Abstract
The medaka, Oryzias latipes, has an XX/XY sex-determination system, and a Y-linked DM-domain gene, DMY, is the sex-determining gene in this species. Since DMY appears to have arisen from a duplicated copy of the autosomal DMRT1 gene ∼10 million years ago, the medaka Y chromosome is considered to be one of the youngest male-determining chromosomes in vertebrates. In the screening process of sex-reversal mutants from wild populations, we found a population that contained a number of XY females. PCR, direct sequencing, and RT–PCR analyses revealed two different null DMY mutations in this population. One mutation caused loss of expression during the sex-determining period, while the other comprised a large deletion in putative functional domains. YY females with the mutant-type DMY genes on their Y chromosomes were fully fertile, indicating that the X and Y chromosomes were functionally the same except for the male-determining function. In addition, we investigated the frequencies of the sex chromosome types in this population over four successive generations. The Y chromosomes bearing the mutant-type DMY genes were detected every year with no significant differences in their frequencies. These results demonstrate that aberrant Y chromosomes behaving as X chromosomes have been maintained in this population.
IN vertebrates, only two genes have been identified as master sex-determining genes, namely SRY/Sry in mammals (Gubbay et al. 1990; Sinclair et al. 1990; Koopman et al. 1991) and DMY (DM-domain gene on the Y chromosome) in the medaka, Oryzias latipes (Matsuda et al. 2002, 2007; Nanda et al. 2002). It has been assumed that SRY/Sry evolved before the eutherian radiation ∼80 million years ago (MYA), since humans and rodents, and all other orders of eutherian mammals examined so far, have an SRY/Sry gene (Graves 2002). In contrast to the widespread distribution of SRY/Sry in mammals, DMY is found only in the medaka and O. curvinotus, the closest relative to the medaka (Kondo et al. 2003; Matsuda et al. 2003; Tanaka et al. 2007). The DMY gene appears to have arisen from a duplicated copy of the autosomal DMRT1 gene (Nanda et al. 2002; Kondo et al. 2006) and this DMRT1 duplication event is estimated to have occurred ∼10 MYA in a common ancestor of O. latipes and O. curvinotus (Kondo et al. 2004).
In mammals, sex chromosomes are highly dimorphic. The large gene-rich X and small heterochromatic Y chromosomes are almost completely differentiated. The human X chromosome bears ∼1000 genes with a variety of general and specialized functions (Ross et al. 2005), whereas the human Y chromosome encodes only 45 unique proteins (Skaletsky et al. 2003). In addition, crossovers occur only in a small homologous region, designated the pseudoautosomal region. On the other hand, both X and Y chromosomes are homomorphic in the medaka. Sex chromosomal crossovers occur over almost the entire length of the corresponding linkage groups. In fact, the Y chromosome-specific region is ∼260 kb in length (Kondo et al. 2006). In addition, the genotypic sex can easily be reverted by sex hormone or high-temperature treatments (Kobayashi and Iwamatsu 2005; Sato et al. 2005; Iwamatsu et al. 2006; Hattori et al. 2007). These results indicate that the medaka sex chromosomes are quite undifferentiated and suggest that sex determination in the medaka is at an early stage of evolution and has not reached a similar stability to that in other vertebrates like birds and mammals.
Since the identification of DMY, we have screened sex-reversal mutants from wild populations of the medaka to reveal the molecular functions of DMY and identify other genes involved in sex differentiation. In the process of screening, we found a population in O-bu (Aichi prefecture) that contained a number of XY sex-reversed females. In the present study, we demonstrate that there are two types of Y chromosomes that have loss-of-function-type DMY mutations in this population and found that these Y chromosomes were stably maintained for at least four successive generations. These facts imply that in organisms like the medaka with undifferentiated sex chromosomes, the Y chromosome that has lost the male-determining function can behave as an X chromosome.
MATERIALS AND METHODS
Sexing of wild fish:
Phenotypic sex was judged from secondary sex characters, namely the shapes of the dorsal and anal fins and the papillary processes on the male anal fin rays. Genotypic sex, XY or XX, was determined by the presence or absence of the DMY gene using PCR amplification of caudal fin clip DNA extracted according to Shinomiya et al. (1999). PCR was performed with the primer set PG17.5 and PG17.6 for DMY and DMRT1 at an annealing temperature of 55°. This primer set amplified a 982-bp DMY fragment and a 1245-bp DMRT1 fragment. The PCR products were analyzed by 1% agarose gel electrophoresis. Individuals with only the DMRT1 fragment were judged to be XX, while those with both the DMY and DMRT1 fragments were judged to be XY.
