Abstract
Sirtuins are conserved proteins implicated in myriad key processes including gene control, aging, cell survival, metabolism, and DNA repair. In Saccharomyces cerevisiae, the sirtuin Silent information regulator 2 (Sir2) promotes silent chromatin formation, suppresses recombination between repeats, and inhibits senescence. We performed a genomewide screen for factors that negatively regulate Sir activity at a reporter gene placed immediately outside a silenced region. After linkage analysis, assessment of Sir dependency, and knockout tag verification, 40 loci were identified, including 20 that have not been previously described to regulate Sir. In addition to chromatin-associated factors known to prevent ectopic silencing (Bdf1, SAS-I complex, Rpd3L complex, Ku), we identified the Rtt109 DNA repair-associated histone H3 lysine 56 acetyltransferase as an anti-silencing factor. Our findings indicate that Rtt109 functions independently of its proposed effectors, the Rtt101 cullin, Mms1, and Mms22, and demonstrate unexpected interplay between H3K56 and H4K16 acetylation. The screen also identified subunits of mediator (Soh1, Srb2, and Srb5) and mRNA metabolism factors (Kem1, Ssd1), thus raising the possibility that weak silencing affects some aspect of mRNA structure. Finally, several factors connected to metabolism were identified. These include the PAS-domain metabolic sensor kinase Psk2, the mitochondrial homocysteine detoxification enzyme Lap3, and the Fe-S cluster protein maturase Isa2. We speculate that PAS kinase may integrate metabolic signals to control sirtuin activity.
SIRTUINS are a conserved family of proteins found in all domains of life. In eukaryotic cells, they have been characterized as deacetylases, ADP-ribosylases, or both (Tanny et al. 1999; Imai et al. 2000; Landry et al. 2000). Work in a variety of systems has shown that they play roles in many key cellular processes including gene regulation, aging, cell survival, metabolic control, and DNA repair (Brachmann et al. 1995; Kaeberlein et al. 1999; Lin et al. 2000; Langley et al. 2002). Small molecule inhibitors and activators of sirtuin activity have received considerable attention recently as potential therapeutic agents for aging-associated diseases including Parkinson's disease and type II diabetes (Smith and Denu 2007). Thus, there is substantial general interest in understanding how this family of enzymes is regulated.
The founding member of the sirtuin family is the budding yeast Saccharomyces cerevisiae gene-silencing factor Silent information regulator 2 (Sir2), which is required for the formation of virtually all silent chromatin in budding yeast reviewed in Rusche et al. (2003). Sir2 acts in conjunction with the other Sir proteins to promote silencing. It forms a NAD-dependent histone deacetylase complex with Sir3 and Sir4 and is recruited to the silent mating-type cassettes through interactions with Sir1. Additionally, silencing at telomeres is mediated by the recruitment of the Sir proteins by Rap1 protein. Sir2 acts at the rDNA to promote silencing and to suppress recombination between rDNA repeats (Smith and Boeke 1997). Sir2 also inhibits the senescence of mother cells; it has been suggested that this is related to its anti-recombination activity at the rDNA (Sinclair and Guarente 1997).
The Sir complex appears to spread laterally from its nucleation points (Morris and Moazed 2007). This is thought to be accomplished through a cycle of histone-tail deacetylation by Sir2 that enables binding of additional copies of the Sir complex through the histone-tail binding sites in Sir3 and Sir4, whose binding to nucleosomes is inhibited by histone-tail acetylation. This mechanism of Sir protein spreading presents a potential problem in that ectopic spread of silencing activity to regions designated to be transcriptionally active would presumably be deleterious. Recent work has identified several mechanisms that prevent the local ecoptic spread of silent chromatin in budding yeast. A tRNA gene at the right (telomere-proximal) border of the HMRa acts as a boundary element that blocks the lateral spread of silent chromatin (Donze et al. 1999). Subsequent work has identified similar boundary elements in other species (Noma et al. 2006; Scott et al. 2007). Substitution of histone H2A with the H2A.Z variant in euchromatin also antagonizes silencing, as do three distinct methylations in histone H3 (Van Leeuwen et al. 2002; Meneghini et al. 2003; Tompa and Madhani 2007; Venkatasubrahmanyam et al. 2007), and acetylation on lysine 16 on the H4 tail (Kimura et al. 2002; Suka et al. 2002). In at least one case, these factors act redundantly to block the global spread of silencing: we recently reported that H2A.Z substitution and the Set1 complex act together to prevent the global spread of silencing (Venkatasubrahmanyam et al. 2007).
A forward insertional mutagenesis screen aimed at identifying factors that prevent the spread of silencing from the HMRa locus has been reported (Jambunathan et al. 2005). This work utilized a rearranged HMRa locus containing a partial duplication in which the previously mentioned tRNA boundary element at HMRa was moved to delete a segment of the HMRa1–a2 region, an intact copy of the HMRa1 gene was placed to the right of this element, and the HMR-I silencer was deleted. Using mating as an assay for the repression of the a1 gene, this screen identified the known anti-silencing factors SAS4, SAS5, and RPD3, as well as one protein not previously linked to silencing, the bromodomain protein YTA7.
To identify novel negative regulators of sirtuin activity in this context, we sought to extend this analysis to the whole-genome level using the yeast nonessential deletion collection. We crossed the collection to a strain containing a simple insertion of a sensitized URA3 reporter gene just to the right of the tRNA boundary element that flanks the HMRa locus.
MATERIALS AND METHODS
SGA screen:
Colonies were transferred using a Virtek colony arrayer as described in Tong et al. (2001). The yeast MATa knockout library (http://www.openbiosystems.com/GeneExpression/Yeast/YKO/) was grown for 2 days on rich media (YPAD) and pinned onto lawns of the reporter (bait) strain, and then grown for another day at 30° to cross. Diploid progeny were selected on rich media plus G418 (120 mg/liter) and ClonNat (60 mg/liter) drugs and grown for 2 days. Diploids were transferred to presporulation media (GNA) and grown at 30° for 1 day, and then transferred to sporulation media (NGS) and incubated at 23° in an open-air humidified chamber for 5 days. Colonies were transferred to SGA –His –Arg +canavanine plates and grown for 2 days at 30° to select for MATa haploid progeny. Colonies were then transferred to SGA –His –Arg +canavanine +G148 plates and grown at 30° for 1 day to select for recombinant progeny. Colonies were then transferred to SGA –His –Arg +canavanine +G148 +Hygromycin B (180 mg/liter) plates and grown at 30° for 2 days, followed by 1 day of growth on SGA –His –Arg +canavanine +G148 +Hygromycin B +clonNat plates to select for MATa progeny containing a gene knockout allele and the reporter gene. These colonies were transferred to synthetic complete (SC) media +5-FOA (2 g/liter) plates and grown for 3–5 days prior to phenotypic scoring. Positively scored colonies were streaked to SC –Ura plates to verify maintenance of the Candida albicans URA3 reporter gene and subsequently replica plated to 5-FOA media to confirm the resistance.
