Abstract
The appaloosa coat spotting pattern in horses is caused by a single incomplete dominant gene (LP). Homozygosity for LP (LP/LP) is directly associated with congenital stationary night blindness (CSNB) in Appaloosa horses. LP maps to a 6-cM region on ECA1. We investigated the relative expression of two functional candidate genes located in this LP candidate region (TRPM1 and OCA2), as well as three other linked loci (TJP1, MTMR10, and OTUD7A) by quantitative real-time RT–PCR. No large differences were found for expression levels of TJP1, MTMR10, OTUD7A, and OCA2. However, TRPM1 (Transient Receptor Potential Cation Channel, Subfamily M, Member 1) expression in the retina of homozygous appaloosa horses was 0.05% the level found in non-appaloosa horses (R = 0.0005). This constitutes a >1800-fold change (FC) decrease in TRPM1 gene expression in the retina (FC = −1870.637, P = 0.001) of CSNB-affected (LP/LP) horses. TRPM1 was also downregulated in LP/LP pigmented skin (R = 0.005, FC = −193.963, P = 0.001) and in LP/LP unpigmented skin (R = 0.003, FC = −288.686, P = 0.001) and was downregulated to a lesser extent in LP/lp unpigmented skin (R = 0.027, FC = −36.583, P = 0.001). TRP proteins are thought to have a role in controlling intracellular Ca2+ concentration. Decreased expression of TRPM1 in the eye and the skin may alter bipolar cell signaling as well as melanocyte function, thus causing both CSNB and LP in horses.
COAT color has been a fascinating topic of genetic discussion and discovery for over a century. The pigment genes of mice were one of the first genetic systems to be explored through breeding and transgenic studies. To date, at least 127 loci involved in pigmentation have been described (Silvers 1979; Bennett and Lamoreux 2003). The genes that affect pigmentation in the skin and hair influence other body systems, and many of these genes have been studied in different mammals. One of the most extensively studied examples is oculocutaneous albinism type 1, a developmental disorder in humans that affects pigmentation in the skin and hair, as well as eye development. This disease is caused by mutations in the tyrosinase gene (TYR), which is involved in the first step of melanin production (Toyofuko et al. 2001; Ray et al. 2007).
Horses (Equus caballus) are valued by breeders and enthusiasts for their beauty and variety of coat color and patterns. The genetic mechanisms involved in several different variations of coloration and patterning in horses have been reported, including chestnut, frame overo, cream, black, silver dapple, sabino-1 spotting, tobiano spotting, and dominant white spotting (Marklund et al. 1996; Metallinos et al. 1998; Mariat et al. 2003; Rieder et al. 2001; Brooks and Bailey 2005; Brunberg et al. 2006; Brooks et al. 2007; Haase et al. 2007). The mechanism behind appaloosa spotting, a popular coat pattern occurring in several breeds of horses, remains to be elucidated. Likewise, although there are several inherited ocular diseases reported in the horse (cataracts, glaucoma, anterior segment dysgenesis, and congenital stationary night blindness), the modes of inheritance, genetic mutations, and the pathogenesis of these ocular disorders remain unknown.
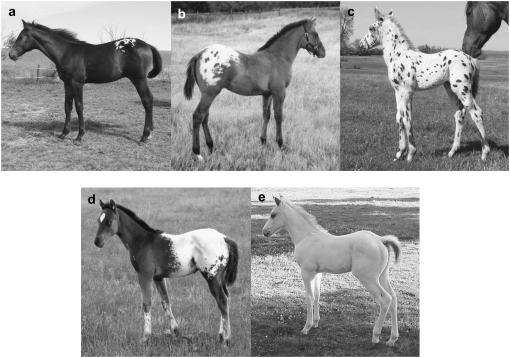
Appaloosa spotting is characterized by patches of white in the coat that tend to be symmetrical and centered over the hips. In addition to the patterning in the coat, appaloosa spotted horses have three additional pigmentation traits: striped hooves, readily visible nonpigmented sclera around the eye, and mottled pigmentation around the anus, genitalia, and muzzle (Sponenberg and Beaver 1983). The extent of spotting varies widely among individuals, resulting in a collection of patterns that are termed “the leopard complex” (Sponenberg et al.1990). The spectrum of patterns with the leopard complex includes very minimal white patches on the rump (known as a “lace blanket”), a white body with many oval or round pigmented spots dispersed throughout (known as “leopard,” from which the genetic locus is named), and nearly complete depigmentation (known as “fewspot”) (Figure 1). A single autosomal dominant gene, leopard complex (LP), is thought to be responsible for the inheritance of these patterns and associated traits, while modifier genes are thought to play a role in determining the amount of white patterning that is inherited (Miller 1965; Sponenberg et al. 1990; S. Archer and R. R. Bellone, unpublished data). Horses that are homozygous for appaloosa spotting (LP/LP) tend to have fewer spots than heterozygotes on the white patterned areas; these horses are known as “fewspots” (largely white body with little to no spots) and “snowcaps” (white over the croup and hips with little to no spots) (Sponenberg et al. 1990; Lapp and Carr 1998; Figure 1).
Figure 1.—
Horses displaying different Appaloosa coat color patterns. (a) Lace blanket (LP/lp). (b) Spotted blanket (LP/lp). (c) Leopard (LP/lp). (d) Snowcap blanket (LP/LP). (e) Fewspot (LP/LP).
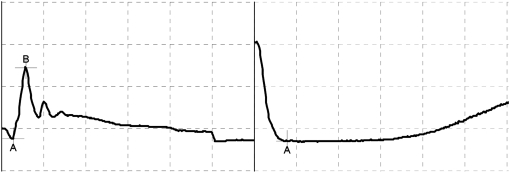
We have recently reported an association between homozygosity for LP and congenital stationary night blindness (CSNB) (Sandmeyer et al. 2007). CSNB is characterized by a congenital and nonprogressive scotopic visual deficit (Witzel et al. 1977, 1978; Rebhun et al. 1984). Affected horses may exhibit apprehension in dimly lit conditions and may be difficult to train and handle in phototopic (light) and scotopic (dark) conditions (Witzel et al. 1977, 1978; Rebhun et al. 1984). Affected animals occasionally manifest a bilateral dorsomedial strabismus (improper eye alignment) and nystagmus (involuntary eye movement) (Rebhun et al. 1984; Sandmeyer et al. 2007). CSNB is diagnosed by an absent b-wave and a depolarizing a-wave in scotopic (dark-adapted) electroretinography (ERG) (Figure 2). This ERG pattern is known as a “negative ERG” (Witzel et al. 1977). No morphological or ultrastructural abnormalities have been detected in the retinas of horses with CSNB (Witzel et al. 1977; Sandmeyer et al. 2007). A similar “negative ERG” is seen in the Schubert–Bornshein type of human CSNB (Schubert and Bornshein 1952; Witzel et al. 1978). This type of CSNB is thought to be caused by a defective neural transmission within the retinal rod pathway (Witzel et al. 1977, 1978; Sandmeyer et al. 2007). Neural transmission is complex and the mechanism of the transmission defect in CSNB is not reported. Rod photoreceptors are most sensitive under scotopic conditions. In the dark, these cells exist in a depolarized state. They hyperpolarize in response to light, and signaling occurs through reductions in glutamate release (Stryer 1991). This hyperpolarization is responsible for the a-wave of the electroretinogram. Normally this results in stimulation of a population of bipolar cells, the ON bipolar cells. The glutamate receptor of the ON bipolar cells is a metabotropic glutamate receptor (MGluR6) and this receptor is expressed only in the retinal bipolar cell layer (Nomura et al. 1994; Nakanishi et al. 1998). The MGluR6 receptors sense the reduction in synaptic glutamate and produce a response that depolarizes the ON bipolar cell (Nakanishi et al. 1998). This depolarization is responsible for the b-wave of the electroretinogram. The ERG characteristics of the Schubert–Bornshein type of CSNB are consistent with a failure in depolarization of the ON bipolar cell (Sandmeyer et al. 2007).
