Abstract
The kidney filter represents a unique assembly of podocyte epithelial cells that tightly enwrap the glomerular capillaries with their foot processes and the interposed slit diaphragm. So far, very little is known about the guidance cues and polarity signals required to regulate proper development and maintenance of the glomerular filtration barrier. We now identify Par3, Par6, and atypical protein kinase C (aPKC) polarity proteins as novel Neph1-Nephrin-associated proteins. The interaction was mediated through the PDZ domain of Par3 and conserved carboxyl terminal residues in Neph1 and Nephrin. Par3, Par6, and aPKC localized to the slit diaphragm as shown in immunofluorescence and immunoelectron microscopy. Consistent with a critical role for aPKC activity in podocytes, inhibition of glomerular aPKC activity with a pseudosubstrate inhibitor resulted in a loss of regular podocyte foot process architecture. These data provide an important link between cell recognition mediated through the Neph1-Nephrin complex and Par-dependent polarity signaling and suggest that this molecular interaction is essential for establishing the three-dimensional architecture of podocytes at the kidney filtration barrier.
Mammalian kidney function requires the coordinate development of specific cell types within a precise architectural framework so that body fluid composition can be monitored and regulated (1). One of the major functions of the kidney is filtration of plasma and the production of urine. Ultrafiltration in the kidney occurs in hundreds of thousands of small microvascular units, the renal glomeruli that consist of three layers: a fenestrated endothelium, the intervening glomerular basement membrane, and podocytes. These podocytes elaborate long, regularly spaced, interdigitating foot processes that envelop the glomerular capillaries and are connected through the slit diaphragm, a membrane-like, highly specialized cell junction. The slit diaphragm has recently been appreciated as a signaling platform that regulates podocyte survival, endocytosis, and cytoskeletal organization (2). Mutations in genes encoding slit diaphragm proteins contribute to hereditary glomerular diseases in patients. Nephrin and Neph proteins, members of the immunoglobulin superfamily of adhesion molecules, are essential components of this protein complex (3–6). They are indispensable for the formation of the cell junction between adjacent podocyte foot processes, form the molecular sieve for glomerular filtration, and help to establish and maintain the architecture of the podocyte. However, the signal mechanisms of this specialized Neph-Nephrin protein complex to establish and maintain the orientation and polarization of the foot process network are unknown.
Polarization is characterized by the asymmetrical partitioning of proteins to specific membrane domains. In recent years, major advances have been made in the identification of proteins that regulate cellular polarity. Numerous multiprotein polarity complexes have been shown to control processes such as epithelial apical-basal polarity, cell migration, and asymmetric cell division. One of the evolutionarily conserved protein complexes required for determination of polarity is composed of the PDZ domain proteins Par3, Par6, and atypical protein kinase C (aPKC).4 These proteins are interdependent, and loss of any one leads to mislocalization of the others (7). In epithelia, Par3, Par6, and aPKC are usually recruited to tight junctions. However, podocytes lack tight junctions and instead rely on a Neph-Nephrin-based slit diaphragm protein complex, and a role for the Par3, Par6, and aPKC complex in regulating the polarized structure of podocytes has not been demonstrated.
In this study, we identify the Par3-Par6-aPKC complex as a novel slit diaphragm component that interacts with Neph-Nephrin proteins. Our results suggest that the Neph-Nephrin-Par multiprotein complex is crucial for the maintenance of podocyte foot process architecture and integrity of the glomerular filtration barrier.
EXPERIMENTAL PROCEDURES
Reagents and Plasmids—Neph1, Neph2, and Neph3 and Nephrin were described previously (8, 9). To create amino-terminally V5-tagged constructs of Neph1 and Nephrin, the leader sequence of CD5 followed by the V5 tag was fused to the 5′-end of the coding sequence without the original leader sequence. Membrane-bound fusion proteins of the carboxyl-terminal cytoplasmic domain of Neph1, Neph2, and Neph3 were generated using a pCDNA6 cassette that contained the leader sequence of CD5 fused to the CH2 and CH3 domain of human IgG1 followed by the transmembrane region of CD7 (10). Full-length cDNA of rat Par3, mouse aPKCλ, and dominant-negative mouse aPKCλK273E were kindly provided by Shigeo Ohno (Yokohama City University School of Medicine, Yokohama, Japan). Mouse Par6 was cloned from a mouse fetal kidney library and has been described previously (11). The Neph1 and Nephrin antisera have been described (5, 8). Antibodies were obtained from Sigma (anti-FLAG M2 mAb, anti-FLAG pAb), Serotec (anti-V5 mAb), Chemicon (anti-V5 pAb, anti-MBP mAb), Zymed Laboratories Inc. (anti-ZO-1 mAb), Upstate Biotechnology (anti-Par3 pAb), and Santa Cruz Biotechnology (anti-hemagglutinin pAb, anti-aPKCλ mAb).