Strains:
All strains used in this study are listed with their genotype in supplemental Table S1. The reporter strains used were generated by integrating the C. albicans URA3 gene at position 295,754 of S. cerevisiae chromosome III by homologous recombination. The dominant drug markers NatMX4 and HphMX4 (Hygromycin B resistance) (Goldstein and McCusker 1999) were integrated at positions 299,553 and 290,254 of chromosome III, respectively. Strain genotypes of single knockouts recovered from the screen were confirmed by sequencing. Strains containing deletions of the SIR3 gene were created by homologous recombination using either HphMX4 or the LYS2 gene from C. albicans and were confirmed by PCR for presence of the integrated deletion cassette and loss of the wild-type SIR3 allele. Primers used for strain construction are listed in supplemental Table S2.
Chromatin immunoprecipitation:
All chromatin immunoprecipitation (ChIP) experiments were performed according to the protocol used in Raisner et al. (2005). For each sample, we used 2.5 μl of the H3K56-ac antibody, which was generously donated by the lab of Michael Grunstein. We used 20 μl of the Sir3 antibody, which was used as serum and generated against a GST-tagged C-terminal fragment of the protein. We used 2.5 μl of the H4K16-ac antibody (Upstate no. 07-329) and 1 μl of the H3 antibody (Abcam no. Ab1791) per sample.
Plate assays:
For all plate assays, cells were pregrown on rich (YPAD) media and then resuspended in water and plated in fivefold dilutions. The same dilutions were plated at the same time to rich media (SC), rich media (SC) containing 5-FOA (1 g/liter), and media lacking uracil (SC –Ura). Plates were incubated for 3 days at 30° prior to photography.
RESULTS
Genomewide screen to identify anti-silencing factors:
To identify factors responsible for antagonizing the spread of Sir activity to proximal regions, we devised the following strategy to screen the entire yeast nonessential gene deletion library using a reporter-based assay. The reporter strain contains a promoter-truncated allele of the C. albicans URA3 homolog, which is able to complement S. cerevisiae ura3 mutants (Figure 1A). Several promoter-truncation alleles of varying length were integrated outside of HMR, 200 bp to the telomere-proximal side of its characterized boundary element, and tested for activity. The goal of performing these integrations was to obtain a low-expressing allele of URA3 that was sensitized to spread of silencing events in mutants such as the htz1Δ mutant (Meneghini et al. 2003). Our criteria for selecting the reporter strain were threefold: (1) a reproducible 5-fluoroorotic acid resistance (FOAR) phenotype in the htz1Δ mutant but not in wild-type cells to demonstrate sensitivity to the spread of silencing, (2) the smallest possible promoter fragment, and (3) a Ura+ phenotype when plated on –Ura media indicating that the reporter has sufficient expression of C. albicans URA3 that allows for assaying on these media. The strain containing the allele with 70 bp of its endogenous promoter (Figure 1B) best fit these criteria and was chosen as the bait strain in our screen.
Figure 1.—
Reporter strain for assaying spread of silencing activity from HMRa. (A) Promoter-truncated alleles of the Candida albicans URA3 gene were integrated adjacent to the right boundary element of the silenced HMRa locus. Reporter alleles were integrated by homologous recombination of PCR-amplified genomic C. albicans URA3 of varying length, containing 50 bp of homology upstream and downstream of the region 200 bp to the right of HMRa. (B) Fivefold serial dilutions to test growth rate of 70-bp promoter bearing reporter construct on rich media (SC), 5-FOA, and media lacking uracil (SC –Ura).
We adapted the SGA method developed by others (Tong et al. 2001) to cross our reporter strain with the available gene deletion library to generate a library of yeast colonies that were MATa haploids, bearing the reporter gene along with a single gene deletion. In addition, the parent strain contained dominant drug selection markers to track the segregation of the reporter allele, independent of potential silencing effects. These strains were arrayed on plates in 768-colony format in which each of 384 strains was arrayed in duplicate. They were then transferred to 5-FOA-containing plates for phenotypic testing. Because this automated method transfers a relatively large percentage of the colony to the new plate, and also because 5-FOA will select for spontaneous C. albicans ura3 mutants that produce colony papillations, we were selective about scoring positives. Specifically, only colonies that were symmetric and lacked obvious papillations and were present as pairs (meaning that they had to arise from independent meioses) were chosen. These putative positive colonies were tested for URA3 expression by assaying their ability to grow on synthetic media lacking uracil (SD −Ura) to screen for undesired Ura− mutants. Our original reporter strain contained a single dominant selectable drug marker, NatMX4, integrated 3800 bp telomere-proximal to the reporter at coordinate 299,553 of chromosome III (Figure 1A). Because of the large number of meioses that are potentially screened by this method, 5-FOA-resistant colonies arose. Upon further investigation, these were determined to be Ura− strains that likely arose from recombination events between the reporter and the drug marker. The first pass of the screen with this strain had a 5-FOAR rate of ∼5% (∼220/4600). However, the majority of these strains were found to have undergone recombination between the reporter and the positive drug selection marker and were therefore Ura−. To avoid this problem, a second reporter strain was generated with an additional drug marker (HphMX4) integrated 5300 bp centromere proximal to the reporter at coordinate 290,254 of chromosome III. The probability of a double-recombination event leading to a drug-resistant strain lacking the reporter allele is considerably less likely than the single crossover event, reducing the likelihood of recovering 5-FOA-resistant colonies due to loss of the reporter gene. The screen was repeated accordingly and an additional 100 mutants that fit our criteria were scored as positives.