Figure 2.—
Scotopic electroretinogram from an lp/lp Appaloosa (left) and an LP/LP Appaloosa with CSNB (right). Note the absence of a b-wave in the ERG tracing from the LP/LP horse. (50 msec, 100 mV).
A whole-genome scanning panel of microsatellite markers was used to map LP to a 6-cM region on ECA1 (Terry et al. 2004). Prior to the sequencing of the equine genome, two candidate genes—Transient Receptor Potential Cation Channel, Subfamily M, Member 1 (TRPM1) and Oculoctaneous Albinism Type II (OCA2)—were suggested on the basis of comparative phenotypes in humans and mice (Terry et al. 2004). Both TRPM1 and OCA2 were FISH mapped to ECA1, to the same interval as LP (Bellone et al. 2006a). One SNP in the equine OCA2 gene has been ruled out as the cause for appaloosa spotting (Bellone et al. 2006b).
TRPM1, also known as Melastatin 1 (MLSN1), is a member of the transient receptor potential (TRP) channel family. Channels in the TRP family may permit Ca2+ entry into hyperpolarized cells, producing intracellular responses linked to the phosphatidylinositol and protein kinase C signal transduction pathways (Clapham et al. 2001). TRPs are important in cellular and somatosensory perception (Nilius 2007). Defects in a light-gaited TRP channel results in a loss of phototransduction in Drosophila (reviewed in Kim 2004). Although the specific function of TRPM1 has yet to be described, cellular sensation and intercellular signaling are vital for normal melanocyte migration (reviewed in Steingrímsson et al. 2006). In mice and humans, the promoter region of this gene contains four consensus binding sites for a melanocyte transcription factor, MITF (Hunter et al. 1998; Zhiqi et al. 2004). One of these sites, termed an M-box, is unique to melanocytic expression (Hunter et al. 1998). TRPM1 is downregulated in highly metastatic melanoma cells, suggesting that this protein plays an important role in normal melanogenesis (Duncan et al.1998).
Mutations in the OCA2 gene (also P, or pink-eyed dilution) cause hypopigmentation phenotypes in mice (Gardner et al. 1992). Similarly, in humans, mutations in OCA2 cause the most common form of albinism (Lee et al. 1994). Additionally, other mutations in this gene are thought to be responsible for the variation in human eye color (Duffy et al. 2007; Eiberg et al. 2008). It is believed that during melanogenesis this protein functions to control intramelanasomal pH and aids in tryosinase processing (Sturm et al. 2001; Ni-Komatsu and Orlow 2006).
The objectives of this investigation included determining if differential gene expression could be the cause of LP and CSNB. We have evaluated the relative expression of candidate genes by quantitative real-time RT–PCR. We further investigated whether a local regulatory phenomenon exists by measuring the expression of three additional nearby genes. These included two genes positioned on either side of TRPM1—the OTU domain containing 7A (OTUD7A) and the myotubularin-related protein 10 (MTMR10)—and one gene more distal—tight junction protein 1 (TJP1)—according to the first assembly of the equine genome (http://www.genome.ucsc.edu/cgi-bin/hgGateway?org=Horse&=equCab1) (Figure 3).
Figure 3.—
Genomic map highlighting those genes tested for differential expression within LP candidate region.
MATERIALS AND METHODS
Horses and genotype categories:
Horses were categorized according to genotype and phenotype for LP, which was diagnosed by coat color assessment, breeding records, and, for those horses used in the retinal study, also by ocular examination, including scotopic ERG. Horses were included in the LP/LP group if they had a “fewspot” or “snowcap blanket” pattern and a scotopic ERG consistent with CSNB (Figure 1a). Horses in the LP/lp group all displayed white patterning with dark spots and/or had breeding records consistent with heterozygosity (“leopard,” “spotted blanket,” or “lace blanket” patterns) and a normal scotopic ERG. Horses were included in the non-appaloosa (lp/lp) group if they were solid colored and showed no other traits associated with the presence of LP (striped hooves, white sclera, and mottled skin) and a normal scotopic ERG. The non-appaloosa horses were from the Thoroughbred and American Quarter Horse breeds, two breeds that are not known to possess any appaloosa spotted individuals. Due to the invasive nature of some of the experiments performed, it was impossible to obtain a significant number of samples from age, sex, and base-coat-color-matched horses. Both male and female horses were used in this study, horses ranged in age from <1 year to 23 years old, and the base coat colors of black, bay, and chestnut were all represented (Table 1).
TABLE 1.