Co-Immunoprecipitation—Co-immunoprecipitations were performed as described (12). Briefly, transiently transfected human embryonic kidney (HEK) 293T cells, glomeruli isolated from adult C57BL/6 mice using Dynabead perfusion (13), or mouse total kidneys were lysed in a 1% Triton X-100 lysis buffer (containing 20 mm CHAPS for endogenous immunoprecipitation). After centrifugation (15,000 × g, 15 min, 4 °C) and ultracentrifugation at 100,000 × g (30 min, 4 °C) and preclearing with 10 μl of protein G-Sepharose beads, lysates containing equal amounts of total protein were incubated for 1 h at 4 °C with the appropriate antibody followed by incubation with 20 μl of protein G-Sepharose beads for ∼1 h. The beads were washed extensively with lysis buffer, and bound proteins were resolved by 10% SDS-PAGE.
Pull-down Assay—HEK 293T cells were transiently transfected with plasmid DNA as indicated. Cells were lysed in a 1% Triton X-100 lysis buffer. Following centrifugation, the supernatant was incubated for 1 h at 4°C with 4–8 μg of recombinant purified fusion proteins of glutathione S-transferase (GST) and the three PDZ domains of Par3 (Par3256–370, Par3448–667, Par3561–717) prebound to glutathione-Sepharose beads (GE Healthcare). Bound proteins were separated by 10% SDS-PAGE, and precipitated proteins were visualized with anti-V5 antibody. Equal loading of recombinant proteins was confirmed by Coomassie Blue staining of the gels.
In Vitro Interaction—5 μg of recombinant purified fusion proteins of GST and two PDZ domains of Par3 (Par3256–370, Par3448–667) prebound to glutathione-Sepharose beads (GE Healthcare) were incubated for 1 h with 5 μg of recombinant purified fusion protein of MBP and Neph1742–789 in 1% Triton X-100 lysis buffer. The beads were washed extensively with lysis buffer, and bound proteins were resolved by 10% SDS-PAGE.
Immunofluorescence Staining—Rat kidneys of adult and 1-day-old Wistar rats (of note, newborn rats display various stages of glomerular development since glomerular development is asynchronous in rats) were frozen in OCT compound and sectioned at 6 μm (Leica Kryostat). The sections were fixed with 4% paraformaldehyde, blocked in phosphate-buffered saline containing 5% bovine serum albumin, and incubated for 1 h with primary antibodies as indicated. After phosphate-buffered saline rinse for several times, fluorophore-conjugated secondary antibodies (Invitrogen) were applied for 30 min. Confocal images were taken using a Zeiss laser scan microscope equipped with a ×63 water immersion objective.
Immunoelectron Microscopy—Fixed samples of rat kidney were embedded in Lowicryl K4M resin (Electron Microscopy Sciences), and ultrathin sections were labeled by an indirect immunogold protocol, as described (14).
aPKC Inhibitor Assay—Glomeruli isolated from adult C57BL/6 mice were incubated with a cell-permeable myristoylated pseudosubstrate inhibitor of atypical PKCλ/ζ (myr-SIYRRGARRWRKL; Quality Controlled Biochemicals, Hopkington, MA) (15) or with scrambled peptide as control (myr-RLYRKRIWRSAGR; Quality Controlled Biochemicals) at a concentration of 10 μm in Hanks' balanced salt solution. Glomeruli were incubated in a 24-well plate shaking gently in 37 °C for 120 min, harvested, fixed in 2.5% glutaraldehyde (Sigma) and subjected to electron microscopy.