Secondary screenings to confirm phenotypic linkage:
The two passes of the screen yielded 320 total candidates. Because this large-scale screening technique allowed for a variety of events that would lead to false detections, we imposed additional criteria:
The colonies needed to grow when streaked as singles on SD –Ura, while retaining the ability to yield FOAR colonies when replica-plated, a hallmark of anti-silencing factors.
The phenotype was tightly linked to the knockout. For this, a minimum of 100 random spores were plated for each mutant and subsequently tested for linkage between the knockout, the reporter genotype, and 5-FOA resistance.
The identity of the knockout allele was verified by amplification and sequencing of the bar codes present in each deletion allele. This was necessary due to potential errors in the knockout collection and possible cross-contamination events arising in the course of the automated strain generation and scoring.
After removal of false positives, 46 nonredundant positively scoring mutants remained. The final results of both screens are summarized in Table 1. While we have confirmed the identity of the knockouts in all the strains, as well as the linkage of the knockout alleles with the FOAR phenotype, we cannot of course rule out the possibility that a spontaneous mutation in a gene tightly linked to the marked gene deletion is responsible for a given phenotype.
TABLE 1.
Genes identified in this study that display an antisilencing function
| Systematic name | Gene name | Cellular localization | GO terms | FOA plating efficiency | FOA plating efficiency of double mutant with sir3Δ | SIR dependent? |
|---|---|---|---|---|---|---|
| YNL134C | ynl134c | Cytoplasm/nucleus | Alcohol dehydrogenase (NADP+) activity | 6.40E-05 | 2.56E-06 | Yes |
| YML102W | cac2 | Nucleus | Chromatin assembly, DNA repair, nucleosome assembly | 3.20E-04 | 2.56E-06 | Yes |
| YOR144C | elg1 | Cytoplasm/nucleus | Telomere maintenance, DNA replication, DS-break repair | 1.60E-03 | 2.56E-06 | Yes |
| YMR106C | yku80 | Nucleus | Chromatin assembly, chromatin silencing, telomere maintenance | 3.20E-04 | 2.56E-06 | Yes |
| YER088C | dot6 | Cytoplasm/nucleus | Chromatin silencing at rDNA, chromatin silencing at telomere | 8.00E-03 | 2.56E-06 | Yes |
| YBR275C | rif1 | Nucleus | Chromatin silencing, telomere maintenance | 3.20E-04 | 2.56E-06 | Yes |
| YGL127C | soh1 | Nucleus | Mediator complex, telomere maintenance, DNA repair | 1.60E-03 | 2.56E-06 | Yes |
| YHR041C | srb2 | Cytoplasm/nucleus | Mediator complex, telomere maintenance | 3.20E-04 | 2.56E-06 | Yes |
| YGR104C | srb5 | Nucleus | Mediator complex, telomere maintenance | 3.20E-04 | 2.56E-06 | Yes |
| YDL070W | bdf2 | Cytoplasm/nucleus | Bromodomain protein, redundant with Bdf1 | 1.60E-03 | 1.28E-05 | Partial |
| YLR399C | bdf1 | Nucleus | Swr1 complex, chromatin remodeling | 6.40E-05 | 1.28E-05 | Partial |
| YDR334W | swr1 | Nucleus | Swr1 complex, chromatin remodeling, Swi2/Snf2-related ATPase | 3.20E-04 | 2.56E-06 | Yes |
| YOR213C | sas5 | Cytoplasm/nucleus | H3/H4 histone acetyltransferase activity, chromatin silencing at telomere | 4.00E-02 | 2.56E-06 | Yes |
| YMR127C | sas2 | Cytoplasm/nucleus | H3/H4 histone acetyltransferase activity, chromatin silencing at telomere | 4.00E-02 | 2.56E-06 | Yes |
| YDR181C | sas4 | Cytoplasm/nucleus | H3/H4 histone acetyltransferase activity, chromatin silencing at telomere | 4.00E-02 | 2.56E-06 | Yes |
| YLL002W | rtt109 | Nucleus | Histone acetyltransferase activity, negative regulation of transposition, DNA damage reponse | 3.20E-04 | 2.56E-06 | Yes |
| YJL115W | asf1 | Nucleus | Chromatin assembly, chromatin silencing, histone exchange, histone acetylation | 3.20E-04 | 2.56E-06 | Yes |
| YKR048C | nap1 | Cytoplasm | Histone binding, nucleosome assembly | 6.40E-05 | 2.56E-06 | Yes |
| YOL004W | sin3 | Mitochondrion | Rpd3S, Rpd3L, histone deacetylase | 4.00E-02 | 2.56E-06 | Yes |
| YIL084C | sds3 | Nucleus | Histone deacetylation, Rpd3L complex | 4.00E-02 | 2.56E-06 | Yes |
| YMR263W | sap30 | Nucleus | Histone deacetylation, Rpd3L complex | 8.00E-03 | 2.56E-06 | Yes |
| YNL097C | pho23 | Nucleus | Histone deacetylation, Rpd3L complex | 8.00E-03 | 2.56E-06 | Yes |
| YPL181W | cti6/rxt1 | Nucleus | Histone deacetylation, Rpd3L complex | 8.00E-03 | 2.56E-06 | Yes |
| YBR095C | rxt2 | Nucleus | Histone deacetylation, Rpd3L complex | 8.00E-03 | 2.56E-06 | Yes |
| YAL013W | dep1/fun54 | NA | Histone deacetylation, Rpd3L complex | 4.00E-02 | 2.56E-06 | Yes |
