Abstract
We report the development and characterization of three new microsatellite markers in the tsetse fly, Glossina pallidipes (Diptera: Glossinidae). Fifty-eight alleles were scored in 192 individuals representing six natural populations. Allelic diversity ranged from 9 to 28 alleles per locus (mean 19.3 ± 5.5). Averaged across loci, observed heterozygosity was 0.581 ± 0.209, and expected heterozygosity was 0.619 ± 0.181. Cross-species amplifications of the G. pallidipes loci in other tsetse fly taxa are reported.
Keywords: tsetse flies, Glossina, microsatellites
INTRODUCTION
Tsetse flies (Diptera: Glossinidae) are discontinuously distributed throughout sub-Saharan Africa (Rogers and Robinson, 2004). They feed exclusively on blood and transmit hemoflagellate protozoan parasites, trypanosomes that cause sleeping sickness in humans and nagana in livestock. Tsetse taxa are subdivided into three subgenera, Glossina (morsitans group), Nemorhina (palpalis group), and Austenina (fusca group). Glossina pallidipes belongs to the morsitans group and is among the most important vectors of trypanosomes.
Earlier genetic studies on tsetse flies were based largely on cytological and allozyme methods (Krafsur and Griffiths, 1997; Gooding and Krafsur, 2005). Adequately sampling geographically diverse tsetse fly populations is difficult and expensive. Recent progress in tsetse fly population genetics includes the Solano et al. (1997) examination of microsatellite variation in Glossina palpalis gambiensis and G. p. palpalis natural populations. Baker and Krafsur (2001) and Ouma et al. (2003) uncovered microsatellite loci in the morsitans group of tsetse flies, and the population genetics of diverse natural morsitans group populations have now been investigated (reviews in Krafsur, 2003; Gooding and Krafsur, 2005). Additional genomic DNA markers would be most useful, and here we report three new microsatellite loci and their homologs in other tsetse fly taxa.
METHODS AND MATERIALS
Genomic DNA was extracted from Glossina pallidipes obtained from a colony established at the International Atomic Energy Agency, Seibersdorf, Austria. Approximately 100 µg of DNA was used to construct four genomic libraries enriched for CA, GA, ATG, and CAG microsatellites. Libraries were constructed by Genetic Identification Services (http://www.genetic-id-services.com; Chatsworth, CA). Enriched G. pallidipes DNA fragments were ligated into the HindIII cut site of pUC19 plasmid and electroporated into Escherichia coli strain DH5α (Electro-Max, Gibco). After transformation, E. coli cells were screened for inserts between 350 and 700 bp by polymerase chain reaction (PCR) amplification. PCR and sequencing of individual clones was performed using forward and reverse universal M13 primers and ABI Prism BigDye Terminator Chemistry as previously described (Ouma et al., 2003).
Fifty microsatellite clones were sequenced. PCR primers were designed for 28 of these clones by using the software DesignerPCR, version 1.03 (Research Genetics, Inc.). Three to eight primer pairs were designed for each locus, and the pair yielding a specific PCR as determined by a single band on 1% agarose gel was selected for use. Oligonucleotides were synthesized by Integrated DNA Technologies, Coralville, Iowa.
To evaluate presumptive loci for polymorphisms, G. pallidipes were sampled from six natural populations in Kenya and Tanzania. In all, 24–48 individuals were genotyped per population for a total of 192 flies (136 females and 56 males). PCR was carried out on a PTC-100 thermocycler (MJ Research). Reaction volumes were 10 µL and contained 25 ng template DNA, 1 × Biolase PCR buffer, 2.5 MgCl2, 0.4 mM dNTPs, 0.4 units Biolase polymerase (Bioline USA, Springfield, NJ), 0.5 µM each of forward and reverse primers. The forward primers were fluorescently labeled with FAM or HEX. Primers were also tested for cross-amplification of DNA from nine species representing the three Glossina subgenera.
FStat version 2.9.3.2 was used to estimate genetic diversity (Goudet, 1995). Arlequin version 2.0 (Schneider et al., 2000) was used to test for Hardy–Weinberg equilibrium. Micro-Checker (van Oosterhout et al., 2004) was used to test for causes of departures from Hardy–Weinberg equilibrium. Genotypic disequilibrium was tested using the log-likelihood ratio G-statistic. Here FIT estimates departures from random mating from all causes, and FIS the departure from random mating within populations.
