5 and TP6 transgenes appeared roughly similar; thus, different insertions of these transgenes appeared to be expressed more or less equivalently. By contrast, the native telomeric P elements were much less effectively expressed. The TP5 element did not yield a distinct germ-line cDNA product at all, and the TP6 element yielded very little of this product. The rarity of the TP6 germ-line product was confirmed in another PCR using the primers PΔ0/1-d and PΔ2/3-u, which are specific for fully spliced germ-line cDNA (supplemental Figure S2). Collectively, these experiments indicate that in the female germ line the native telomeric P elements are not as effectively expressed as the transgenic P elements.
We also tested the RT samples for P cDNA that retains the 2/3 intron by performing PCR with primers in exons 2 and 3 (Figures 2C and 3C). All the RT samples yielded the expected 748-bp DNA product, although it was less abundant in the samples from the native telomeric elements than in the samples from the transgenic elements; the inferior expression of the native TP6 element was confirmed in another PCR specific for cDNA that lacked the first P intron (supplemental Figure S3). Figure 2C also shows faint traces of a 558-bp product, which represents cDNA lacking the 2/3 intron (i.e., germ-line cDNA). This product was seen in the samples from the TP5 transgenes A and B, but not in any of the other samples. Its rarity suggests that fully spliced P-element RNA is a minor component of the total P-element RNA in these samples.
This RT–PCR analysis indicates that each of the H(hsp/TP) transgenes expresses germ-line-specific and nonspecific P RNA more abundantly than the native telomeric P elements. On the repressor polypeptide model of cytotype regulation, these transgenes would logically be expected to produce more repressor P polypeptides than the native elements. We therefore carried out genetic tests for repressor function in transgenic flies. The positive controls in these experiments had a transgene with KP, an incomplete P element that produces a 22-kDa repressor polypeptide (Lee et al. 1998), or they had a native telomeric P element. The negative controls had a transgene containing SP, a small P element that does not produce a repressor polypeptide (Rasmusson et al. 1993), or they had no transgene at all.
Effects of H(hsp/P) transgenes on GD:
Hybrid dysgenesis occurs in the offspring of crosses between M-cytotype females and P males, but not in the offspring of crosses between P-cytotype females and P males. Failure of the gonads to develop—gonadal dysgenesis—is the most dramatic manifestation of hybrid dysgenesis. We therefore tested the H(hsp/P) transgenes and native telomeric P elements for repression of GD induced in crosses between females carrying these transgenes or elements and males from the strong P strain Harwich (Table 1); the tests were spread over two separate experiments.
TABLE 1.
Gonadal dysgenesis in the offspring of control, TP, and H(hsp/P) stocks
| Stocka | Experimentb | No. of vials | No. of ♀♀ | GD ± SEc (%) |
|---|---|---|---|---|
| y w | 1 | 25 | 460 | 100.0 ± 0.0 |
| 2 | 14 | 176 | 99.6 ± 0.4 | |
| M5B#1 | 1 | 25 | 433 | 86.4 ± 3.2 |
| 2 | 18 | 200 | 90.9 ± 2.0 | |
| TP5-N | 1 | 25 | 489 | 51.5 ± 5.4 |
| 2 | 23 | 399 | 60.1 ± 4.2 | |
| TP6-N | 1 | 29 | 289 | 44.2 ± 6.8 |
| 2 | 16 | 161 | 58.1 ± 8.6 | |
| SP-A | 2 | 20 | 266 | 100.0 ± 0.0 |
| SP-B | 1 | 25 | 420 | 100.0 ± 0.0 |
| KP-3 | 1 | 25 | 462 | 99.4 ± 0.5 |
| KP-7 | 2 | 20 | 268 | 96.1 ± 1.7 |
| KP-14 | 2 | 18 | 209 | 99.7 ± 0.4 |
| TP5-A | 2 | 24 | 137 | 98.9 ± 0.8 |
| TP5-B | 2 | 16 | 241 | 99.7 ± 0,3 |
| TP5-C | 1 | 24 | 331 | 99.7 ± 0.4 |
| TP5-D | 2 | 23 | 331 | 99.3 ± 0.5 |
| TP6-A | 1 | 26 | 373 | 100.0 ± 0.0 |
| TP6-B | 1 | 25 | 490 | 100.0 ± 0.0 |
| TP6-C | 1 | 22 | 287 | 100.0 ± 0.0 |
y w is an M strain and M5B#1 is an M′ strain (Simmons et al. 2007a). TP5-N and TP6-N are strains with the native telomeric P elements on the X chromosome and are the same strains analyzed by Simmons et al. (2007a); other entries are strains with H(hsp/P) transgenes that contain the indicated P element.