Progeny test:
The DMY gene of the northern population had 21 nucleotide deletions in intron 2 compared to that of the southern population (Shinomiya et al. 2004). In order to distinguish between Y chromosomes derived from XY females and that derived from inbred strain XY males, O-bu XY females were mated with XY Hd-rR.YHNI males, which had the HNI (northern population)-derived DMY gene on the genetic background of Hd-rR (southern population) (Matsuda et al. 1998), since the O-bu population belongs to the southern population. Four genotypes were distinguished in the F1 progeny (XX, XYm, XYp, and YmYp; where Ym and Yp are maternal and paternal Y chromosomes, respectively) by separating the DMY PCR products in 10% vertical polyacrylamide slab gels according to a previous report (Shinomiya et al. 2004).
PCR and direct sequencing:
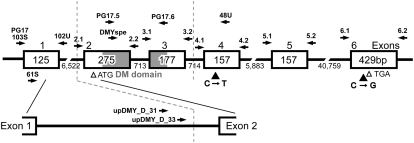
To screen for mutations in the amino acid coding sequence of DMY, exons 2–6 of DMY were PCR amplified from caudal fin clip DNA using the following primer sets: exon 1: PG17_103S, 5′-GGA AAC AAT TTT GCC TTG GA-3′ and PG17_102U, 5′-ACA CAA CGC ACG CAT AAA AA-3′; exon 2: PG17ex2.1, 5′-GGA GTC ACG TGA CCC TCT TTC TTG GG-3′ and PG17ex2.2, 5′-TTT CGG GTG AAC TCA CAT GGT TGT CG-3′; exon 3: PG17ex3.1, 5′-GCA ACA GAG AGT TGG ATT TAC GTC TCA-3′ and PG17ex3.2, 5′-CTT TTG ACT TCA GTT TGA CAC ATC AAT G-3′; exon 4: PG17ex4.1, 5′-CTC AGG TTT GAC TTG GAT GCT GAC CTG A-3′ and PG17ex4.2, 5′-CAA ACC AGG CCA TGA CCA TTC CGA-3′; exon 5: PG17ex5.1, 5′-CCG ATT CTA GCG GAT GAT GCC ACC-3′ and PG17ex5.2, 5′-GGG AGC CAA AAA TGC GCC ACA TAA-3′; and exon 6: PG17ex6.1, 5′-GTC ATT AAC ACA ACG CAC AAC AAC TT-3′ and PG17ex6.2, 5′-AAA AAC CAG AAG ACC CGA GAG GAA G-3′ (Figure 1). The PCR products were sequenced directly in an ABI Prism 310 automated sequencer. Two additional primers in intron 1, upDMY_D_31 (5′-GAG TGT GTG TGA GCG CAA GT-3′) and upDMY_D_33 (5′-TTG AAA TCC GAG CTT CTG AAA-3′), were used for PCR and direct sequencing of mutant DMY genes.
Figure 1.—
DMY structure of the Hd-rR.YHNI congenic strain. Open boxes, shaded boxes, and the horizontal line indicate exons, the DM domain, and introns, respectively. Open arrowheads indicate the translation start site, ATG, and stop site, TGA. The numbers represent the nucleotide sequence lengths (bp). Solid arrows indicate the primer positions. Solid arrowheads indicate the positions of substitutions in the DMY of YwObu1. The region between the two dotted vertical lines was deleted in the DMY of YwObu2.
RNA extraction and RT–PCR:
Total RNA was extracted from embryos or fry at 9.5 days after fertilization in the F1 generation using an RNeasy Mini kit (QIAGEN) and subjected to RT–PCR using a OneStep RT–PCR kit (QIAGEN). Aliquots (20 ng) of the total RNA samples were used as templates in 25-μl reaction volumes. The PCR conditions were 30 min at 55°; 15 min at 95°; 35 cycles of 20 sec at 96°, 30 sec at 55°, and 60 sec at 72°; and 5 min at 72°. The primers for DMY (DMYspe, 5′-TGC CGG AAC CAC AGC TTG AAG ACC-3′ and 48U, 5′-GGC TGG TAG AAG TTG TAG TAG GAG GTT T-3′) amplified a 404-bp DNA fragment. The primers for β-actin (3b, 5′-CMG TCA GGA TCT TCA TSA GG-3′, and 4, 5′-CAC ACC TTC TAC AAT GAG CTG A-3′) amplified a 322-bp DNA fragment. A primer in exon 1, PG17_61S (5′-CGT CTG GCT TCA CCG TTG GA-3′), was used to amplify the mutant-type DMY of YwObu2.