Base coat color, proposed LP genotype, disease status, age, sex, and tissue sampled for each horse used in qRT–PCR experiments
| Horse sample no. | Base color | Proposed LP genotype | CSNB phenotype | Age at sampling | Sex | Tissue sampled |
|---|---|---|---|---|---|---|
| 05-10 | Bay dun | LP/LP | CSNB | 5 | Mare | Skin |
| 05-12 | Black | LP/LP | CSNB | 13 | Mare | Skin |
| 05-13 | Chestnut | LP/LP | CSNB | 5 | Mare | Skin |
| 06-261 | Black | LP/LP | Not examined | 15 | Stallion | Skin |
| 06-222 | Bay | LP/LP | CSNB | 5 mo | Mare | Skin |
| 07-51 | Liver chestnut | LP/LP | CSNB | 4 | Gelding | Skin/retina |
| 07-54 | Chestnut | LP/LP | CSNB | 1 | Stallion | Skin/retina |
| 07-53 | Chestnut | LP/LP | CSNB | 1 | Stallion | Retina |
| 07-52 | Chestnut | LP/LP | CSNB | 1 | Stallion | Retina |
| 05-14 | Black | LP/lp | Normal | 2 | Stallion | Skin |
| 05-15 | Dark bay | LP/lp | Not examined | 2 | Stallion | Skin |
| 05-18 | Bay dun | LP/lp | Normal | 5 | Gelding | Skin |
| 07-49 | Chestnut | LP/lp | Normal | Unknown | Gelding | Skin/retina |
| 07-50 | Bay | LP/lp | Normal | 3 | Gelding | Skin/retina |
| 06-275 | Chestnut | LP/lp | Not examined | 11 | Mare | Skin |
| 06-268 | Black | LP/lp | Normal | 1 | Gelding | Skin/retina |
| 06-269 | Bay dun | LP/lp | Normal | 1 | Gelding | retina |
| 05-48 | Red dun | lp/lp | Not examined | 3 | Gelding | Skin |
| 05-49 | Dark bay | lp/lp | Not examined | 23 | Mare | Skin |
| D052 | Bay | lp/lp | Not examined | 4 | Stallion | Skin |
| 06-270 | Chestnut | lp/lp | Normal | 6 mo | Stallion | Skin |
| 06-271 | Dark bay | lp/lp | Normal | 7 | Mare | Skin/retina |
| 07-46 | Chestnut | lp/lp | Normal | 1 | Stallion | Skin/retina |
| 07-48 | Bay | lp/lp | Normal | 2 | Mare | Skin/retina |
| 07-47 | Buckskin | lp/lp | Normal | 1 | Mare | Retina |
| 07-44 | Bay | lp/lp | Normal | 17 | Mare | Retina |
| 07-45 | Chestnut | lp/lp | Normal | 1 | Stallion | Retina |
Ophthalmic examinations:
Horses used in this study were categorized by ocular examination, which included. neurophthalmic examination, slit-lamp biomicroscopy (SL-14, Kowa, Japan), indirect ophthalmoscopy (Heine Omega 200, Heine Instruments), and electroretinography (Cadwell Sierra II, Cadwell Laboratories, Kenewick, WA). For electroretinography, horses were sedated with 10 μg/kg detomidine hydrochloride (Dormosedan, Orion Pharma, Pfizer Animal Health, Kirkland, QC, Canada) by intravenous bolus. Pharmacological mydriasis was achieved with 0.2 ml 1% tropicamide (1% mydriacyl, Alcon, Mississauga, ON, Canada). Auriculopalpebral nerve blocks were performed using 2 ml of a 2% lidocaine hydrochloride injectable solution (Bimeda-MTC Animal Health, Cambridge, ON, Canada). Scotopic ERGs were completed bilaterally to identify nyctalopia and CSNB. A corneal DTL microfiber electrode (DTL Plus Electrode, Diagnosys, Littleton, MA) was placed on the cornea, and platinum subdermal needle electrodes (Cadwell Low Profile Needle electrodes, Cadwell Laboratories) were used as reference and ground. The reference electrode was placed subdermally 3 cm from the lateral canthus and the ground electrode was placed subdermally over the occipital bone. The ERGs were elicited with a white xenon strobe light and recorded with a Cadwell Sierra II (Cadwell Laboratories) with the bandwidth set at 0.3–500 Hz; eyelids were held open manually for each test and a pseudo-Ganzfeld was used to attempt even stimulation of the entire retina (Komarómy et al. 2003). Horses were dark adapted for 25 min and dark-adapted ERG responses were stimulated using maximum light intensity with each recording representing the average of 20 responses. An a-wave dominated ERG or “negative ERG” was considered diagnostic of CSNB (Witzel et al. 1977; Sandmeyer et al. 2007). Horses included in the LP/LP (n = 4) group had a “negative ERG,” and those in the LP/lp group (n = 4) and lp/lp group (n = 6) had normal scotopic and phototopic electroretinograms (Figure 2, Table 2).
TABLE 2.
Scotopic ERG results for sample horses used in retinal study
| LP/LP | LP/lp | lp/lp | |
|---|---|---|---|
| Number | 4 | 4 | 6 |
| Normal scotopic ERG | 0 | 4 | 6 |
| “Negative” scotopic ERG | 4 | 0 | 0 |
Retina and collection and RNA isolation:
Horses were humanely euthanized by intravenous overdose of barbiturate (Euthanyl, MTC Pharmaceuticals) following the Canadian Council on Animal Care Guidelines for Experimental Animal Use and approved by the University of Saskatchewan Animal Care Committee. The eyes were removed immediately and placed on ice. The posterior segment of the globes were isolated by removing the anterior segment via a 360° incision posterior to the limbus. The vitreous was removed by gentle traction. In one eye from each horse, the retina was detached from the periphery and was transected at the optic nerve with Vannas scissors. For the second eye from each horse, the posterior segment was transected with a scalpel blade and one-half was prepared for histology. The retina was removed from the remaining posterior segment and added to the entire retina of the first eye. Retina was then centrifuged and suspended in the appropriate volume of Trizol (Invitrogen) and homogenized in a Polytron mechanical homogenizer (Brinkman Instruments, Westbury, NY). Total retinal RNA was isolated according to the manufacturer's instructions and stored at −80° until used.
Skin collection and RNA isolation:
Skin samples from seven homozygous appaloosa spotted horses (LP/LP), seven heterozygotes (LP/lp), and seven non-appaloosa (lp/lp) were obtained. Samples were taken from live horses (with appropriate consent of owner) and from those euthanized as described above. Donor skin sites of the live horses were infiltrated with a local anesthetic (2% lidocaine hydrochloride, Bimeda-MTC Animal Health, Cambridge, ON, Canada). Following hair removal by shaving the sample area, five 6-mm dermal punch biopsies were collected and immediately snap frozen in liquid nitrogen. Samples were placed at −80° until processing. From each horse in the LP/LP group and LP/lp group, two sample areas were collected for RNA extraction: one sample area that was pigmented (i.e., a darkly pigmented body spot) and one area where skin and hair where completely unpigmented. Skin samples from euthanized horses were collected in a similar fashion; however, punch biopsies were not used. Instead 10 × 1-cm2 sections of skin were harvested from each site by sharp incision with a sterile no. 22 scalpel blade (Paragon, Sheffield, England). A new scalpel blade and a new pair of sterile gloves were worn to perform the harvest from each site to avoid transfer of genetic material. Prior to RNA isolation, skin samples were first powdered by crushing under liquid nitrogen. Total RNA was isolated from 0.5 g of tissue in a buffer of 4 m guanidinium isothiocyanate, 0.1 m Tris–HCl, 25 mm EDTA (pH 7.5), and 1% (v/v) 2-mercaptoethanol, followed by differential alcohol and salt precipitations (Chomczynski and Sacchi 1987; MacLeod et al. 1996). All samples were stored at −80°.