The two experiments were carried out at different times.
Unweighted mean percentage of GD ± standard error.
None of the transgenic stocks (SP, KP, TP5, or TP6) showed any ability to repress gonadal dysgenesis. In all cases, the frequency of GD was at or near 100%, as it was in tests with the y w stock, an M-cytotype strain that served as a negative control. In contrast, the telomeric P stocks TP5 and TP6 showed moderate repression (GD frequencies 44–60%). Thus, although the native telomeric P elements could repress GD, the transgenic stocks—even those carrying the KP transgene—could not.
To extend this analysis, we tested if any of the transgenes could repress GD in combination with P elements from M5B#1, a P-bearing strain that by itself has weak repression ability (GD frequency 86–91%, Table 1) but no ability to induce GD. The rationale for these tests comes from Simmons et al. (2007a), who showed that the native telomeric P elements TP5 and TP6 interact synergistically with P elements from M5B#1 to repress GD induced by Harwich. We used the same experimental schemes as Simmons et al. (2007a). Each transgenic strain was reciprocally crossed with M5B#1, and the F1 hybrid females were then crossed to Harwich males to induce GD in their offspring. In the same way, we tested the y w stock as a negative control and the telomeric P stocks TP5 and TP6 as positive controls.
The data from these interaction tests (supplemental Tables S2 and S3) were collected in two experiments carried out in conjunction with the primary tests for repression of GD (Table 1). No interactions between any of the transgenes and the P elements from M5B#1 were observed. In all cases, the frequency of GD in the F2 females was at or near 100%—similar to the results obtained with the negative control stock (y w). However, in both experiments, the native telomeric P elements showed moderate to strong abilities to repress GD when they were combined with P elements from M5B#1. Repression was more effective in the offspring of F1 hybrids produced by crossing TP females with M5B#1 males (denoted cross I) than in the offspring of the reciprocal hybrids (cross II). It was also more effective in offspring from TP6 hybrids (<4% GD in cross I and 65–68% GD in cross II) than in those from TP5 hybrids (35–59% GD in cross I and 82–87% GD in cross II). However, in all cases, the level of repression was not significantly affected by the presence or absence of a telomeric P element in the F2 females themselves. Simmons et al. (2007a) made similar observations.
To ascertain if the repression seen in the offspring from the TP-M5B#1 hybrids was due to an interaction between the telomeric and M5B#1 P elements, we assessed the level of repression in offspring from hybrids between each of the native TP strains and a wild-type M-cytotype strain (Samarkand). The results (supplemental Table S4) indicate that in this hybrid context, TP6 was a weak repressor and TP5 was a negligible repressor. Thus, the moderate to strong repression seen in offspring of the TP-M5B#1 hybrids must be due to a synergism between the telomeric and M5B#1 P elements, as observed by Simmons et al. (2007a). These results show that the native telomeric P elements, TP5 and TP6, interact synergistically with the M5B#1 P elements to repress GD, whereas the H(hsp/P) transgenes do not.