Gene copy number quantification using real-time PCR:
Wild medaka DNA was extracted according to Shinomiya et al. (1999). The purity and concentration of the DNA were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). The DNA concentrations of all samples were adjusted to give a total of 20 ng for each real-time quantitative PCR assay. Primers were designed to amplify a 106-bp fragment of DMY exon 4 (DMY_RT_4-1, 5′-GAG GAA GCG TCT GAC TGC-3′ and DMY_RT_4-2, 5′-CCT GGT ACT GCT GGT AGT TGT G-3′). SYBR Green PCR master mix (Takara) was used according to the manufacturer's protocol with an ABI 7000 real-time PCR instrument (Applied Biosystems). The PCR conditions were 1 min at 94°; 40 cycles of 5 sec at 95°, and 30 sec at 65°. To confirm that the amplified PCR products were specific for the DMY gene, their melting temperatures were determined from dissociation curves generated by the real-time quantitative PCR instrument. The fact that the melting temperature was the same for each product indicated that the same product was amplified from all the DNA samples. Direct sequencing confirmed that each product was amplified from the DMY gene (data not shown). Quantification of the copy number of DMY was achieved by comparison of the real-time quantitative PCR results to a standard curve generated from the inbred strain Hd-rR. Hd-rR DNA was serially diluted by 1:2 from 80 ng to 5 ng. The real-time quantitative PCR instrument software was used to plot the threshold cycle (cycle number of each sample that intersected the threshold line) of each sample on the standard curve of Hd-rR DNA and thereby determine the quantity. Since each sample had an identical amount of DNA as the initial template (20 ng), the DMY gene copy number could be calculated as a ratio of the quantity of each sample to that of Hd-rR. The real-time quantitative PCR experiments were repeated three times.
Statistical analysis:
The significance of differences among the frequencies of sex chromosomes was tested by the G-test.
RESULTS AND DISCUSSION
Frequent appearance of XY females:
Previously, we performed genotypic sexing of 2274 wild-caught medaka from 40 localities and found 12 XY females from 8 localities (Shinomiya et al. 2004). The average frequency of XY sex-reversed females was 1.1% (12 of 1089 XY individuals). In 2003, we performed genotypic sexing of 113 fishes in the O-bu population (Aichi prefecture) and found 17 XY females (Table 1). The frequency of XY sex-reversed females in this population was 21.8% (17 of 78 XY individuals). This value was significantly higher than those in the other populations examined in our previous study.
TABLE 1.
Phenotypic and genotypic sexes of the wild medaka in the O-bu population (2003)
| Female
|
Male
|
|||
|---|---|---|---|---|
| Total | XX | XY | XX | XY |
| 113 | 35 | 17 | 0 | 61 |
Y chromosomes bearing DMY with no expression during the sex-determining period:
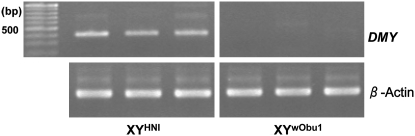
To clarify the cause of the XY sex reversals, three XY females were mated with XY males of Hd-rR.YHNI, and the genotypic and phenotypic sexes of the F1 progeny were analyzed (Table 2). All XX individuals in the F1 progeny were female and all XYp and YmYp individuals were male, while all XYm individuals were female (where Ym and Yp are maternal and paternal Y chromosomes, respectively). In other words, the occurrence of XY sex reversals in the F1 progeny was perfectly linked to the presence of the maternal Y chromosome. Since DMY is considered to be the sole functional gene in the Y-specific region (Kondo et al. 2006), this finding suggests that the sex-reversal mutants had a mutation at DMY or a gene tightly linked to DMY. A previous study demonstrated that all XY sex-reversal mutants in wild populations were associated with defective DMY and could be classified into two types. One had mutations in the amino acid coding sequence of DMY, while the other had a normal coding sequence but exhibited depressed or no DMY expression during the sex-determining period (Otake et al. 2006). Therefore, we sequenced exons 2–6 of DMY to identify possible mutations in the amino acid coding sequence. We found a C-to-T substitution in exon 4 and a C-to-G substitution in exon 6 (Figure 1), both of which were synonymous substitutions. Next, to examine DMY expression during the sex-determining period, we performed RT–PCR on the hatching day. DMY transcripts were not detected in F1 XYm progeny (Figure 2), suggesting that the cause of the sex reversal in the XY females was severe suppression or elimination of DMY expression during the sex-determining period, similar to the case for sex-reversal mutants from Oura (Otake et al. 2006). We designated this Y chromosome YwObu1. This mutated DMY may contain regulatory mutations in the flanking region of this gene.