| YER072W | vtc1 | ER | Microautophagy, vacuole transport | 8.00E-03 | 2.56E-06 | Yes |
| YLR436C | ecm30 | Cytoplasm | Cell wall organization and biogenesis, cytoplasm | 6.40E-05 | 2.56E-06 | Yes |
| YPR067W | isa2 | Mitochondrion | Biotin biosynthesis, mitochondrion | 3.20E-04 | 2.56E-06 | Yes |
| YIL055C | yil055c | NA | NA | 3.20E-04 | 2.56E-06 | Yes |
| YNL239W | lap3 | Cytoplasm/mitochondrion | Cysteine-type peptidase activity, nucleic acid binding, mitochondrion | 3.20E-04 | 2.56E-06 | Yes |
| YCL005W | ldb16 | Lipid particles | Mitochondrion | 3.20E-04 | 2.56E-06 | Yes |
| YKL116C | prr1 | Cytoplasm | Receptor signaling protein, serine/threonine kinase activity | 3.20E-04 | 6.40E-05 | Partial |
| YOL045W | psk2 | Cytoplasm | PAS domain protein, serine/threonine kinase | 3.20E-04 | 6.40E-05 | Partial |
| YLR079W | sic1 | Cytoplasm/nucleus | CDK inhibitor | 1.60E-03 | 2.56E-06 | Yes |
| YGL173C | kem1 | Cytoplasm | 5′–3′ exoribonuclease activity, telomere maintenance | 6.40E-05 | 2.56E-06 | Yes |
| YIL122W | pog1 | Nucleus | RNA Pol II transcription factor activity | 3.20E-04 | 2.56E-06 | Yes |
| YDR293C | ssd1 | Cytoplasm | RNA binding, cell wall organization | 1.60E-03 | 2.56E-06 | Yes |
| YGL081W | ygl081w | NA | Unknown protein, FHA domain | 3.20E-04 | 2.56E-06 | Yes |
| YPR028W | yop1 | ER | Vesicle-mediated transport, cell membrane | 6.40E-05 | 2.56E-06 | Yes |
| YML095C | rad10 | Cytoplasm/nucleus | DS break repair, nucleotide-excision repair factor 1 complex | 4.00E-02 | 1.60E-03 | Partial |
| YGL163C | rad54 | Cytoplasm/nucleus | Chromatin remodeling, DS break repair, telomere maintenance | 2.00E-01 | 2.00E-01 | No |
| YOR305W | YOR305w | Mitochondrion | Unknown protein, mitochondrion localization | 2.00E-01 | 2.00E-01 | No |
| YMR007W | YMR007w | NA | ORF, dubious | 8.00E-03 | 1.60E-03 | No |
| YBR078W | ecm33 | NA | GPI-anchored protein, cell membrane, mitochondrion | 3.20E-04 | 3.20E-04 | No |
| YLR337C | vrp1 | Punctate/actin | Actin binding, actin organization | 1.60E-03 | 1.60E-03 | No |
| YMR206W | YMR206W | NA | ORF, dubious | 3.20E-04 | 3.20E-04 | No |
Phenotypic characterization of anti-silencing mutants:
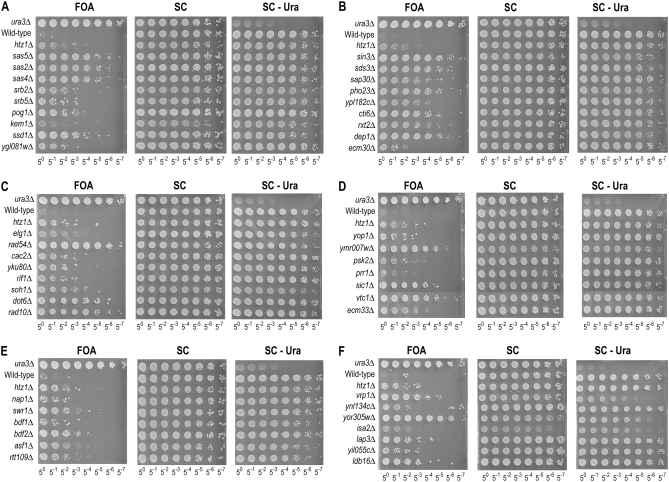
To quantitatively assess the anti-silencing phenotypes of the mutants, we plated serial dilutions of each mutant on 5-FOA media (Figure 2). Each plate contained the parental strain (wild type containing reporter construct), a ura3Δ strain, and an htz1Δ harboring the reporter as controls. Mutants in known pathways or complexes, such as the SAS-I histone acetyl transferase genes SAS2, SAS4, and SAS5, were plated side by side (Figure 2A). Other mutants were grouped together on the basis of available gene ontology (GO) assignments curated at the Saccharomyces Genome Database (http://www.yeastgenome.org). Additionally, all of the strains were plated on SC media and media lacking uracil (SC –Ura) to control for plating efficiency and URA3 expression. As shown in Figure 2, genes belonging to common functional categories tended to display similar phenotypic strength. For example, mutants in the Rpd3-L complex provide one example (Figure 2B). Phenotypic strength varied from a strong anti-silencing defect such as that of the sas2Δ mutant, in which nearly 10% of the colonies grow on 5-FOA (Figure 2A; compared to SC media), to weak ones such as the nap1Δ mutant (Figure 2E), which had an approximate relative plating efficiency of 1/55. As a reference, the wild-type reporter strain was generally observed to have less than 1/58 relative plating efficiency, while the ura3Δ control strain displayed ∼100% efficiency. FOA plating efficiency for strains was determined by comparing the titer for which several colonies grew on FOA to the titer on SC plates for which a similar number of colonies was observed. The quantitative relative plating efficiencies for the mutants are listed in Table 1.
Figure 2.—
Quantitative reporter plating assay for spread of silencing. Serial platings of mutant strains show increased growth on 5-FOA media, assembled into subcategories. Each plate contains a ura3Δ strain as a positive control, and a reporter strain bearing no knockouts (wild type) as a negative control, and an htz1Δ mutation in the reporter strain as an example of an ectopic silencing phenotype. (A) SAS-I complex and Mediator components. (B) Rpd3-L complex components. (C) Telomere maintenance genes. (D) Genes affecting protein phosphorylation and others. (E) Swr1 and Rtt109 complex components. (F) Metabolism-linked genes.