RESULTS AND DISCUSSION
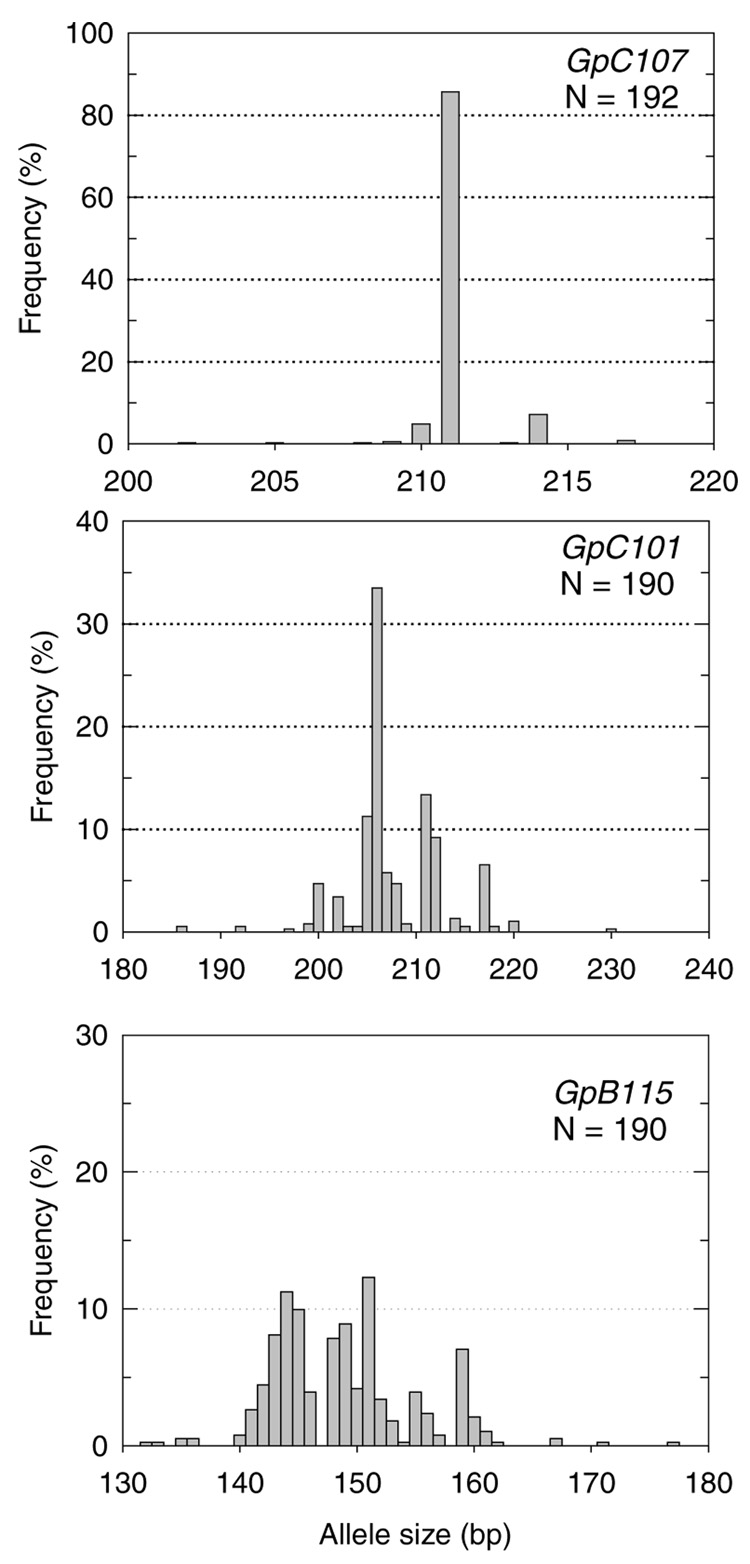
All loci were autosomal. Two were trinucleotide repeats, one of which, GpC107, was compound (Table I). The loci were highly polymorphic in Glossina pallidipes. The number of alleles per locus ranged from 9 to 28, with a mean of 19.3 (Table I). Mean observed and expected heterozygosity was 0.581 and 0.619, respectively, leading to a modest FIT estimate of 0.06. The least diverse locus was GpC107; it did not occur in Hardy–Weinberg proportions in any population. Rules postulated in Micro-Checker software indicated that large allele dropouts and allelic stutterings were unlikely causes of the deficit, thereby pointing to a null allele frequency of 0.125. Allele frequency distributions showed 43 rare (<5%) of a total of 58 alleles summed over loci, for an overall mean of 74% (Fig. 1). No significant genotypic linkage was detected in 18 locus–population combination tests.