Effects of H(hsp/P) transgenes on snw mutability:
In the germ line P elements are excised and transposed through the action of the P transposase. Previous studies have shown that both the KP and CP transgenes encode repressors of this activity (Simmons et al. 2002a,b). These studies made use of the snw allele, which is unusually sensitive to transposase attack. In the presence of P(ry+, Δ2-3)99B, a P transgene that encodes the P transposase but not the 66-kDa P repressor, either of the two P elements inserted in this allele can be excised to produce singed alleles with different phenotypes. However, when either a KP or a CP transgene is also present, this mutability is partially repressed.
To determine if the TP5 or TP6 transgenes could repress snw mutability, we tested snw; H(hsp/TP)/+ males that were heterozygous for the P(ry+, Δ2-3)99B transposase source. For comparison, we also tested males that carried the native telomeric P elements or the other H(hsp/P) transgenes. The results of all these tests, which were spread over two separate experiments, are summarized in Table 2.
TABLE 2.
Repression of Δ2-3-induced snw mutability by telomeric P elements and H(hsp/P) transgenes
| Experiment 1
|
Experiment 2
|
|||||
|---|---|---|---|---|---|---|
| TP element or transgenea | No. of vials | No. of flies | Mutation rate ± SEb | No. of vials | No. of flies | Mutation rate ± SEb |
| None | 48 | 1155 | 0.557 ± 0.021 | 49 | 1576 | 0.471 ± 0.019 |
| TP5-N | 38 | 534 | 0.061 ± 0.015 | 47 | 1826 | 0.054 ± 0.011 |
| TP6-N | 43 | 1194 | 0.313 ± 0.032 | 50 | 1748 | 0.287 ± 0.023 |
| SP-A | 48 | 1016 | 0.576 ± 0.026 | 50 | 1667 | 0.553 ± 0.018 |
| SP-B | 49 | 1056 | 0.591 ± 0.021 | 45 | 1023 | 0.463 ± 0.028 |
| KP-3 | 50 | 1273 | 0.218 ± 0.018 | 49 | 1925 | 0.227 ± 0.017 |
| KP-11 | 49 | 1526 | 0.351 ± 0.020 | 49 | 1367 | 0.381 ± 0.019 |
| KP-14 | 46 | 1096 | 0.272 ± 0.024 | 42 | 1052 | 0.291 ± 0.022 |
| TP5-A | 49 | 1033 | 0.445 ± 0.023 | |||
| TP5-B | 50 | 1268 | 0.439 ± 0.024 | |||
| TP5-C | 48 | 961 | 0.424 ± 0.023 | |||
| TP5-D | 48 | 1047 | 0.540 ± 0.025 | |||
| TP6-A | 47 | 1468 | 0.443 ± 0.022 | |||
| TP6-B | 49 | 1531 | 0.450 ± 0.023 | |||
| TP6-C | 50 | 1863 | 0.421 ± 0.019 | |||
TP5-N and TP6-N are the native telomeric P elements on the X chromosome; other entries are H(hsp/P) transgenes containing the indicated P element.
Unweighted mean mutation rate ± standard error.
In the absence of any transgene, the germ-line mutation rate of the snw allele was high: 0.557 in experiment 1 and 0.471 in experiment 2. This level of snw mutability was markedly reduced by the native TP5 element (mutation rate = 0.061 in experiment 1 and 0.054 in experiment 2), and it was moderately reduced by the native TP6 element (mutation rate = 0.313 in experiment 1 and 0.287 in experiment 2). Among the control transgenes, neither insertion of H(hsp/SP) reduced snw mutability in either experiment, but all three insertions of H(hsp/KP) reduced it significantly in both experiments (mutation rates ranging from 0.218 to 0.381). The repressing effects of the KP transgene were therefore comparable to those of the native TP6 element. In experiment 1, the mutation rates for the flies that carried a TP5 transgene ranged from 0.424 to 0.540; three of these rates are significantly less than the rates seen in the negative controls (no transgene and both insertions of the SP transgene). Thus, there is evidence that the TP5 transgene represses snw mutability, although less effectively than the KP transgene and much less effectively than the native TP5 element. In experiment 2, the mutation rates for the flies that carried a TP6 transgene ranged from 0.421 to 0.450—not significantly different from two of the negative controls in that experiment (no transgene and SP-B). Thus, the TP6 transgene does not appear to repress snw mutability.