Quantitative real-time RT–PCR:
RNA was quantified using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE) and sample concentrations were adjusted to 50 ng/μl with RNAse free water (Ambion, Austin, TX). RNA integrity and purity was verified using a Bioanalyzer (Agilent Technologies, Santa Clara, CA). All skin and retinal samples isolated were of high purity and integrity, and all samples used had RNA integrity numbers >8.
Equine homologs for TRPM1, OCA2, TJP1, MTMR10, and OTUD7A were identified from the Entrez Trace Archive using a Discontiguous Megablast (http://www.ncbi.nih.gov/BLAST) or by a BLAT search against the horse (January 2007) (equCab1) assembly (http://www.genome.ucsc.edu/). Taqman primers and probes were designed as previously described (Murphy et al. 2006). Preliminary experiments revealed that β-actin was the most stable reference gene among those tested in our samples. The PCR efficiency of primer/probe combinations were calculated using serial dilutions of RNA spanning a magnitude of eightfold (or greater) by the REST analysis program (Pfaffl et al. 2002). R2 values for standard curves were ≥0.98 for all products tested (Table 3). All primer pairs were tested to ensure that genomic DNA was not being amplified by using a minus reverse transcription control in each assay.
TABLE 3.
Primer and probe sequences and PCR efficiency used in quantitative real-time RT–PCR
| Gene | Primer/probe | Sequence | Exon no. | PCR efficiency | R2 |
|---|---|---|---|---|---|
| β-actin | Forward | 5′-GCCGTCTTCCCCTCCAT-3′ | 2 | 2.07 | 1 |
| Reverse | 5′-GCCCACGTATGAGTCCTTCTG-3′ | 3 | |||
| Probe | 5′-GGCACCAGGGCGTGATGGTGGGC-3′ | 2 and 3 | |||
| TRPM1 | Forward | 5′-GACGACATCTCCCAGGATCT-3′ | 16 | 2.09 | 0.99 |
| Reverse | 5′-TGCTCGTCGTGCTTATAGGA-3′ | 17 | |||
| Probe | 5′-ATTCAAAAGACTTTGGCCAGCTGGC-3′ | 16 and 17 | |||
| OCA2 | Forward | 5′-AGATCAAGGAAAGTTCTGGCAGT-3′ | 6 | 2.19 | 0.99 |
| Reverse | 5′-CTGGAGCAGCGTGGAATC-3′ | 7 | |||
| Probe | 5′-AAGCTACTCTGTGAACCTCAGCAGCCAT-3′ | 6 and 7 | |||
| TJP1 | Forward | 5′-ATATGGGAACAACACACAGTGA-3′ | 2 | 2.18 | 0.98 |
| Reverse | 5′-GGTCCTCCTTTCAGCACATC-3′ | 3 | |||
| Probe | 5′-CTTCACAGGGCTCCTGGATTTGGAT-3′ | 2 and 3 | |||
| MTMR10 | Forward | 5′-TGTCAGATTTCGCTTTGATGA-3′ | 5 | 2.28 | 0.98 |
| Reverse | 5′-GGTCTGTTGGCTGGGAATAA-3′ | 6 | |||
| Probe | 5′-TCAGGTCCTGAAAGTGCCAAAAAGG-3′ | 5 and 6 | |||
| OTUD7A | Forward | 5′-CAGACTTTGTTCGGTCCACA-3′ | 3 | 2.27 | 0.98 |
| Reverse | 5′-AGTCACTCAGAGCGGCTGTC-3′ | 4 | |||
| Probe | 5′-AGAACCTGGTCTGGCCAGAGACCTG-3′ | 4 |
Taqman quantitative real-time RT–PCR was performed using a Smart Cycler real-time thermal cycler (Cepheid, Sunnyvale, CA). Each 25-μl reaction contained 250 ng of RNA, 1× EZ buffer (Applied Biosystems, Foster City, CA), 300 μm of each dNTP, 2.5 mm manganese acetate, 200 nm forward and reverse primer, 125 nm fluorogenic probe, 40 units RNasin (Roche, Indianapolis), and 2.5 units rTth (Applied Biosystems). Cepheid also recommends the addition of an “additive reagent” to prevent binding of polymerases and nucleic acids to the reaction tubes. This reagent was added to give a final concentration of 0.2 mg/ml bovine serum albumin (nonacetylated), 0.15 m trehalose, and 0.2% Tween 20. Thermocycler parameters for all assays consisted of a 30-min reverse transcription (RT) step at 60°, 2 min at 94°, and 45 cycles of 94° for 15 sec (denaturation) and 60° for 30 sec (annealing and extension). The threshold crossing cycle (Ct) values generated by the Smart Cycler were used to calculate the relative expression ratios and statistical significance between each group of horses for each tissue tested using REST-MCS version-2. The relative mean expression ratios were calculated according to the following mathematical model: relative expression ratio (R) = ( (Pfaffl 2001), where E represents the calculated efficiencies for the corresponding genes, Ct is the crossing threshold cycle number, and ΔCt(target) and ΔCt(reference) represent the Ct difference between the control group (non-appaloosa horses lp/lp) and the experimental group (either LP/LP or LP/lp) for the target and the reference (β-actin) transcripts, respectively. Given the variability that may occur among individual samples, REST was used to analyze the data to make group-wise comparisons within our populations. REST makes no assumptions about the distribution of observations in the population and thus has been shown to be an appropriate statistical model for analyzing gene expression population data (Pfaffl et al. 2002). This gene expression software tool calculates mean expression ratios for each of the sample groups being tested and then runs permutation tests to determine if the results are due to random allocation or to the effects of treatment (which in this case is the genotype at the LP locus). Gene expression was analyzed with the pairwise fixed reallocation randomization test using REST software to compare gene expression of homozygotes (LP/LP) and heterozygotes (LP/lp) relative to non-appaloosa skin (lp/lp) and to compare CSNB affected (LP/LP) and CSNB unaffected (LP/lp) relative to unaffected (lp/lp) retina. Data are expressed as both relative expression ratios (R) and as fold changes (FC). Data are log transformed for graphical representation so that large relative expression differences can be easily visualized on a graph.