Sir dependence of increased repression of C. albicans URA3 reporter in FOA+ mutants:
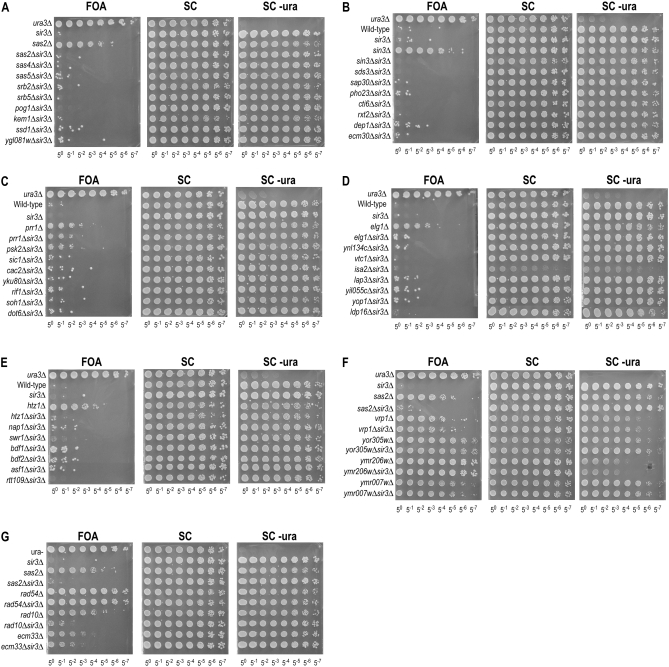
The screen was designed to identify any mutant strain for which there was a large enough decrease in URA3 expression to allow for increased growth on 5-FOA. Mutants were predicted to fall into two categories: genes that are essential for full URA3 expression independently of silencing and genes that normally prevent a spread of silencing activity from HMRa. Because we were interested only in the latter, we sought to identify mutants that have phenotypes dependent on Sir activity. To do this, we deleted SIR3, which encodes an essential subunit of the Sir complex. In a sir3Δ background, bona fide anti-silencing mutants should revert to a 5-FOA-sensitive phenotype in the absence of SIR3. Conversely, mutants in processes such as uracil biosynthesis, transcription per se, or drug resistance should not display a sir3Δ-suppressible phenotype.
For each mutant isolated in the screen, we deleted SIR3 and tested for 5-FOA resistance. Deletion of SIR3 by itself did not confer any fitness benefit compared to wild type (Figure 3A). As in Figure 2, each plate displayed in Figure 3 includes wild-type and ura3Δ strains as controls. Additionally, each plate assays a single paired example of a 5-FOA-resistant deletion strain next to its respective double mutant with sir3Δ as a reference. As shown in Figure 3, A–C, a large fraction (35/46) of the genes from the screen displayed 5-FOA phenotypes that were fully suppressed by deletion of SIR3. Phenotypes of mutants in PRR1 and RAD10 showed partial suppression of the phenotype, as evidenced by intermediate 5-FOA resistance (Figure 3, C and G, and supplemental Figure S1), whereas mutants in BDF1 and BDF2 had weak suppression (Figure 3E and supplemental Figure S1). Mutants in PSK2 showed variable degrees of suppression (Figure 3C vs. supplemental Figure S1). Intermediate and weak suppression suggests dual roles for these genes in anti-silencing and transcription. Finally, the genes RAD54, VRP1, ECM33 YOR206W, YMR007W, and YOR305W (Figure 3, F and G) displayed phenotypes that were unaffected by deletion of SIR3, suggesting they affect expression or genetic mutability of the URA3 reporter gene. Quantitative estimations of the relative 5-FOA plating efficiencies for the double deletions are listed in Table 1.
Figure 3.—
Sir-suppressible and nonsuppressible anti-silencing phenotypes of mutant strains. Serial platings of mutant strains show increased growth on 5-FOA media, assembled into subcategories. Each plate contains ura3Δ as a positive control for 5-FOA resistance, and a reporter strain bearing no knockouts (wild type) as a control for 5-FOA sensitivity, and sir3Δ in the reporter strain as a negative control for Sir-independent effects. (A–D) Cases of robust suppression of 5-FOA phenotypes by sir3Δ. (E and G) Cases of weak suppression. (F and G) Cases of nonsuppression.
Rtt109/Asf1 have a novel function to antagonize silencing of the reporter gene:
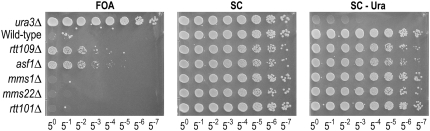
Our screen identified Rtt109 and Asf1 as sirtuin-dependent anti-silencing factors. These proteins form a complex that acetylates the lysine 56 core residue of histone H3 (H3K56) (Driscoll et al. 2007; Han et al. 2007). The complex acetylates non-chromatin-associated H3 during DNA replication. Together with the Hst3 deacetylase that acts outside of S phase, this mechanism restricts H3K56 acetylation to S phase (Celic et al. 2006; Maas et al. 2006). For unknown reasons, cells defective in this modification are sensitive to DNA damage. On the basis of genetic interaction maps, it has been suggested that the role of this mechanism in DNA repair is facilitated by the proteins Asf1, Mms1, Mms22, Rtt101, and Rtt109 (Collins et al. 2007). Interestingly however, only RTT109 and ASF1 share additional synthetic interactions with the Swr1 complex, which deposits H2A.Z (Collins et al. 2007). This is consistent with the results of the screen, which identified anti-silencing phenotypes only for asf1Δ and rtt109Δ mutants (Figure 2). However, it remained possible that mutants in MMS1, MMS22, and RTT101 were false negatives. Therefore, we used homologous recombination to generate deletions of these genes in the reporter strain background and tested them for growth on 5-FOA media (Figure 4). We observed that mms1Δ, mms22Δ, and rtt101Δ did not display resistance to 5-FOA, in contrast to asf1Δ and rtt109Δ mutants.
Figure 4.—
Rtt109 and Asf1 have phenotypes distinct from the Rtt101 cullin complex. Indicated genotypes were analyzed for reporter gene expression as described in Figures 2 and 3.