Effects of telomeric P elements on transposase mRNA:
The preceding molecular and genetic analyses of the telomeric and transgenic P elements have shown that the native telomeric P elements express much less mRNA in the germ line than their transgenic counterparts, but they repress hybrid dysgenesis much more strongly. This paradox is difficult to explain on the polypeptide repressor model of cytotype regulation. However, it can be explained on the piRNA model if RNA transcribed from the native telomeric P elements is diced into small RNAs that interact with Piwi-type proteins, and the resulting piRNA/Piwi complexes repress dysgenesis through RNA interference. The simplest way for this process to work would be for the piRNAs to target transposase mRNA for destruction. On this idea, we would expect to find less transposase mRNA in flies that carry a native telomeric P element.
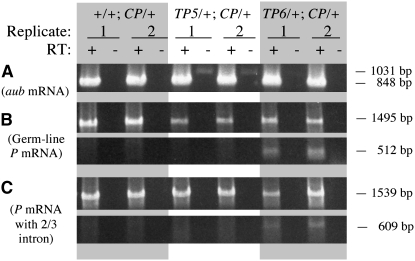
Evidence supporting this idea was obtained from an experiment in which transposase mRNA levels were assessed by RT–PCR (Figure 4). The RNA for this experiment was extracted from the daughters of crosses between females homozygous for the native TP5 or TP6 elements and homozygous H(hsp/CP) males. The native TP5 element in the females came from the stock that had been used as a positive control in the snw mutability experiment (see Table 2) after removing the snw mutation by recombination, and the native TP6 element came from the stock that had been used as a positive control in the gonadal dysgenesis experiment (see Table 1). For comparison, we analyzed RNA from the daughters of crosses between M-cytotype females devoid of any P elements and homozygous H(hsp/CP) males. Two independently isolated RNA samples from each genotype were reverse transcribed using an oligo-dT primer and the cDNA products were subsequently amplified by PCR. Even though amplification with primers for the aubergine gene indicated that aub cDNAs were equally abundant in all the RT samples (Figure 4A), amplification with primers for germ-line P cDNA showed that transposase mRNA from the CP transgene was less abundant in the samples from the flies with the native telomeric P elements and least abundant in the two samples from the TP5-bearing flies (Figure 4B). This last observation is noteworthy because as this molecular analysis was underway, we tested the native TP5 and TP6 strains for their ability to repress gonadal dysgenesis induced by the P strain Harwich and found that TP5 was a much stronger repressor (GD frequency 6% compared to 64% for TP6 and 99% for the M-cytotype control; supplemental Table S5. Note that the TP5 strain used here is also a stronger repressor of GD than the TP5 strain used in the experiment summarized in Table 1). A final PCR using primers for cDNA from the complete P element that retained the 2/3 intron (i.e., not specific to the germ line) showed no consistent reduction of product in the presence of the native telomeric P elements (Figure 4C). Thus, the diminution of CP RNA by the native telomeric P elements was apparently confined to the germ line.
Figure 4.—
RT–PCR analysis of transposase mRNA expression in flies carrying the CP transgene and the native TP5 or TP6 elements. Replicate samples (1 and 2) with (+) and without (−) reverse transcription (RT) were analyzed by PCR amplification with appropriate primers. (A) Amplification using primers Aub-d and Aub-u to detect aubergine mRNA. (B) Amplification using primers PΔ0/1-d and PΔ2/3-u to detect germ-line P-transposase mRNA from the CP transgene (top) and fully spliced germ-line mRNA from the TP6 element (bottom). (C) Amplification using primers PΔ0/1-d and P2075-u to detect partially spliced mRNA from the CP transgene (top) and the TP6 element (bottom); this mRNA retains the 2/3 intron and is therefore not germ-line specific.