In Situ Hybridization—A mouse kidney cDNA library served as template to clone fragments of the coding sequence and 3′-untranslated region. The following primer were used: Nephrin.fp (5′-CAGACCACACCAACATCCAG-3′), Nephrin.rp (5′-CCCCTTGGGTCCTCATATTT-3′); mPar3.fp (5′-TGGCAAACATCGAAAAGATG-3′); mPar3.rp (5′-ATACCTGGGGTTCTCCTTGG-3′); mPar6.fp (5′-GGACCAGGTGACTGACATGA-3′); mPar6.rp (5′-GGCCACCACAAGGTTAAGAA-3′); mPKCλ.fp (5′-TATGGCTTCAGCGTTGACTG-3′), mPKCλ.rp (5′-CTCGAATCCTGCCTCTGAAC-3′). Neph1 was a kind gift from A. Kispert (Medizinische Hochschule Hannover, Hannover, Germany). PCR products were cloned into pBluescript SK+, linearized, and transcribed with T3 and T7 RNA polymerases (Promega) to generate sense and antisense digoxigenin-labeled probes (digoxigenin RNA labeling mix; Roche Applied Science). Embryos and kidneys at embryonic day 16.5 were fixed overnight at 4 °C in 4% paraformaldehyde, embedded in paraffin, and sectioned at 8 μm. For mRNA detection, slides were treated with H202 and proteinase K, refixed with 4% paraformaldehyde, and hybridized at 65 °C in hybridization buffer (40% formamide, 5× SSC, 1× Denhardt's, 1% tRNA, 1% herring sperm DNA, 1% probe). Stringency washes were performed with wash I (5× SSC), wash II (20% formamide, 0.5× SSC), and wash III (2× SSC) and were followed by RNase treatment. For detection, slides were incubated with alkaline phosphatase-conjugated anti-digoxigenin antibody (Roche Applied Science) 1:4000 at 4 °C overnight followed by nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Roche Applied Science). Digital photographs were captured on an Axioplan2 microscope (Zeiss).
Gene Delivery—All animal studies were approved by the Subcommittee on Research Animal Care of the Massachusetts General Hospital. C57BL/6 mice were obtained from The Jackson Laboratory (Bar Harbor. ME). The pcDNA6-FLAG.aPKCλK273E plasmid was introduced using the TransIT in vivo gene delivery system (Mirus) essentially as described before (16). In brief, 48 μg of endotoxin-free plasmid DNA was mixed with 48 μl of Mirus polymer solution (1 μl/μg plasmid DNA) and brought to a total volume of 200 μl with endotoxin-free H2O. 1.8 ml of Mirus delivery solution was then added before injection of the entire volume through the tail vein. The empty vector and aPKCλ wild type cDNA served as negative controls. Before gene delivery (baseline) and 16 h after gene delivery, mouse urine was collected and clarified by centrifugation (2000 × g, 5 min, 4 °C). Total protein in the urine was determined using the Bio-Rad protein assay (Bio-Rad) according to the manufacturer's instructions. For detection of FLAG.aPKCλK273E protein, kidneys were harvested 16 h after injection and fixed in 3% paraformaldehyde, and immunogold analysis was performed as described (17) using anti-FLAG pAb antibody (Sigma). Statistical analyses were performed by using the unpaired t test, and the null hypothesis was rejected at the 0.05 level. Values are presented as mean ± S.D.
RESULTS
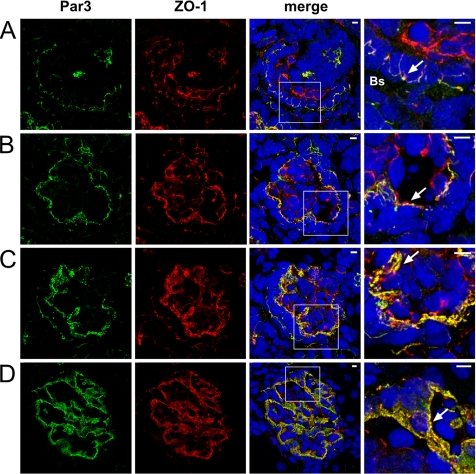
Par3 and ZO-1 Colocalize during Glomerular Development—We hypothesized that the slit diaphragm is actively involved in polarity signaling. Interestingly, Nephrin, Neph1, and the Par3-Par6-aPKC polarity complex exhibited similar glomerular expression patterns by in situ hybridization during kidney development (supplemental Fig. 1). Confocal microscopy revealed that Par3 colocolized with the known junctional and slit diaphragm marker ZO-1 (18) at all stages of differentiating glomeruli. Colocalization of Par3 and ZO-1 could be demonstrated early in development when the apical junctional complexes of adjacent presumptive podocytes are composed of typical tight and adherens junctions (Fig. 1A), and the colocolization was maintained when these junctions maturated to form slit diaphragm junctional complexes (Fig. 1D).
FIGURE 1.
Par3 and ZO-1 colocalize during glomerular development. Frozen kidney sections of new born Wistar rat (day 1) were stained using antibodies against Par3 and ZO-1 and subjected to confocal laser microscopy. Par3 signal colocalized with ZO-1 at all stages of differentiating glomeruli (arrows), S-shaped body with open Bowman's space (Bs)(A), and capillary-loop stage (B) and during foot process development and glomerular maturation (C and D). Scale bars represent 5 μm.