Silencing outside of HMRa in the rtt109Δ mutant results in decreased H4K16 acetylation without a detectable increase in Sir3 binding:
We investigated the phenotype of the rtt109Δ mutant further using ChIP, using the probes shown in Figure 5A. Probes A–F span the HMRa silent cassette, probes G–J span the C. albicans URA3 reporter gene, probes K–M correspond to the promoter regions of the three genes to the right (telomere-proximal) of the reporter gene, and probe N corresponds to the promoter of HMLα-proximal gene MRC1. We also examined the promoter regions of several genes in euchromatic segments of chromosome III (probes O–S) and two probe sets (probes T and U) in the middle of the large ORF of the BUD3 gene (used for normalization in Figure 5B). We first performed ChIP using polyclonal antibodies against Sir3 in the wild-type reporter strain, as well as in the mutant strains rtt109Δ and the previously characterized anti-silencing mutant sas2Δ as a control. Consistent with previous reports, Sir3 is dramatically enriched within HMRa, while regions outside have very little or no observable Sir3 (Figure 5B). The data in all of our ChIP experiments were normalized to values obtained using ChIP performed with antibodies to histone H3 to control for nucleosome density. Additionally, for the Sir3 ChIP data, we normalized the data to the signal obtained from the BUD3 ORF region, which shows no detectable Sir3 binding in wild-type cells. Notably, the promoter regions of the reporter construct (probe G) displayed less Sir3 association than the immediately adjacent HMRa region, but more than that observed at control regions. Strikingly, when we examined Sir3 localization in the rtt109Δ and sas2Δ strains, no increased association of Sir3 within the reporter gene region was apparent, suggesting in this context that Rtt109 and Sas2 act downstream of Sir binding to antagonize silencing. Since a key function of the Sir complex is to deacetylate lysine 16 of histone H4 (H4K16), we examined the acetylation status of this residue (Figure 5C). We observed decreased acetylation on H4K16 using probes that cover the reporter gene in the rtt109Δ mutant (Figure 5C), which supports the idea that the increased activity of the Sir complex is responsible for the change in reporter gene expression in this mutant. Consistent with previous reports, we observed a dramatic depletion of this acetylation from HMRa and we observed that its presence was dependent on the H4K16 HAT Sas2 at many, but not all, locations (Figure 5C). We also examined H3K56 acetylation by ChIP (Figure 5D). Although its removal has been suggested to be important for silencing (Xu et al. 2007), we observed only a modest decrease of this modificiation within HMRa compared to euchromatic sites and only a slight decrease at the reporter gene in the sas2Δ mutant. Importantly, the signal we observed in this region was dependent on RTT109.
Figure 5.—
ChIP data for Sir3, H3K56 acetylation, and H4K16 acetylation. (A) Schematic of HMRa and the surrounding region with locations of PCR amplicons used for quantitation. Primer set N corresponds to the promoter of the HMLα-proximal gene MRC1, and primer sets O, P, Q, R, and S correspond to the promoters of the euchromatic genes CDC10, CWH43, NFS1, DCC1, and RBK1. Primer sets T and U are to the middle of the ORF region of BUD3. All data shown are averages of three independent ChIP experiments with standard error of the mean error bars. (B–D) t-Tests were applied to data for probes G–J only. (B) Sir3 ChIP data for wild-type, sas2Δ, and rtt109Δ strains. Data are normalized to H3 ChIP values for each locus, and all loci are normalized to the average of primer sets T and U. t-Tests for the null hypothesis that Sir3 levels did not change between wild type and the sas2Δ and rtt109Δ mutants were passed by loci denoted by *. (C) H4K16-ac ChIP values for wild type, sas2Δ, and rtt109Δ, normalized to H3 enrichment. t-Tests for the hypothesis that K16 acetylation levels decreased between the rtt109Δ mutant and wild type were passed by loci denoted by *. (D) H3K56-ac ChIP values for wild type, sas2Δ, and rtt109Δ, normalized to H3 enrichment. A t-test for the hypothesis that there is a significant difference in H3K56 acetylation between wild-type and sas2Δ cells was passed by the locus that is denoted by +.
DISCUSSION
Systematic screen for negative regulators of sirtuin activity:
We present a strategy and results for identification of genes that negatively affect the Sir2-dependent repression of a reporter gene placed just outside a chromatin boundary element in S. cerevisiae. Our screen is similar in overall design to that reported by Jambunathan et al. (2005), which identified the bromodomain/AAA+ ATPase-encoding gene YTA7 as a new negative regulator of silencing (Jambunathan et al. 2005). The major differences are that (1) we used a sensitized C. albicans URA3 reporter gene and growth on FOA as output instead of a rearranged HMRa1 locus and mating and (2) we used an SGA approach and the yeast deletion collection instead of mTn insertional mutagenesis. Although our screen did not identify YTA7, it did identify a number of previously described anti-silencing factors that were identified in the previous screen including RPD3, SAS4, and SAS5. As discussed above, given the potential for threshold and reporter-specific effects, false negatives are difficult to avoid in any genomewide reporter-based screen. Nonetheless, we identified 20 new loci that negatively regulate silencing outside of the tRNA boundary element. These assignments were based on individual mutants passing tests of linkage to the deletion collection marker, verification of the deletion barcodes, and dependency on SIR2 for the increased-repression FOA phenotype.
Negative regulation of Sir2 by regulators of chromatin and DNA metabolism:
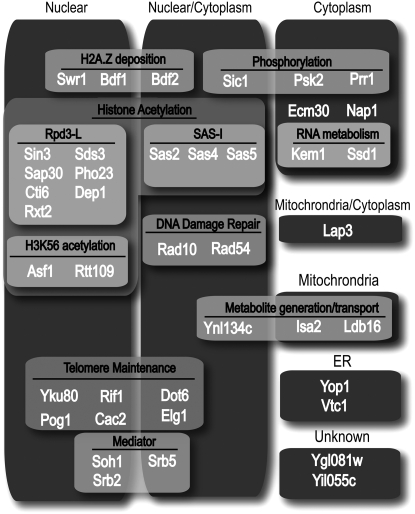
In our effort to take an unbiased approach to identify novel negative regulators of silencing, we uncovered a disparate range of protein activities. Figure 6 describes the reported cellular locations, complexes, and functions of the 40 proteins identified in our screen that displayed SIR-dependent increased repression of the C. albicans URA3 reporter gene on the basis of available annotation curated at the Saccharomyces Genome Database (http://www.yeastgenome.org). The majority of the chromatin modifying factors that we identified have been implicated previously in regulation of SIR activity. These include Bdf1 (a component of the Swr1 complex that deposits both H2A.Z and the TFIID complex), the SAS-I H4K16 acetyltransferase complex, the Rpd3-L HDAC complex, and a number of factors reported to control telomere length (Yku80, Rif1, Dot1, Pog1, Cac2, Elg1, and Mediator). The latter mutants may act by producing a longer telomere near HMRa, which lies ∼35 kb from the right telomere of chromosome III. Increased Sir complex nucleation might then lead to increased Sir-dependent repression of the reporter gene. Alternatively, some members of this group might act by distinct mechanisms as suggested previously for Yku (Maillet et al. 2001). The negative role for Rpd3-L in silencing remains a mystery. Despite deacetylating histone tail residues in a similar fashion to the Sir HDAC complex, Rpd3-L appears to oppose the activity of the Sir HDAC complex (De Rubertis et al. 1996). One possibility is that Rpd3-L antagonizes direct acetylation of the Sir complex. Indeed, N-terminal acetylation of Sir3 has been reported to promote silencing (Wang et al. 2004).