To confirm these findings, we performed a similar RT–PCR analysis in which eight independently isolated RNA samples from TP5/+; H(hsp/CP)/+ females were randomly paired with eight RNA samples from females that carried an H(hsp/TP5) transgene instead of the native TP5 element. Each of the four insertions of the H(hsp/TP5) transgene was represented by two independent RNA samples. After reverse transcribing the RNA with an oligo-dT primer, we amplified the cDNAs with PCR using different sets of primers and then compared the abundance of the PCR products between pairs of samples, one from flies that had the native TP5 element and the other from flies that had a TP5 transgene. In these comparisons, we noted the number of pairs in which the native TP5 sample had at least 20% less PCR product than the transgene sample. This conservative approach ignores nominal differences between the paired native and transgene samples. The original data are shown in supplemental Figure S4 and the results of the pairwise comparisons are summarized in Table 3. There was no tendency for the native TP5 samples to have less product than the transgene samples in the PCRs for the control aubergine mRNA or for CP mRNA that retained the 2/3 intron (i.e., CP mRNA not specific to the germ line); however, in the PCRs for the germ-line TP5 and CP mRNA, the native TP5 samples had less product than the transgene samples in all eight comparisons. Assuming equal representation of these mRNAs in the native and transgene samples, the probability of this result is 0.0039 in each case. On the basis of this nonparametric statistical test, we therefore reject the assumption of equal representation and conclude that in the germ line, the TP5 and CP mRNAs are significantly less abundant when the native TP5 element is present.
TABLE 3.
Paired comparisons of RT–PCR products from flies with the native TP5 element and H(hsp/TP5) transgenes
| Primers | mRNA detected | Scorea |
|---|---|---|
| Aub-d; Aub-u | aubergine | 2 |
| TP5-d; PΔ2/3-u | Germ-line TP5 | 8 |
| PΔ0/1-d; PΔ2/3-u | Germ-line CP | 8 |
| PΔ0/1-d; P2075-u | CP with 2/3 intron | 4 |
Number of pairs in which the sample with the native TP5 element had at least 20% less PCR product than the sample with the TP5 transgene. A total of 8 pairs were tested.
DISCUSSION
The standard model for the P cytotype postulates that P activity is regulated by P-encoded repressor polypeptides—in particular, the 66-kDa repressor produced from complete P elements by alternate splicing of their transcripts in the germ line (Rio 1990; Roche et al. 1995; Ashburner et al. 2005; Lewin 2007). However, this polypeptide cannot account for repression of hybrid dysgenesis by the telomeric P elements TP5 and TP6 because neither of these elements has the coding potential to produce it. The P cytotype of the TP5 and TP6 strains must therefore have some other explanation.
One possibility is that like the KP element, TP5 and TP6 produce smaller polypeptides that repress P-element activity. However, transgenes containing either TP5 or TP6 have little or no ability to repress hybrid dysgenesis—certainly less than that of the KP transgene. In addition, the TP5 and TP6 transgenes do not affect the phenotypes of repressor-sensitive P-insertion mutations, either in the germ line or in the soma, whereas the KP transgene does (Simmons et al. 2004; K. Newman and M. J. Simmons, unpublished data). These observations cast doubt on a model in which cytotype regulation of the P-element family is mediated by repressor polypeptides produced by elements such as TP5 and TP6.