(Pfaffl 2001), where E represents the calculated efficiencies for the corresponding genes, Ct is the crossing threshold cycle number, and ΔCt(target) and ΔCt(reference) represent the Ct difference between the control group (non-appaloosa horses lp/lp) and the experimental group (either LP/LP or LP/lp) for the target and the reference (β-actin) transcripts, respectively. Given the variability that may occur among individual samples, REST was used to analyze the data to make group-wise comparisons within our populations. REST makes no assumptions about the distribution of observations in the population and thus has been shown to be an appropriate statistical model for analyzing gene expression population data (Pfaffl et al. 2002). This gene expression software tool calculates mean expression ratios for each of the sample groups being tested and then runs permutation tests to determine if the results are due to random allocation or to the effects of treatment (which in this case is the genotype at the LP locus). Gene expression was analyzed with the pairwise fixed reallocation randomization test using REST software to compare gene expression of homozygotes (LP/LP) and heterozygotes (LP/lp) relative to non-appaloosa skin (lp/lp) and to compare CSNB affected (LP/LP) and CSNB unaffected (LP/lp) relative to unaffected (lp/lp) retina. Data are expressed as both relative expression ratios (R) and as fold changes (FC). Data are log transformed for graphical representation so that large relative expression differences can be easily visualized on a graph.
RESULTS AND DISCUSSION
TRPM1 as the gene for CSNB in Appaloosa horses:
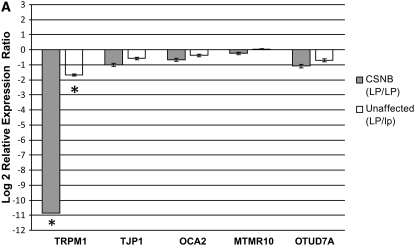
TRPM1 was the only gene of those investigated that was differentially expressed in the retina. In the retina of CSNB (LP/LP) horses, expression was 0.05% of the level found in non-appaloosa horses (R = 0.0005). This constitutes an FC decrease >1800. (FC = −1870.637, P = 0.001). TRPM1 was marginally downregulated in horses heterozygous for appaloosa spotting (LP/lp) (R = 0.312, FC = −3.201, P = 0.005) (Figure 4A; Table 4). It is possible that the downregulation of TRPM1 in the retina of LP/LP horses is the etiology of CSNB. TRPM1 may play a role in neural transmission in the retina through changing cytosolic free Ca2+ levels in the retinal ON bipolar cells. The MGluR6 receptors of the ON bipolar cells are coupled to Gαo proteins, the most abundant heteromeric G protein in the brain. However, there are no known downstream targets of Gαo proteins (Duvoisin et al. 2005). Our observations lead to speculation that TRPM1 is a cation channel that is a downstream target of the Gαo protein in the ON bipolar cell. In dark adaptation, the cation channel activity of TRPM1 would be turned off by glutamate binding to the MGluR6 receptor. Light-induced decreases in synaptic glutamate concentration could remove a negative Gαo signal from TRPM1, leading to cation currents that depolarize the ON bipolar cell. Most recently, expression of TRPM1 has been detected specifically in retinal bipolar cells, further supporting the possibility that lack of TRPM1 is responsible for the failure of b-wave perpetuation (Koike et al. 2007).
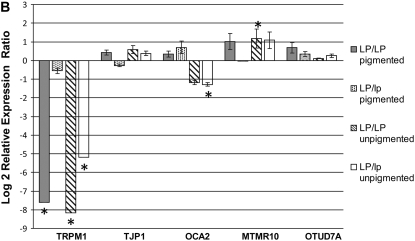
Figure 4.—
Retinal and skin gene expression for five genes in the LP candidate region normalized to β-actin. Relative mRNA expression is represented as a log 2 relative expression ratio (means ± SE). (A) CSNB affected (LP/LP) and CSNB unaffected (LP/lp) retinal RNA samples. Data are expressed as relative to CSNB unaffected (lp/lp) mRNA levels. (B) Pigmented and unpigmented skin samples of homozygous (LP/LP) and heterozygous (LP/lp) horses. Data are expressed as relative to non-appaloosa (lp/lp) mRNA levels. An asterisk indicates significant results (P < 0.05).
TABLE 4.
Statistically significant results from qRT–PCR of retinal tissue samples (normalized to β-actin) relative to expression for non-appaloosa horses (lp/lp)
| Sample group | n (control, sample)a | TRPM1 Rb = | Direction | Significancec |
|---|---|---|---|---|
| CSNB (LP/LP) | 6, 4 | 0.0005 | Down | P = 0.001 |
| Normal (LP/lp) | 6, 4 | 0.312 | Down | P = 0.005 |
Only statistically significant loci are presented.
RNA isolated from lp/lp retina samples with normal night vision as diagnosed by ERGs were used as controls. Data are expressed relative to these controls.
R, relative expression ratio.
Statistically significant results (P ≤ 0.05).
Alterations in TRPM1 may cause appaloosa spotting:
Compared to skin from non-appaloosa horses (lp/lp), TRPM1 was significantly downregulated (P = 0.001) in both pigmented (R = 0.005, FC = −193.963, P = 0.001) and unpigmented (R = 0.003, FC= −288.686, P = 0.001) skin from homozygous (LP/LP) horses. In unpigmented skin from heterozygous (LP/lp) horses, TRPM1 was downregulated to a lesser extent (R = 0.027, FC = −36.583, P = 0.001) (Figure 4B, Table 5). However, gene expression values for heterozygotes were not half the difference between appaloosa homozygotes and non-appaloosa horses, indicating that the difference is not a simple dosage effect. Relative expression differences at or near this magnitude were not detected for any of the other genes tested from this chromosome region (Figure 4B). When compared to mRNA from non-appaloosa skin samples, small changes with less stringent P-values were detected for OCA2 and MTMR10 in LP/lp and LP/LP unpigmented skin samples, respectively (Table 5). These small changes are likely due to the generalized difference between pigmented and unpigmented skin rather than a direct effect of LP.
TABLE 5.
Statistically significant results from qRT–PCR of skin tissue samples (normalized to β-actin) relative to expression for non-appaloosa horses (lp/lp)
| Sample group | n (control, sample)a | TRPM1 Rb = | Direction | Significancec | OCA2 Rb = | Direction | Significancec | MTMR10 Rb = | Direction | Significancec |
|---|---|---|---|---|---|---|---|---|---|---|
| Pigmented LP/LP | 7, 7 | 0.005 | Down | P = 0.001 | 1.267 | Up | P = 0.591 | 2.027 | Up | P = 0.078 |
| Pigmented LP/lp | 7, 7 | 0.681 | Down | P = 0.465 | 1.629 | Up | P = 0.285 | 0.977 | Down | P = 0.946 |
| Unpigmented LP/LP | 7, 7 | 0.003 | Down | P = 0.001 | 0.436 | Down | P = 0.090 | 2.267 | Up | P = 0.031 |
| Unpigmented LP/lp | 7, 7 | 0.027 | Down | P = 0.001 | 0.411 | Down | P = 0.031 | 2.117 | Up | P = 0.091 |
Only statistically significant loci are presented.