Table I.
Characterization of New Microsatellite Loci in 192 Glossina pallidipes Representing Six Populations
| Locus | Repeat motif | No. alleles | Allele size (bp) | HO | HE | FIT | Primer sequence (5′–3′) | GenBank accession no. |
|---|---|---|---|---|---|---|---|---|
| GpB115 | (CT)10 | 28 | 132–177 | 0.916 | 0.825 | −0.110 | F:AGCGATAGAAAGGGTCAATC | DQ168823 |
| R:CGTAGAGATAGCGAGAGTGTG | ||||||||
| GpC101 | (TGA)11 | 21 | 186–230 | 0.630 | 0.773 | 0.185 | F:CCTCAATACAGCAGCAGATG | DQ168824 |
| R:CAAGGTGTGTTGTCGTCTTC | ||||||||
| GpC107 | (CAG)5(CAA)5 | 9 | 202–217 | 0.197 | 0.259 | 0.239*** | F:CAATCGCAACAACATCAAAC | DQ168825 |
| R:GGCAATAACAACTGTCTGTGG | ||||||||
| Mean | 19.3 | 0.581 | 0.619 | 0.105 | ||||
| S.E | 5.5 | 0.209 | 0.181 | 0.072 |
γ2 (36) = 86.8, P < 0.001.
Fig. 1.
Allele frequency distribution in G. pallidipes microsatellite loci.
Cross-amplification of species within a genus is commonly observed, and a pronounced ascertainment bias was recorded. The mean number of alleles among the fusca and palpalis group was 2.78, and in the morsitans group (apart from G. pallidipes) it was 4.8 alleles per locus.
Our primers (Table I) did not amplify GpB115 in Glossina brevipalpis, G. longipennis, or G. palpalis gambiensis (Table II). GpC101 and GpC107 were monomorphic in G. fuscipes fuscipes; GpC101 was monomorphic in G. brevipalpis. No homozygotes were recorded at GpB115 in G. m. morsitans and G. austeni (Table II), the reason for which is obscure. Significant departures from random mating (FIS) were observed at GpC101 in G. longipennis and G. swynnertoni, GpB115 in G. austeni, and GpC107 in G. m. submorsitans. The best amplifications were in morsitans group flies. The three loci can be used in G. m. morsitans and G. m. centralis. There now is at least one new and well-behaved locus in G. m. submorsitans, G. austeni, and G. swynnertoni.
Table II.
Cross-Amplification and Variability in Microsatellite Loci in Tsetse Fly Taxa
| Group (subgenus) | Locus | N | No. alleles | Allele size range | ho | he | FIS | |
|---|---|---|---|---|---|---|---|---|
| Fusca (Austenina) | G. brevipalpis | GpB115 | – | – | – | – | – | – |
| GpC101 | 15 | 1 | 187 | 0.000 | 0.000 | NA | ||
| GpC107 | 8 | 2 | 205–208 | 0.438 | 0.514 | 0.148 | ||
| G. longipennis | GpB115 | 15 | 6 | 213–224 | 0.467 | 0.818 | 0.429 | |
| GpC101 | 16 | 3 | 173–193 | 0.063 | 0.280 | 0.775*** | ||
| GpC107 | 16 | 2 | 208–211 | 0.375 | 0.315 | −0.190 | ||
| Palpalis (Nemorhina) | G. f. fuscipes | GpB115 | – | – | – | – | – | – |
| GpC101 | 16 | 1 | 208 | 0.000 | 0.000 | NA | ||
| GpC107 | 16 | 1 | 211 | 0.000 | 0.000 | NA | ||
| G. p. gambiensis | GpB115 | – | – | – | – | – | – | |
| GpC101 | 15 | 5 | 199–211 | 0.800 | 0.733 | −0.091 | ||
| GpC107 | 16 | 4 | 205–214 | 0.563 | 0.569 | 0.011 | ||
| Morsitans (Glossina) | G. austeni | GpB115 | 14 | 3 | 194–209 | 1.000 | 0.553 | −0.808*** |
| GpC101 | 14 | 6 | 210–234 | 0.714 | 0.812 | 0.121 | ||
| GpC107 | 16 | 1 | 211 | 0.000 | 0.000 | NA | ||