However, one of these elements—TP5—does seem to be capable of encoding a repressor polypeptide because three of the four insertions of the TP5 transgene repressed snw mutability modestly. The putative TP5 polypeptide would be 113 amino acids long, and the first 95 of these amino acids would be identical to those of the P transposase. This polypeptide would therefore be shorter than the KP polypeptide, which has 207 amino acids and contains the first 199 amino acids of the transposase. In vitro studies have shown that the KP polypeptide represses transposase activity in a concentration-dependent manner by binding competitively to specific sequences in a target P element (Lee et al. 1996, 1998). A DNA-binding domain near the amino terminus of the polypeptide is essential for this repression, and two dimerization domains located between amino acids 101 and 207 facilitate it. Furthermore, a polypeptide that consists of the first 88 amino acids of the KP polypeptide—and therefore that has the DNA-binding domain but lacks both of the dimerization domains—represses in vitro transposase activity only about one-sixth as effectively as the entire KP polypeptide (Lee et al. 1998). Thus, the TP5 polypeptide, which would have the DNA-binding domain but lack the dimerization domains, would be expected to be a weaker repressor than KP—a prediction borne out by the in vivo data, which show a 25% reduction in snw mutability by the TP5 transgene compared to a 40–60% reduction by the KP transgene.
It is unlikely that the weak repressor function of the putative TP5 polypeptide explains the strong repression abilities of the native TP5 element. In its telomeric position, TP5 reduces P excisions from snw by 90%. It also represses gonadal dysgenesis—something not done by any of the transgenes, even H(hsp/KP), and this repression is strengthened when TP5 is combined with P elements from the M5B#1 strain; however, none of the TP5 transgene insertions shows this synergism with M5B#1. It might be argued that in its telomeric position, TP5 produces more repressor polypeptide than it does in a H(hsp/TP5) transgene. However, our RT–PCR analysis indicates that less germ-line mRNA is produced by the native TP5 element than by any of the transgenic TP5 elements. This expression paradox, also seen with the TP6 element, is inconsistent with a model of cytotype on the basis of vigorous production of repressor polypeptides by telomeric P elements. It is, however, consistent with a model in which the telomeric P elements generate piRNAs at the expense of mRNAs. This shift to piRNA generation might be so great that little or no mRNA is produced by the telomeric P element. Indeed, the piRNAs might be derived primarily from antisense transcription of the P element.
Many loci in the Drosophila genome produce piRNAs, and the TAS array at the left end of the X chromosome is one of the major producers (Brennecke et al. 2007). A P element inserted in the TAS would therefore be expected to contribute to piRNA production, as would P's inserted in other piRNA loci, and once made, these RNAs could target transposase mRNA for destruction by an RNA interference mechanism. Indeed, we have shown that the abundance of this mRNA is decreased in flies that carry P elements inserted in the TAS. The efficiency of this targeting/destruction process is not known, although it is not 100% efficient because some transposase mRNA survives. This surviving mRNA might, however, not be translated because it is bound by Piwi-type proteins that are associated with P piRNAs. Thus, transposase synthesis—and ultimately, P-element activity—would still be repressed.
The piRNA model is consistent with the establishment and maintenance of cytotype regulation in the female germ line—paternally inherited telomeric P elements do not confer it (Simmons et al. 2004)—and the prominent expression of the Piwi proteins, which bind piRNAs, in that tissue (Brennecke et al. 2007). In addition, genetic studies have shown that cytotype regulation is disrupted by mutations in aubergine, a gene that encodes one of the Piwi proteins involved in RNA interference (Reiss et al. 2004; Simmons et al. 2007b). Related genetic studies have demonstrated that P-lacZ transgenes inserted in the TAS silence the germ-line expression of P-lacZ transgenes inserted elsewhere in the genome (Roche and Rio 1998; Ronsseray et al. 1998), and, importantly, that this silencing is suppressed by mutations in several genes whose products have been implicated in RNA interference (Josse et al. 2007). Other analyses have implicated repeat-associated small interfering RNAs, which may be the same as piRNAs, in the regulation of diverse transposons, including those that insert specifically at the ends of Drosophila chromosomes (Vagin et al. 2006). Cytotype regulation of P elements may therefore be a special case of a general mechanism that represses transposable elements by RNA interference.