Figure 6.—
Chart depicting functional and localization categories of Sir-dependent genes in this study. Genes are assigned functional categories on the basis of available functional or biochemical annotation (boxes with light shading), and those are overlaid on cellular localization annotations.
Function of H3K56 acetylation:
Another protein complex identified in our screen is the Rtt109 histone H3K56 acetyltransferase complex (Figure 6). H3K56 acetylation is cell-cycle regulated and peaks in S phase due to the downregulation of the Hst3 Sir2-related HDAC that removes the modification (Celic et al. 2006; Maas et al. 2006). Mutations of H3K56 have been shown to disrupt silencing (Xu et al. 2007). Specifically, mutation of this residue to a state that mimics either the acetylated (K > Q) or deacetylated (K > R) lysine resulted in decreased silencing of a subtelomeric silencing reporter gene as did a mutation (K > G) that removed the side chain altogether. This loss of silencing was not associated with a decrease in silencing-protein binding; however, there is an apparent increase in chromatin accessibility in that region (Xu et al. 2007). These results, however, do not clarify what state of K56 is favored for complete silencing. Mutation of SIR2 resulted in increased K56 acetylation in silenced regions, and it has been suggested that deacetylation of this residue by Sir2 is necessary for silencing (Xu et al. 2007). Our data indicate a fairly modest reduction in K56 acetylation at HMRa in the wild-type strain relative to euchromatic sites (Figure 5D). Thus, it may be that acetylation antagonizes silencing at a post-Sir-binding step, but its complete removal is not essential for silencing. As described above, it has been suggested that Rtt101, a cullin homolog, cooperates with Mms1 and Mms22 to affect the function of H3K56 acetylation in genome stability. However, our results indicate that these factors are dispensable for the anti-silencing function of the Rtt109 complex, indicating a branch in the pathway.
In contrast, two other factors implicated in DNA repair, Rad10 and Elg1, affect silencing. These do not seem to act by increasing the mutability of the C. albicans URA3 reporter gene since their effects were SIR3 dependent. Elg1, which functions in an alternative RFC-like clamp loader complex, has been implicated previously in both genome stability and silencing (Ben-Aroya et al. 2003). Rad10 is a single-stranded DNA endonuclease involved in nucleotide excision repair (Prakash 1977). Precisely how these factors control silencing remains unknown, but they suggest an intriguing link between DNA repair factors and the regulation of sirtuin function. Such links are not without precedent. For example, Sir2 itself suppresses recombination between the rDNA repeats (Gottlieb and Esposito 1989).
Negative regulation of Sir2 by factors linked to metabolism:
Given the established role of Sir2 and its orthologs in aging and metabolism in a variety of organisms, there is a great deal of interest in understanding how Sir2 activity is globally regulated. Current thinking focuses mostly on the fact that sirtuins require NAD for their decetylase and/or ADP-ribosylation activities, but other mechanisms of control have not been ruled out. Three proteins identified in our screen seem notable in this respect. First, we identify the PAS domain kinase Psk2 as a negative regulator of Sir2. This factor has been shown to phosphorylate a number of proteins involved in metabolism and translation (Rutter et al. 2002). Therefore, it is possible that Psk2 could regulate Sir2 in response to metabolic conditions. However, what the PAS domain binds to or senses and the biologically relevant outputs of this conserved kinase remain poorly understood.
It is intriguing to note that one of the factors identified in our screen, Ygl081w, contains a FHA domain. Such domains in other proteins have been shown to be a binding motif for phosphorylated proteins (Li et al. 2004). Likewise, our screen identified Lap3, a protein originally identified as a bleomycin hydrolase, but that is thought to function to limit the levels of the toxic metabolic side-product homocysteine in cells (Xu and Johnston 1994). One hypothetical possibility is that Lap3 limits silencing by hydrolyzing the o-acetyl-ADP-ribose product of the Sir2 deacetylase reaction, which has been shown in vitro to promote the assembly of the Sir complex. If Lap3 and Psk2 functions are related, perhaps the PAS domain of Psk2 binds o-acetyl-ADP-ribose. Finally, we note that Isa2, identified in our screen, is required for the maturation of Fe-S cluster proteins, again suggesting a link between Sir2 and metabolism. Further analysis of the genetic relationships of these genes should help to elucidate the pathways by which these genes control sirtuin outputs.
Conclusion:
Our screen represents the most comprehensive survey to date for negative regulators of sirtuin activity in any system. Our survey identified 40 genes, including 20 genes not previously known to be involved in regulating Sir. The precise mechanisms by which they function in this context and their relationship to each other remain fertile ground for future investigation. Although we have emphasized potential regulation of Sir2 activity, any of the factors we describe could act by affecting steps in the expression of Sir proteins, ranging from synthesis to degradation of the corresponding mRNAs and proteins. Likewise, given that there are limited pools of Sir2 in the cell and competition for Sir2 association between the silent cassettes, telomeres, and the rDNA, it is possible that some of the mutations described here affect that Sir2 distribution. We note, however, that the rDNA-specific loss-of-silencing mutations described previously (Smith and Boeke 1997) were not identified in our screen with the exception of cac1. Regardless of the mechanism of action of the individual factors, we hope that our results will provide a useful resource for the field for future investigation of sirtuin regulation. In particular, the exploration of links between Sir2 and processes of DNA repair and metabolic control are likely to lead to advances in our understanding of Sir2 function and control. Given the central importance of this family of regulators in biology, understanding how they are controlled in S. cerevisiae may ultimately provide insights into the regulation of the sirtuins in humans, which have been suggested to play roles in common maladies ranging from aging to diabetes to cancer.
Acknowledgments
We thank Marie Bao and Anupama Seshan for critical reading of the manuscript. This work was supported by grant GM071801 from the National Institutes of Health. R.M.R. is a trainee of the Herbert Boyer Program in Biological Sciences and the Tetrad Graduate Program. H.D.M. is a Scholar of the Leukemia and Lymphoma Society.