RNA isolated from lp/lp skin samples were used as controls. Data are expressed as relative to these controls.
R, relative expression ratio.
Data in italics are statistically significant results (P ≤ 0.05).
In humans, TRPM1 is expressed in several isoforms (Fang and Setaluri 2000; Xu et al. 2001). The long isoform, termed MLSN-L, is thought to be responsible for Ca2+ influx (Xu et al. 2001). Primers and probes were designed to specifically detect this long isoform. It is possible the large relative expression difference that we detected for the long isoform of TRPM1 may interfere with Ca2+ signaling in the melanocytes and thus participate in the biological mechanisms of appaloosa spotting.
The specific function of TRPM1 in melanocytes remains unknown. It has been described as a tumor suppressor that may regulate the metastatic potential of melanomas, as its expression declines with increased metastatic potential (Duncan et al. 1998, 2001; Deeds et al. 2000). Treatment of pigmented melanoma cells with a differentiation-inducing agent upregulated the long isoform of this gene (Fang and Setaluri 2000). TRPM1 therefore has potential roles in Ca2+-dependent signaling related to melanocyte proliferation, differentiation, and/or survival.
One potential role of TRPM1 in melanocyte survival is in interaction with the signaling pathway of the cell surface tyrosine kinase receptor KIT and its ligand KITLG. Signaling through the KIT receptor is critical for the growth, survival, and migration of melanocyte precursors (reviewed by Erickson 1993). It has been shown that both phospholipase C activation and Ca2+ influx are important in supporting KIT-positive cells (Berger 2006). Stimulation with KIT ligand while blocking Ca2+ influx led to a novel form of cell death that is termed activation enhanced cell death (AECD) (Gommerman and Berger 1998). It is possible that during melanocyte proliferation and differentiation, when KIT positive cells are stimulated by the ligand in vivo, the absence of TRPM1 expression may result in decreased Ca2+ influx and ultimately result in AECD. Early melanocyte death could therefore explain LP hypopigmentation patterns. Notably, TRPM1 expression in pigmented skin from heterozygous (LP/lp) horses did not differ significantly from that of non-appaloosa horses. TRPM1 expression is likely tissue specific as we found 4000 times greater expression in the retina than in skin (P = 0.001). Similarly, temporal regulatory elements may direct relatively higher expression in migrating melanocyte precursors than in mature melanocytes; thus in the skin we may not be measuring expression at the biologically relevant time point. We have also shown an association between decreased TRPM1 expression and unpigmented LP/lp skin. However, further work is required to rule out the possibility that decreased expression of TRPM1 in unpigmented LP/lp skin when compared to non-appaloosa skin may simply reflect an absence of TRPM1-expressing melanocytes.
Summary and prospects:
LP has been mapped to a 6-cM region on ECA1 containing the candidate genes TRPM1 and OCA2 (Terry et al. 2004; Bellone et al. 2006a). In addition, CSNB has been associated with homozygosity for LP (Sandmeyer et al. 2007). Here we report that TRPM1 is the only gene from this candidate region that is significantly downregulated in the retina and skin of LP/LP horses. The previously published mapping data, in connection with this reported gene expression data, support the hypothesis that TRPM1 is the molecular mechanism for both LP and CSNB.
This report is the first describing a gene expressional mechanism associated with an eye disease and coat color phenotype in the horse. Future work will include investigation of coding and regulatory regions by sequence analysis to identify the basis of the observed TRPM1 differential expression. As previously mentioned, three E-boxes and one M-box have been identified in the proximal promoter of this gene in humans and mice. The newly available assembled equine genome will be used to identify and investigate regions of interest for evidence of mutations in these regulatory elements. Many of the genes involved in melanogenesis have distinct distal regulatory elements that control their expression. For example, TYR has a distal regulatory element specific to melanocytes 15 kb away from the start of transcription (Porter et al. 1991; Ganss et al. 1994; Porter and Meyer 1994). Novel distal regulatory elements of TRPM1 are likely to be identified. Appaloosa spotted horses may serve as an important research tool illustrating the role of TRPM1 in normal night vision and melanogenesis. Although several mutations have been identified as the cause of CSNB in humans (Dryja et al. 2005; Xiao et al. 2006; Zeitz et al. 2006; Szabo et al. 2007), none to date involve TRPM1. Thus, the horse could serve as a model for as-yet-unsolved forms of heritable human CSNB. In addition, mutations in CABP4, a member of the calcium-binding protein family, were recently shown to cause a 30–40% reduction in transcript levels and result in an autosomal recessive form of CSNB in humans (Zeitz et al. 2006).Therefore, studying the molecular interaction of TRPM1 and other genes causing CSNB involved in calcium signaling could lead to a better understanding of signal transduction during night vision.
Acknowledgments
We thank Michael Mienaltowski for his technical assistance in skin RNA extraction. We thank Frank Cook and James MacLeod for their support and the use of their research equipment. This study was supported by the L. David Dube and Heather Ryan Veterinary Health Research Fund, Equine Health Research Fund, Appaloosa Horse Club of Canada, an Albert and Lorraine Clay Fellowship at the University of Kentucky, and a Dana Faculty Development Grant from the University of Tampa.