Acknowledgments
Jordan Schoephoerster provided technical help, Johng Lim kindly made comments on the manuscript, and two reviewers provided helpful suggestions to improve it. Construction of the H(hsp/TP) transgenes was funded by National Institutes of Health grant R01-GM40263. Genetic analysis was supported by the University of Minnesota Foundation. P.A.J. was the recipient of a grant from the University of Minnesota's Undergraduate Research Opportunities Program. Other support came from the Department of Genetics, Cell Biology, and Development and the College of Biological Sciences of the University of Minnesota.
References
- Abramoff, M. D., P. J. Magelhaes and S. J. Ram, 2004. Image processing with ImageJ. Biophotonics Int. 11 36–42. [Google Scholar]
- Ajioka, J. W., and W. F. Eanes, 1989. The accumulation of P-elements on the tip of the X chromosome in populations of Drosophila melanogaster. Genet. Res. 53 1–6. [DOI] [PubMed] [Google Scholar]
- Andrews, J. D., and G. B. Gloor, 1995. A role for the KP leucine zipper in regulating P element transposition. Genetics 141 587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., K. G. Golic and R. S. Hawley, 2005. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Black, D. M., M. S. Jackson, M. G. Kidwell and G. A. Dover, 1987. KP elements repress P-induced hybrid dysgenesis in Drosophila melanogaster. EMBO J. 6 4125–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J., A. A. Aravin, A. Stark, M. Dus, M. Kellis et al., 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128 1089–1103. [DOI] [PubMed] [Google Scholar]
- Engels, W. R., 1979. Hybrid dysgenesis in Drosophila melanogaster: rules of inheritance of female sterility. Genet. Res. 33 219–236. [DOI] [PubMed] [Google Scholar]
- Engels, W. R., 1989. P elements in Drosophila melanogaster, pp. 437–484 in Mobile DNA, edited by D. E. Berg and M. M. Howe. American Society for Microbiology Publications, Washington, DC.
- Josse, T., L. Teysset, A. L. Todeschini, C. M. Sidor, D. Anxolabéhère et al., 2007. Telomeric trans-silencing: an epigenetic repression combining RNA silencing and heterochromatin formation. PLoS Genet. 3 1633–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess, R., and G. M. Rubin, 1984. Analysis of P transposable element function in Drosophila. Cell 38 135–146. [DOI] [PubMed] [Google Scholar]
- Kidwell, M. G., 1981. Hybrid dysgenesis in Drosophila melanogaster: the genetics of cytotype determination in a neutral strain. Genetics 98 275–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell, M. G., J. F. Kidwell and J. A. Sved, 1977. Hybrid dysgenesis in Drosophila melanogaster: a syndrome of aberrant traits including mutation, sterility, and male recombination. Genetics 86 813–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laski, F. A., D. C. Rio and G. M. Rubin, 1986. Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell 44 7–19. [DOI] [PubMed] [Google Scholar]
- Lee, C. C., Y. M. Mul and D. C. Rio, 1996. The Drosophila P-element KP repressor protein dimerizes and interacts with multiple sites on P-element DNA. Mol. Cell. Biol. 16 5616–5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. C., E. L. Beall and D. C. Rio, 1998. DNA binding by the KP repressor protein inhibits P-element transposase activity in vivo. EMBO J. 17 4166–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin, B., 2007. Transposons, pp. 521–549 in Genes IX. Jones & Bartlett, Sudbury, MA.
- Lindsley, D. L., and G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, New York.