TABLE 2.
Progeny test of XY females in the O-bu population
| ID no. of XY female | XX
|
XYm
|
XYp
|
YmYp
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Female | Male | Female | Male | Female | Male | Female | Male | |
| 01 | 54 | 11 | 0 | 10 | 0 | 0 | 23 | 0 | 10 |
| 02 | 34 | 11 | 0 | 7 | 0 | 0 | 8 | 0 | 8 |
| 03 | 22 | 5 | 0 | 4 | 0 | 0 | 5 | 0 | 8 |
Figure 2.—
RT–PCR analysis of DMY in XYwObu1 on the hatching day. The DMY PCR products obtained using the DMYspe and 48U primer set were subjected to 1% agarose gel electrophoresis. β-actin served as a positive control.
Spread of mutant-type Y chromosomes:
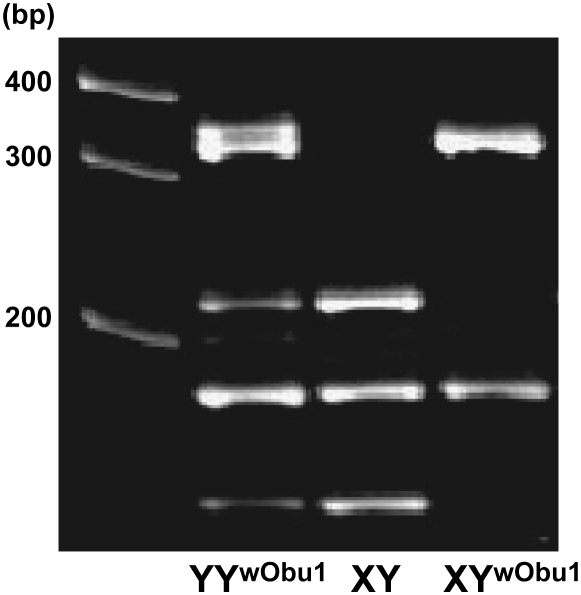
To investigate the frequency of YwObu1, we performed a PCR–RFLP analysis for all 78 XY individuals in the O-bu population. Since the mutant-type DMY on YwObu1 had a C-to-T substitution in exon 4, PCR amplification using the Ex4.1 and Ex4.2 primer set (Figure 1) and restriction enzyme digestion with Hpy8I produced three bands for wild-type DMY, two bands for mutant-type DMY, and four bands for wild-type and mutant-type heterozygote DMY (Figure 3). We found 16 individuals with YwObu1 and 4 individuals with both wild-type Y and Ywobu1. These results demonstrated that YwObu1 existed at a high frequency in the O-bu population and raised the possibility that there were YwObu1YwObu1 individuals in this population since XYwObu1 females were fully fertile. Since the PCR–RFLP method cannot distinguish between XYwObu1 and YwObu1YwObu1 individuals, we performed real-time PCR for quantification of the copy number of mutant-type DMY. By comparison of the threshold cycle of each sample with the standard curve of Hd-rR genomic DNA, which contains one copy of the DMY gene, we determined the copy number (see materials and methods). We examined 16 individuals with YwObu1 and concluded that 14 individuals had a single copy and 2 individuals had two copies of the mutant-type DMY. As a result, PCR–RFLP and real-time PCR analyses could detect 14 XYwObu1, 2 YwObu1YwObu1 and 3 YYwObu1 males, and interestingly one YYwObu1 female in this population.