References
- Bellone, R., T. Lear, D. L. Adelson and E. Bailey, 2006. a Comparative mapping of oculocutaneous albinism type II (OCA2), transient receptor potential cation channel, subfamily M member 1 (TRPM1) and two equine microsatellites, ASB08 and 1CA43, among four equid species by fluorescence in situ hybridization. Cytogenet. Genome Res. 114 93A. [DOI] [PubMed] [Google Scholar]
- Bellone, R., S. Lawson, N. Hunter, S. Archer and E. Bailey, 2006. b Analysis of a SNP in exon 7 of equine OCA2 and its exclusion as a cause for appaloosa spotting. Anim. Genet. 37 525. [DOI] [PubMed] [Google Scholar]
- Bennett, D. C., and M. L. Lamoreux, 2003. The color loci of mice: a genetic century. Pigment Cell Res. 16 333–344. [DOI] [PubMed] [Google Scholar]
- Berger, S. A., 2006. Signaling pathways influencing SLF and c-kit-mediated survival and proliferation. Immunol. Res. 35 1–12. [DOI] [PubMed] [Google Scholar]
- Brooks, S. A., and E. Bailey, 2005. Exon skipping in the KIT gene causes a Sabino spotting pattern in horses. Mamm. Genome 11 893–899. [DOI] [PubMed] [Google Scholar]
- Brooks, S., T. L. Lear, D. Adelson and E. Bailey, 2007. A chromosome inversion near the KIT gene and the Tobiano spotting pattern in horses. Cytogenet. Genome Res. 119 225–230. [DOI] [PubMed] [Google Scholar]
- Brunberg, E., L. Andersson, G. Cothran, K. Sandberg, S. Mikko et al., 2006. A missense mutation in PMEL17 is associated with the silver coat color in the horse. BMC Genet. 7 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P., and N. Sacchi, 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162 156–159. [DOI] [PubMed] [Google Scholar]
- Clapham, D. E., L. W. Runnels and C. Strübing, 2001. The TRP ion channel family. Nat. Rev. Neurosci. 2 387–396. [DOI] [PubMed] [Google Scholar]
- Deeds, J., F. Cronin and L. M. Duncan, 2000. Patterns of melastatin mRNA expression in melanocytic tumors. Hum. Pathol. 31 1346–1356. [PubMed] [Google Scholar]
- Dryja, T. P., T. L. McGee, E. L. Berson, G. A. Fishman, M. A. Sandberg et al., 2005. Night blindness and abnormal cone electroretinogram ON responses in patients with mutations in the GRM6 gene encoding mGluR6. Proc. Natl. Acad. Sci. USA 102 4884–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy, D. L., G. W. Montgomery, W. Chen, Z. Z. Zhao, L. Le et al., 2007. A three-single-nucleotide polymorphism haplotype in intron 1 of OCA2 explains most human eye-color variation. Am. J. Hum. Genet. 80 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, L. M., J. Deeds, J. Hunter, J. Shao, L. M. Homgren et al., 1998. Down-regulation of the novel gene melanstatin correlates with potential for melanoma metastasis. Cancer Res. 58 1515–1520. [PubMed] [Google Scholar]
- Duncan, L. M., J. Deeds, F. E. Cronin, M. Donovan, A. J. Sober et al., 2001. Melastatin expression and prognosis in cutaneous malignant melanoma. J. Clin. Oncol. 19 568–576. [DOI] [PubMed] [Google Scholar]
- Duvoisin, R. M., C. W. Morgans and W. R. Taylor, 2005. The mGluR6 receptors in the retina: analysis of a unique G-protein signaling pathway. Cell Sci. Rev. 2 225–243. [Google Scholar]
- Eiberg, H., J. Troelsen, M. Nielsen, A. Mikkelsen, J. Mengel-From et al., 2008. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located in the HERC2 gene inhibiting OCA2 expression. Hum. Genet. 123 177–187. [DOI] [PubMed] [Google Scholar]
- Erickson, C. A., 1993. From the crest to the periphery: control of pigment cell migration and lineage segregation. Pigment Cell Res. 6 336–347. [DOI] [PubMed] [Google Scholar]
- Fang, D., and V. Setaluri, 2000. Expression and up-regulation of alternatively spliced transcripts of melastatin, a melanoma metastasis-related gene, in human melanoma cells. Biochem. Biophys. Res. Commun. 279 53–61. [DOI] [PubMed] [Google Scholar]
- Ganss, R., L. Montoliu, A. P. Monaghan and G. Schütz, 1994. A cell-specific enhancer far upstream of the mouse tyrosinase gene confers high level and copy number-related expression in transgenic mice. EMBO J. 13 3083–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, J. M., Y. Nakatsu, Y. Gondo, S. Lee, M. F. Lyon et al., 1992. The mouse pink-eyed dilution gene: association with human Prader-Willi and Angelman syndromes. Science 257 1121–1124. [DOI] [PubMed] [Google Scholar]
- Gommerman, J. L., and S. A. Berger, 1998. Protection from apoptosis by steel factor but not interleukin-3 is reversed through blockade of calcium influx. Blood 91 1891–1900. [PubMed] [Google Scholar]
- Haase, B., S. A. Brooks, A. Schlumbaum, P. Azor, E. Bailey et al., 2007. Allelic heterogeneity at the equine KIT locus in dominant white (W) horses. PLoS Genet. 3 e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, J. J., J. Shao, J. S. Smutko, B. J. Dussault, D. L. Nagle et al., 1998. Chromosomal localization and genomic characterization of the mouse melastatin gene (Mlsn1). Genomics 54 116–123. [DOI] [PubMed] [Google Scholar]
- Kim, C., 2004. Transient receptor ion channels and animal sensation: lessons from Drosophila functional research. J. Biochem. Mol. Biol. 37 114–121. [DOI] [PubMed] [Google Scholar]
- Koike, C., R. Sanuki, K. Miyata, T. Koyasu, T. Miyoshi et al., 2007. The functional analysis of TRPM1 in retinal bipolar cells. Neurosci. Res. 58S S41. [Google Scholar]
- Komarómy, A. M., S. E. Andrew, H. L. Sapp, Jr., D. E. Brooks and W. W. Dawson, 2003. Flash electroretinography in standing horses using the DTL microfiber electrode. Vet. Ophthalmol. 6 27–33. [DOI] [PubMed] [Google Scholar]
- Lapp, R. A., and G. Carr, 1998. Applied appaloosa color genetics. Appaloosa J. 52 113–115. [Google Scholar]
- Lee, S. T., R. D. Nicholls, R. E. Schnur, L. C. Guida, J. Lu-Kuo et al., 1994. Diverse mutations of the P gene among African-Americans with type II (tyrosinase-positive) oculocutaneous albinism (OCA2). Hum. Mol. Genet. 3 2047–2051. [PubMed] [Google Scholar]
- MacLeod, J. N., N. Burton-Wurster, D. N. Gu and G. Lust, 1996. Fibronectin mRNA splice variant in articular cartilage lacks bases encoding the V, III-15, and I-10 protein segments. J. Biol. Chem. 271(31): 18954–18960. [DOI] [PubMed] [Google Scholar]
- Mariat, D., S. Taourit and G. Guérin, 2003. A mutation in the MATP gene causes the cream coat colour in the horse. Genet. Sel. Evol. 35 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklund, L., M. J. Moller, K. Sandberg and L. Andersson, 1996. A missense mutation in the gene for melanocyte-stimulating hormone receptor (MC1R) is associated with the chestnut coat color in horses. Mamm. Genome 7 895–899. [DOI] [PubMed] [Google Scholar]
- Metallinos, D. L., A. T. Bowling and J. Rine, 1998. A missense mutation in the endothelin-B receptor gene is associated with Lethal White Foal Syndrome: an equine version of Hirschsprung disease. Mamm. Genome 9 426–431. [DOI] [PubMed] [Google Scholar]
- Miller, R. W., 1965. Appaloosa coat color inheritance. Ph.D. Thesis, Animal Science Department, Montana State University, Bozeman, MT.