- Marin, L., M. Lehmann, D. Nouaud, H. Izaabel, D. Anxolabéhère et al., 2000. P-element repression in Drosophila melanogaster by a naturally occurring defective telomeric P copy. Genetics 155 1841–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra, S., and D. C. Rio, 1990. Cytotype control of Drosophila P element transposition: the 66 kD protein is a repressor of transposase activity. Cell 62 269–284. [DOI] [PubMed] [Google Scholar]
- Rasmusson, K. E., J. D. Raymond and M. J. Simmons, 1993. Repression of hybrid dysgenesis in Drosophila melanogaster by individually naturally occurring P elements. Genetics 133 605–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss, D., T. Josse, D. Anxolabéhère and S. Ronsseray, 2004. aubergine mutations in Drosophila melanogaster impair P cytotype determination by telomeric P elements inserted in heterochromatin. Mol. Genet. Genomics 272 336–343. [DOI] [PubMed] [Google Scholar]
- Rio, D. C., 1990. Molecular mechanisms regulating Drosophila P element transposition. Annu. Rev. Genet. 24 543–578. [DOI] [PubMed] [Google Scholar]
- Rio, D. C., F. A. Laski and G. M. Rubin, 1986. Identification and immunochemical analysis of biologically active Drosophila P element transposase. Cell 44 21–32. [DOI] [PubMed] [Google Scholar]
- Robertson, H. M., C. R. Preston, R. W. Phillis, D. Johnson-Schlitz, W. K. Benz et al., 1988. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche, S. E., and D. C. Rio, 1998. Trans-silencing by P elements inserted in subtelomeric heterochromatin involves the Drosophila Polycomb group gene, Enhancer of zeste. Genetics 149 1839–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche, S., M. Schiff and D. C. Rio, 1995. P-element repressor autoregulation involves germ-line transcriptional repression and reduction of third intron splicing. Genes Dev. 9 1278–1288. [DOI] [PubMed] [Google Scholar]
- Roiha, H., G. M. Rubin and K. O'Hare, 1988. P element insertions and rearrangements at the singed locus of Drosophila melanogaster. Genetics 119 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsseray, S., L. Marin, M. Lehmann and D. Anxolabéhère, 1998. Repression of hybrid dysgenesis in Drosophila melanogaster by combinations of telomeric P-element reporters and naturally occurring P elements. Genetics 149 1857–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, M. J., K. J. Haley, C. D. Grimes, J. D. Raymond and J. B. Niemi, 2002. a A hobo transgene that encodes the P-element transposase in Drosophila melanogaster: autoregulation and cytotype control of transposase activity. Genetics 161 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, M. J., K. J. Haley, C. D. Grimes, J. D. Raymond and J. C. L. Fong, 2002. b Regulation of P-element transposase activity in Drosophila melanogaster by hobo transgenes that contain KP elements. Genetics 161 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, M. J., J. D. Raymond, J. B. Niemi, J. R. Stuart and P. J. Merriman, 2004. The P cytotype in Drosophila melanogaster: a maternally transmitted regulatory state of the germ line associated with telomeric P elements. Genetics 166 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, M. J., J. B. Niemi, D-F. Ryzek, C. Lamour, J. W. Goodman et al., 2007. a Cytotype regulation by telomeric P elements in Drosophila melanogaster: interactions with P elements from M′ strains. Genetics 176 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, M. J., D-F. Ryzek, C. Lamour, J. W. Goodman, N. E. Kummer et al., 2007. b Cytotype regulation by telomeric P elements in Drosophila melanogaster: evidence for involvement of an RNA interference gene. Genetics 176 1945–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, J. R., K. J. Haley, D. Swedzinski, S. Lockner, P. E. Kocian et al., 2002. Telomeric P elements associated with cytotype regulation of the P transposon family in Drosophila melanogaster. Genetics 162 1641–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved, J. A., 1987. Hybrid dysgenesis in Drosophila melanogaster: evidence from sterility and Southern hybridization tests that P cytotype is not maintained in the absence of chromosomal P factors. Genetics 115 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin, V. V., A. Sigova, C. Li, H. Seitz, V. Gvozdev et al., 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313 320–324. [DOI] [PubMed] [Google Scholar]