Figure 3.—
Electrophoretic patterns of PCR–RFLP products for identifying XYwObu1. DMY PCR products digested with Hpy8I were subjected to 10% polyacrylamide gel electrophoresis. The DMY fragments of Hd-rR and XYwObu1 have one and two recognition sites, respectively.
Y chromosome with a second DMY mutation:
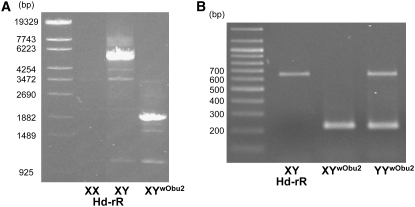
Our previous study showed that all YY individuals with wild-type and mutation-bearing Y chromosomes with low DMY expression developed as males (Otake et al. 2006). In fact, the normal Y chromosome could induce male development in three YYwObu1 individuals in this population. These findings raised the possibility that the YYwObu1 female had another null mutation at the DMY locus. Therefore, we mated the YYwObu1 female with XX males of an Hd-rR inbred strain induced by high-temperature treatment (Sato et al. 2005; Hattori et al. 2007). Not only the F1 progeny with YwObu1 but also the progeny with the other Y chromosome grew as females (Table 3). These results demonstrated that the other Y chromosome had also lost the male-determining function and bore another DMY mutation. We designated this new deficient Y chromosome YwObu2. Next, we carried out PCR and direct sequencing analyses of the DMY gene using the F1 XYwObu2 female progeny. The sequences of exons 1 and 4–6 were normal. However, the primer sets for exons 2 and 3 resulted in no amplification. Thus, we designed a new primer, upDMY_D_31, in intron 1 and performed PCR with 48U in exon 4 (Figure 1). This primer set amplified an ∼2-kb fragment in XYwObu2 females in contrast to a 5.5-kb fragment in normal XY males (Figure 4A). Sequencing analysis of these PCR fragments revealed that an ∼3.5-kb genomic region including exons 2 and 3 was deleted in the DMY gene of YwObu2. RT–PCR and direct sequencing analyses of embryos on the hatching day demonstrated that exons 2 and 3 were deleted in the transcripts (Figure 4B). Exons 2 and 3 of DMY contain the DM domain, a putative functional DNA-binding motif found in DSX of Drosophila melanogaster and MAB-3 of Caenorhabditis elegans (Raymond et al. 1998). Since all the F1 XYwObu2 progeny were females, the DMY transcripts lacking the DM domain appeared to be nonfunctional. In addition, the fact that the DMY gene on YwObu2 did not have a C-to-T substitution in exon 4 (data not shown) suggested that YwObu1 and YwObu2 were generated independently. The DMY gene in YwObu2 carries a deletion of the region bound by the primers for genotyping (PG17.5 and PG 17.6), thereby raising the possibility that XYwObu2 individuals had been judged as XX. To explore this possibility, we examined all 35 individuals judged as XX using the upDMY_D_33 and 48U primer set (Figure 1), which amplified a 1013-bp fragment of the DMY gene in YwObu2. As a result, we found one XYwObu2 female. Collectively, seven combinations of sex chromosomes were found: XX, XYwObu1, YwObu1YwObu1, XYwObu2, YwObu1YwObu2, XY, and YYwObu1 in the 2003 O-bu population (Table 4).
TABLE 3.
Progeny test of YYwObu1 females
| XYwObu1
|
XY
|
|||
|---|---|---|---|---|
| Total | Female | Male | Female | Male |
| 21 | 13 | 0 | 8 | 0 |
Figure 4.—
Genomic and RT–PCR analyses of DMY in XYwObu2. (A) The DMY PCR products obtained using the upDMY_D_33 and 48U primer set were subjected to 1% agarose gel electrophoresis. (B) The DMY RT–PCR products obtained using the 61S and 48U primer set were subjected to 1% agarose gel electrophoresis.
TABLE 4.