- Murphy, B. A., M. M. Vick, D. R. Sessions, R. F. Cook and B. P. Fitzgerald, 2006. Evidence of an oscillating peripheral clock in an equine fibroblast cell line and adipose tissue but not in peripheral blood. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 192 743–751. [DOI] [PubMed] [Google Scholar]
- Nakanishi, S., Y. Nakajima, M. Masu, Y. Ueda, K. Nakahara et al., 1998. Glutamate receptors: brain function and signal transduction. Brain Res. Rev. 26 230–235. [DOI] [PubMed] [Google Scholar]
- Ni-Komatsu, L., and S. J. Orlow, 2006. Heterologous expression of tyrosinase recapitulates the misprocessing and mistrafficking in oculocutaneous albinism type 2: effects of altering intracellular pH and pink-eyed dilution gene expression. Exp. Eye Res. 82 519–528. [DOI] [PubMed] [Google Scholar]
- Nilius, B., 2007. TRP channels in disease. Biochim. Biophys. Acta 1772 805–812. [DOI] [PubMed] [Google Scholar]
- Nomura, M., H. Iwakabe, Y. Tagawa, T. Miyoshi, Y. Yamashita et al., 1994. Developmentally regulated postsynaptic localization of a metabotropic glutamate receptor in rat rod biopolar cells. Cell 77 361–369. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M. W., 2001. A new mathmatical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M. W., G. W. Horgan and L. Dempfle, 2002. Relative Expression software tool (REST) for group-wise comparison and stastistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30 e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, S. D., and C. J. Meyer, 1994. A distal tyrosinase upstream element stimulates gene expression in neural-crest-derived melanocytes of transgenic mice: position-independent and mosaic expression. Development 120 2103–2111. [DOI] [PubMed] [Google Scholar]
- Porter, S., L. Larue and B. Mintz, 1991. Mosaicism of tyrosinase-locus transcription and chromatin structure in dark vs. light melanocyte clones of homozygous chinchilla-mottled mice. Dev. Genet. 12 393–402. [DOI] [PubMed] [Google Scholar]
- Ray, K., M. Chaki and M. Sengupta, 2007. Tyrosinase and ocular diseases: some novel thoughts on the molecular basis of oculocutaneous albinism type 1. Prog. Retin. Eye Res. 26 323–358. [DOI] [PubMed] [Google Scholar]
- Rebhun, W. C., E. R. Loew, R. C. Riis and L. J. Laratta, 1984. Clinical manifestations of night blindness in the Appaloosa horse. Comp. Contin. Edu. Pract. Vet. 6 S103–S106. [Google Scholar]
- Rieder, S., S. Taourit, D. Mariat, B. Langlois and G. Guérin, 2001. Mutations in the agouti (ASIP), the extension (MC1R), and the brown (TYRP1) loci and their association to coat color phenotypes in horses (Equus caballus). Mamm. Genome 12 450–455. [DOI] [PubMed] [Google Scholar]
- Sandmeyer, L., C. B. Breaux, S. Archer and B. H. Grahn, 2007. Clinical and electroretinographic characteristics of congenital stationary night blindness in the Appaloosa and the association with the leopard complex. Vet. Ophthalmol. 10 368–375. [DOI] [PubMed] [Google Scholar]
- Schubert, G., and H. Bornshein, 1952. Beitrag zur A lyse des menschlichen Electroretinogram. Ophthalmolgica 123 396–413. [DOI] [PubMed] [Google Scholar]
- Silvers, W. K, 1979. The Coat Colors of Mice. Springer-Verlag, New York.
- Sponenberg, D. P., and B. V. Beaver, 1983. Horse Color. Texas A&M Press, College Station, TX.
- Sponenberg, D. P., G. Carr, E. Simak and K. Schwink, 1990. The inheritance of the leopard complex of spotting patterns in horses. J. Hered. 81 323–331. [DOI] [PubMed] [Google Scholar]
- Steingrímsson, E., N. G. Copeland and N. A. Jenkins, 2006. Mouse coat color mutations: from fancy mice to functional genomics. Dev. Dyn. 235 2401–2411. [DOI] [PubMed] [Google Scholar]
- Stryer, L., 1991. Visual excitation and recovery. J. Biol. Chem. 266 10711–10714. [PubMed] [Google Scholar]
- Sturm, R. A., R. D. Teasdale and N. F. Box, 2001. Human pigmentation genes: identification, structure and consequences of polymorphic variation. Gene 277 49–62. [DOI] [PubMed] [Google Scholar]
- Szabo, V., H. J. Kreienkamp, T. Rosenberg and A. Gal, 2007. Gln200Glu, a putative constitutively active mutant of rod alpha-transducin (GNAT1) in autosomal dominant congenital stationary night blindness. Hum. Mutat. 28 741–742. [DOI] [PubMed] [Google Scholar]
- Terry, R. B., S. Archer, S. Brooks, D. Bernoco and E. Bailey, 2004. Assignment of the appaloosa coat colour gene (LP) to equine chromosome 1. Anim. Genet. 35 134–137. [DOI] [PubMed] [Google Scholar]
- Toyofuku, K., I. Wada, R. A. Spritz and V. J. Hearing, 2001. The molecular basis of oculocutaneous albinism type 1 (OCA1): sorting failure and degradation of mutant tyrosinases results in a lack of pigmentation. Biochem. J. 355 259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzel, D. A., J. R. Joyce and E. L. Smith, 1977. Electroretinography of congenital night blindness in an Appaloosa filly. J. Eq. Med. Surg. 1 226–229. [Google Scholar]
- Witzel, D. A., E. L. Smith, R. D. Wilson and G. D. Aguirre, 1978. Congenital stationary night blindness: an animal model. Invest. Ophthalmol. Vis. Sci. 1978(117): 788–793. [PubMed] [Google Scholar]
- Xiao, X., X. Jia, X. Guo, S. Li, Z. Yang et al., 2006. CSNB1 in Chinese families associated with novel mutations in NYX. J. Hum. Genet. 51 634–640. [DOI] [PubMed] [Google Scholar]
- Xu, X. Z., F. Moebius, D. L. Gill and C. Montell, 2001. Regulation of melastatin, a TRP-related protein, through interaction with a cytoplasmic isoform. Proc. Natl. Acad. Sci. USA 98 10692–10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitz, C., B. Kloeckener-Gruissem, U. Forster, S. Kohl, I. Magyar et al., 2006. Mutations in CABP4, the gene encoding the Ca2+-binding protein 4, cause autosomal recessive night blindness. Am. J. Hum. Genet. 79 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhiqi, S., M. H. Soltani, K. M. Bhat, N. Sangha, D. Fang et al., 2004. Human melastatin 1 (TRPM1) is regulated by MITF and produces multiple polypeptide isoforms in melanocytes and melanoma. Melanoma Res. 14 509–516. [DOI] [PubMed] [Google Scholar]