References
- Ben-Aroya, S., A. Koren, B. Liefshitz, R. Steinlauf and M. Kupiec, 2003. ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc. Natl. Acad. Sci. USA 100 9906–9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C. B., J. M. Sherman, S. E. Devine, E. E. Cameron, L. Pillus et al., 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9 2888–2902. [DOI] [PubMed] [Google Scholar]
- Celic, I., H. Masumoto, W. P. Griffith, P. Meluh, R. J. Cotter et al., 2006. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr. Biol. 16 1280–1289. [DOI] [PubMed] [Google Scholar]
- Collins, S. R., K. M. Miller, N. L. Maas, A. Roguev, J. Fillingham et al., 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446 806–810. [DOI] [PubMed] [Google Scholar]
- De Rubertis, F., D. Kadosh, S. Henchoz, D. Pauli, G. Reuter et al., 1996. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature 384 589–591. [DOI] [PubMed] [Google Scholar]
- Donze, D., C. R. Adams, J. Rine and R. T. Kamakaka, 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll, R., A. Hudson and S. P. Jackson, 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315 649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 1541–1553. [DOI] [PubMed] [Google Scholar]
- Gottlieb, S., and R. E. Esposito, 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56 771–776. [DOI] [PubMed] [Google Scholar]
- Han, J., H. Zhou, B. Horazdovsky, K. Zhang, R. M. Xu et al., 2007. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315 653–655. [DOI] [PubMed] [Google Scholar]
- Imai, S., C. M. Armstrong, M. Kaeberlein and L. Guarente, 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403 795–800. [DOI] [PubMed] [Google Scholar]
- Jambunathan, N., A. W. Martinez, E. C. Robert, N. B. Agochukwu, M. E. Ibos et al., 2005. Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics 171 913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein, M., M. McVey and L. Guarente, 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, A., T. Umehara and M. Horikoshi, 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32 370–377. [DOI] [PubMed] [Google Scholar]
- Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins et al., 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley, E., M. Pearson, M. Faretta, U. M. Bauer, R. A. Frye et al., 2002. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 21 2383–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., I. J. Byeon, Y. Ju and M. D. Tsai, 2004. Structure of human Ki67 FHA domain and its binding to a phosphoprotein fragment from hNIFK reveal unique recognition sites and new views to the structural basis of FHA domain functions. J. Mol. Biol. 335 371–381. [DOI] [PubMed] [Google Scholar]
- Lin, S. J., P. A. Defossez and L. Guarente, 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289 2126–2128. [DOI] [PubMed] [Google Scholar]
- Maas, N. L., K. M. Miller, L. G. DeFazio and D. P. Toczyski, 2006. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell 23 109–119. [DOI] [PubMed] [Google Scholar]
- Maillet, L., F. Gaden, V. Brevet, G. Fourel, S. G. Martin et al., 2001. Ku-deficient yeast strains exhibit alternative states of silencing competence. EMBO Rep. 2 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini, M. D., M. Wu and H. D. Madhani, 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112 725–736. [DOI] [PubMed] [Google Scholar]
- Morris, C. A., and D. Moazed, 2007. Centromere assembly and propagation. Cell 128 647–650. [DOI] [PubMed] [Google Scholar]
- Noma, K., H. P. Cam, R. J. Maraia and S. I. Grewal, 2006. A role for TFIIIC transcription factor complex in genome organization. Cell 125 859–872. [DOI] [PubMed] [Google Scholar]
- Prakash, L., 1977. Defective thymine dimer excision in radiation-sensitive mutants rad10 and rad16 of Saccharomyces cerevisiae. Mol. Gen. Genet. 152 125–128. [DOI] [PubMed] [Google Scholar]
- Raisner, R. M., P. D. Hartley, M. D. Meneghini, M. Z. Bao, C. L. Liu et al., 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123 233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72 481–516. [DOI] [PubMed] [Google Scholar]
- Rutter, J., B. L. Probst and S. L. McKnight, 2002. Coordinate regulation of sugar flux and translation by PAS kinase. Cell 111 17–28. [DOI] [PubMed] [Google Scholar]
- Scott, K. C., C. V. White and H. F. Willard, 2007. An RNA polymerase III-dependent heterochromatin barrier at fission yeast centromere 1. PLoS ONE 2 e1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair, D. A., and L. Guarente, 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91 1033–1042. [DOI] [PubMed] [Google Scholar]
- Smith, B. C., and J. M. Denu, 2007. Mechanism-based inhibition of sir2 deacetylases by thioacetyl-lysine peptide. Biochemistry 46 14478–14486. [DOI] [PubMed] [Google Scholar]
- Smith, J. S., and J. D. Boeke, 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11 241–254. [DOI] [PubMed] [Google Scholar]
- Suka, N., K. Luo and M. Grunstein, 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32 378–383. [DOI] [PubMed] [Google Scholar]
- Tanny, J. C., G. J. Dowd, J. Huang, H. Hilz and D. Moazed, 1999. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99 735–745. [DOI] [PubMed] [Google Scholar]
- Tompa, R., and H. D. Madhani, 2007. Histone H3 lysine 36 methylation antagonizes silencing in Saccharomyces cerevisiae independently of the Rpd3S histone deacetylase complex. Genetics 175 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294 2364–2368. [DOI] [PubMed] [Google Scholar]
- van Leeuwen, F., P. R. Gafken and D. E. Gottschling, 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109 745–756. [DOI] [PubMed] [Google Scholar]
- Venkatasubrahmanyam, S., W. W. Hwang, M. D. Meneghini, A. H. Tong and H. D. Madhani, 2007. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc. Natl. Acad. Sci. USA 104 16609–16614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., J. J. Connelly, C. L. Wang and R. Sternglanz, 2004. Importance of the Sir3 N terminus and its acetylation for yeast transcriptional silencing. Genetics 168 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, F., Q. Zhang, K. Zhang, W. Xie and M. Grunstein, 2007. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol. Cell 27 890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. E., and S. A. Johnston, 1994. Yeast bleomycin hydrolase is a DNA-binding cysteine protease. Identification, purification, biochemical characterization. J. Biol. Chem. 269 21177–21183. [PubMed] [Google Scholar]