Phenotypic sex and sex chromosome types in the O-bu population
| Female
|
Male
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Year | Total | XX | XYwObu1 | XYwObu2 | YwOub1YwObu1 | YwObu1YwObu2 | XY | YYwObu1 | YYwObu2 |
| 2003 | 113 | 34 | 14 | 1 | 2 | 1 | 58 | 3 | 0 |
| 2004 | 131 | 46 | 12 | 0 | 0 | 0 | 59 | 14 | 0 |
| 2005 | 55 | 10 | 14 | 0 | 1 | 0 | 27 | 3 | 0 |
| 2006 | 113 | 52 | 9 | 0 | 0 | 0 | 36 | 13 | 3 |
Stability of mutant-type Y chromosomes:
Four types of sex chromosomes, namely two aberrant Y chromosomes with loss-of-function DMY mutations and normal X and Y chromosomes, were observed in the O-bu population. To explore whether the mutant-type Y chromosomes were maintained in subsequent generations, we analyzed the frequency of each sex chromosome type in this population over four successive generations (Table 4). We found the YwObu1 chromosome in each year and the YwObu2 chromosome in 2006. The differences in the frequencies of each sex chromosome over 4 years were not significant (G-test, Gadj = 12.542, d.f. = 9, P = 0.1844; Table 5). These results demonstrated that the mutant-type Y chromosomes have been maintained in this population. In addition, the facts that XYwObu1, YwObu1YwObu1, XYwObu2, and YwObu1YwObu2 are viable in the wild and XYwObu1 and YwObu1YwObu2 females are fully fertile in the laboratory condition suggest that X and mutant-type Y chromosomes were functionally the same and these Y chromosomes appear selectively neutral.
TABLE 5.
Estimated number and frequency of each sex chromosome in the O-bu population
| Year | Total | X | YwObu1 | YwObu2 | Y |
|---|---|---|---|---|---|
| 2003 | 226 | 141 (0.624) | 22 (0.097) | 2 (0.009) | 61 (0.270) |
| 2004 | 262 | 163 (0.622) | 26 (0.099) | 0 (0.000) | 73 (0.279) |
| 2005 | 110 | 61 (0.555) | 19 (0.173) | 0 (0.000) | 30 (0.273) |
| 2006 | 226 | 149 (0.659) | 22 (0.097) | 3 (0.013) | 52 (0.230) |
The frequency of XY sex-reversed females in the O-bu population was considerably high compared with the average frequency of other wild populations (∼1%) described in Shinomiya et al. (2004). High frequency of XY females has been observed in Goshogawara (Aomori prefecture). We found 14 XY females (18.2%) of 77 XY individuals from this population in 2005 (M. Sakaizumi, unpublished data). All these XY females had a single nucleotide insertion in the poly (C) tract in exon 3 of DMY identical to the case for XY sex-reversal mutants from Awara (Matsuda et al. 2002; Otake et al. 2006). Both populations appear to have small population sizes and to be isolated from other populations. These facts imply that the mutant-type Y chromosomes could have become frequent by means of genetic drift due to population bottlenecks.
On the other hand, the following three results indicated that the XX/XY genetic sex-determination system controlled by the DMY gene appeared to be active. First, all male individuals had a normal Y chromosome (Table 4). Second, no significant deviation from the expected 1:1 ratio was found for the sex ratio in each year (χ2 = 0.72 in 2003, 1.72 in 2004, 0.45 in 2005, and 1.1 in 2006). Third, the frequency of the normal Y chromosome was ∼25% (Table 5).
Taken together, our data demonstrated that the Y chromosome lacking the male-determining function behaved as an X chromosome in this population. This phenomenon appears to be characteristic of organisms like the medaka that have quite undifferentiated sex chromosomes.
Acknowledgments
We thank Ai Shinomiya, Masaru Matsuda, and Kazunori Yamahira for their helpful technical advice and invaluable suggestions. This work was supported in part by EXTEND 2005 from the Ministry of the Environment of Japan to S.H.