| G. m. centralis | GpB115 | 14 | 4 | 129–163 | 0.500 | 0.476 | −0.050 | |
| GpC101 | 12 | 6 | 200–212 | 0.750 | 0.685 | −0.095 | ||
| GpC107 | 12 | 4 | 214–220 | 0.667 | 0.772 | 0.136 | ||
| G. m. morsitans | GpB115 | 15 | 6 | 137–162 | 1.000 | 0.697 | −0.435 | |
| GpC101 | 14 | 10 | 202–228 | 0.643 | 0.860 | 0.252 | ||
| GpC107 | 15 | 5 | 205–217 | 0.267 | 0.308 | 0.133 | ||
| G. m. submorsitans | GpB115 | 16 | 9 | 144–168 | 0.813 | 0.784 | −0.037 | |
| GpC101 | 15 | 5 | 200–206 | 0.267 | 0.595 | 0.551* | ||
| GpC107 | 16 | 4 | 211–217 | 0.063 | 0.413 | 0.847*** | ||
| G. swynnertoni | GpB115 | 16 | 4 | 131–140 | 0.688 | 0.534 | −0.085 | |
| GpC101 | 16 | 3 | 208–210 | 0.000 | 0.476 | 1.000*** | ||
| GpC107 | 16 | 2 | 205–211 | 0.063 | 0.063 | 0 |
P ~ 0.05.
P < 0.001
γ2 = F2N (k − 1), df = k (k − 1)/2, where k = number of alleles and N = sample size.
This report brings to 12 the number of microsatellite loci isolated from Glossina pallidipes.
ACKNOWLEDGMENT
This research was funded by USPHS-NIH grant AI-052456.
REFERENCES
- Baker MD, Krafsur ES. Identification and properties of microsatellite markers in tsetse flies Glossina morsitans sensu lato (Diptera: Glossinidae) Mol. Ecol. Notes. 2001;1:234–236. doi: 10.1046/j.1471-8278.2001.00087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding RH, Krafsur ES. Tsetse genetics: Contributions to biology, systematics, and control of tsetse flies. Ann. Rev. Entomol. 2005;50:101–123. doi: 10.1146/annurev.ento.50.071803.130443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. FStat: A computer program to calculate F-statistics. J. Heredity. 1995;86:485–486. [Google Scholar]
- Krafsur ES. Tsetse fly population genetics: An indirect approach to dispersal. Trends Parasitol. 2003;19:162–166. doi: 10.1016/s1471-4922(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Krafsur ES, Griffiths N. Genetic variation at structural loci in the Glossina morsitans species group. Biochem. Genet. 1997;35:1–11. doi: 10.1023/a:1022252311715. [DOI] [PubMed] [Google Scholar]
- Ouma JO, Cummings MA, Jones KC, Krafsur ES. Characterization of microsatellite markers in the tsetse fly, Glossina pallidipes (Diptera: Glossinidae) Mol. Ecol. Notes. 2003;3:450–453. doi: 10.1046/j.1471-8286.2003.00480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers DJ, Robinson T. Tsetse distribution. In: Maudlin I, Holmes PH, Miles MA, editors. The Trypanosomes. Wallingford: CABI Publishing; 2004. pp. 139–179. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Arlequin, version 2.000: A software for population genetics data analysis. Switzerland: Genetics and Biometry Laboratory, University of Geneva; 2000. [Google Scholar]
- Solano P, Duvallet G, Dumas V, Cuisance D, Cuny G. Microsatellite markers for genetic population studies in Glossina palpalis (Diptera: Glossinidae) Acta Tropica. 1997;65:175–180. doi: 10.1016/s0001-706x(97)00663-3. [DOI] [PubMed] [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-Checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004;4:535–538. [Google Scholar]