References
- Graves, J.A., 2002. Evolution of the testis-determining gene–the rise and fall of SRY. Novartis Found. Symp. 244 86–97. [PubMed] [Google Scholar]
- Gubbay, J., J. Collignon, P. Koopman, B. Capel, A. Economou et al., 1990. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 346 245–250. [DOI] [PubMed] [Google Scholar]
- Hattori, R. S., R. J. Gould, T. Fujioka, T. Saito, J. Kurita et al., 2007. Temperature-dependent sex determination in Hd-rR medaka Oryzias latipes: gender sensitivity, thermal threshold, critical period, and DMRT1 expression profile. Sex. Dev. 1 138–146. [DOI] [PubMed] [Google Scholar]
- Iwamatsu, T., H. Kobayashi and M. Yamashita, 2006. Sex reversal in medaka treated in vitro with 17alpha-methyldihydrotestosterone during oocyte maturation. Dev. Growth Differ. 48 59–64. [DOI] [PubMed] [Google Scholar]
- Kobayashi, H., and T. Iwamatsu, 2005. Sex reversal in the medaka Oryzias latipes by brief exposure of early embryos to estradiol-17beta. Zoolog. Sci. 22 1163–1167. [DOI] [PubMed] [Google Scholar]
- Kondo, M., I. Nanda, U. Hornung, S. Asakawa, N. Shimizu et al., 2003. Absence of the candidate male sex-determining gene dmrt1b(Y) of medaka from other fish species. Curr. Biol. 13 416–420. [DOI] [PubMed] [Google Scholar]
- Kondo, M., I. Nanda, U. Hornung, M. Schmid and M. Schartl, 2004. Evolutionary origin of the medaka Y chromosome. Curr. Biol. 14 1664–1669. [DOI] [PubMed] [Google Scholar]
- Kondo, M., U. Hornung, I. Nanda, S. Imai, T. Sasaki et al., 2006. Genomic organization of the sex-determining and adjacent regions of the sex chromosomes of medaka. Genome Res. 16 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman, P., J. Gubbay, N. Vivian, P. Goodfellow and R. Lovell-Badge, 1991. Male development of chromosomally female mice transgenic for Sry. Nature 351 117–121. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., C. Matsuda, S. Hamaguchi and M. Sakaizumi, 1998. Identification of the sex chromosomes of the medaka, Oryzias latipes, by fluorescence in situ hybridization. Cytogenet. Cell Genet. 82 257–262. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., Y. Nagahama, A. Shinomiya, T. Sato, C. Matsuda et al., 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417 559–563. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., T. Sato, Y. Toyazaki, Y. Nagahama, S. Hamaguchi et al., 2003. Oryzias curvinotus has DMY, a gene that is required for male development in the medaka, O. latipes. Zoolog. Sci. 20 159–161. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., A. Shinomiya, M. Kinoshita, A. Suzuki, T. Kobayashi et al., 2007. DMY gene induces male development in genetically female (XX) medaka fish. Proc. Natl. Acad. Sci. USA 104 3865–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda, I., M. Kondo, U. Hornung, S. Asakawa, C. Winkler et al., 2002. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 99 11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otake, H., A. Shinomiya, M. Matsuda, S. Hamaguchi and M. Sakaizumi, 2006. Wild-derived XY sex-reversal mutants in the medaka, Oryzias latipes. Genetics 173 2083–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, C. S., C. E. Shamu, M. M. Shen, K. J. Seifert, B. Hirsch et al., 1998. Evidence for evolutionary conservation of sex-determining genes. Nature 391 691–695. [DOI] [PubMed] [Google Scholar]
- Ross, M. T., D. V. Grafham, A. J. Coffey, S. Scherer, K. McLay et al., 2005. The DNA sequence of the human X chromosome. Nature 434 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, T., T. Endo, K. Yamahira, S. Hamaguchi and M. Sakaizumi, 2005. Induction of female-to-male sex reversal by high temperature treatment in medaka, Oryzias latipes. Zoolog. Sci. 22 985–988. [DOI] [PubMed] [Google Scholar]
- Shinomiya, A., M. Matsuda, S. Hamaguchi and M. Sakaizumi, 1999. Identification of genetic sex of the medaka, Oryzias latipes, by PCR. Fish Biol. J. Medaka 10 31–32. [Google Scholar]
- Shinomiya, A., H. Otake, K. Togashi, S. Hamaguchi and M. Sakaizumi, 2004. Field survey of sex-reversals in the medaka, Oryzias latipes: genotypic sexing of wild populations. Zoolog. Sci. 21 613–619. [DOI] [PubMed] [Google Scholar]
- Sinclair, A. H., P. Berta, M. S. Palmer, J. R. Hawkins, B. L. Griffiths et al., 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346 240–244. [DOI] [PubMed] [Google Scholar]
- Skaletsky, H., T. Kuroda Kawaguchi, P. J. Minx, H. S. Cordum, L. Hillier et al., 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423 825–837. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., Y. Takehana, K. Naruse, S. Hamaguchi and M. Sakaizumi, 2007. Evidence for different origins of sex chromosomes in closely related Oryzias fishes: substitution of the master sex-determining gene. Genetics 177 2075–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]