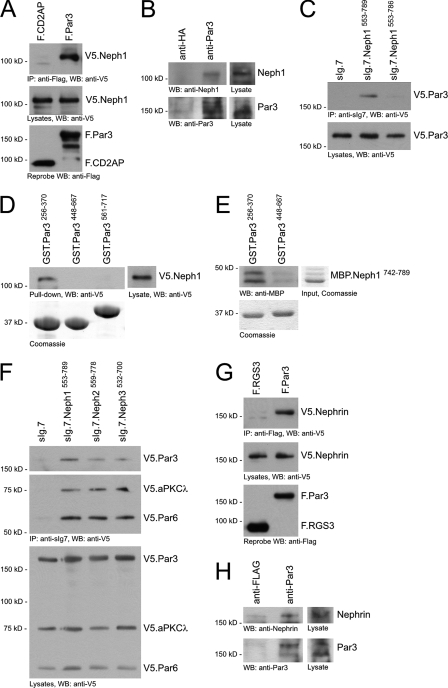
Neph-Nephrin Proteins Bind to the Par3-Par6-aPKC Complex—In recent years, several Par3-interacting transmembrane proteins that target the preformed Par3-Par6-aPKC complex to the plasma membrane and cell-cell contacts have been identified. Therefore, we tested whether Par3 can interact with the Neph-Nephrin complex. Par3 contains three PDZ domains, suggesting that Neph proteins could recruit the Par3-Par6-aPKC complex via their carboxyl terminal PDZ-binding domains. Par3 co-precipitated with Neph1 in lysates of HEK 293T cells and mouse glomeruli (Fig. 2, A and B). Deletion of the PDZ-binding motif (the last three amino acids of the carboxy-terminal cytoplasmic tail) of Neph1 prevented binding of Par3 (Fig. 2C). Neph1 specifically interacted with the first PDZ domain of Par3 (Par3256–370) (Fig. 2D). Direct interaction was confirmed by in vitro interaction of the first PDZ domain of Par3 and the carboxyl terminus of Neph1 (Neph1742–789) (Fig. 2E). The PDZ-binding motif of Neph1 is highly conserved among all Neph molecules. Consequently, all Neph proteins interacted with the Par3-Par6-aPKC complex (Fig. 2F). In addition, Nephrin could also be shown to interact with Par 3 in 293T cells (Fig. 2G). This interaction could also be confirmed with endogenous proteins from mouse kidney lysate, indicating the in vivo association of Nephrin and Par3 (Fig. 2H). The interaction of the Neph, Nephrin, and Par complexes at the slit diaphragm suggested that they form a supramolecular protein complex important for glomerular development and maintenance.
FIGURE 2.
Neph-Nephrin proteins bind to the Par3, Par6, and aPKC complex. A, V5.Neph1 and FLAG-tagged full-length CD2AP or Par3 were expressed in HEK 293T cells. After immunoprecipitation with anti-FLAG antibody, the immobilized Neph1 was detected with anti-V5 antibody in the precipitate containing Par3 but not CD2AP. WB, Western blot. B, lysate from mouse glomeruli was immunoprecipitated with a control polyclonal antibody (rabbit anti-hemagglutinin (anti-HA)) or rabbit anti-Par3 antibody, respectively. Immobilized Neph1 was detected with a Neph1-specific antiserum. C, deletion of the last three amino acids of Neph1, the PDZ domain-binding motif, abrogated binding of Par3 to the cytoplasmic Neph1 tail. D, lysate of HEK 293T cells transfected with V5.Neph1 cDNA was subjected to a pull-down assay with recombinant affinity-purified PDZ domains of Par3 fused to GST. Neph1 specifically interacted with the first PDZ domain of Par3 (Par3256–370). The lower panel shows expression levels of GST fusion proteins on a Coomassie Blue-stained gel. E, this interaction was confirmed by in vitro interaction assay with recombinant affinity-purified carboxyl terminus of Neph1 (MBP.Neph1742–789) and PDZ domains of Par3 fused to GST. F, the Par3-Par6-aPKC polarity complex interacts with all Neph family members in HEK 293T cells. G, V5.Nephrin and FLAG-tagged full-length RGS3 or Par3 were expressed in HEK 293T cells. After immunoprecipitation with anti-FLAG antibody, the immobilized Nephrin was detected with anti-V5 antibody in the precipitate containing Par3 but not RGS3. H, mouse total kidney lysate was immunoprecipitated with a control polyclonal antibody (rabbit anti-FLAG) or rabbit anti-Par3 antibody, respectively. Immobilized Nephrin was detected with a Nephrin-specific antiserum.
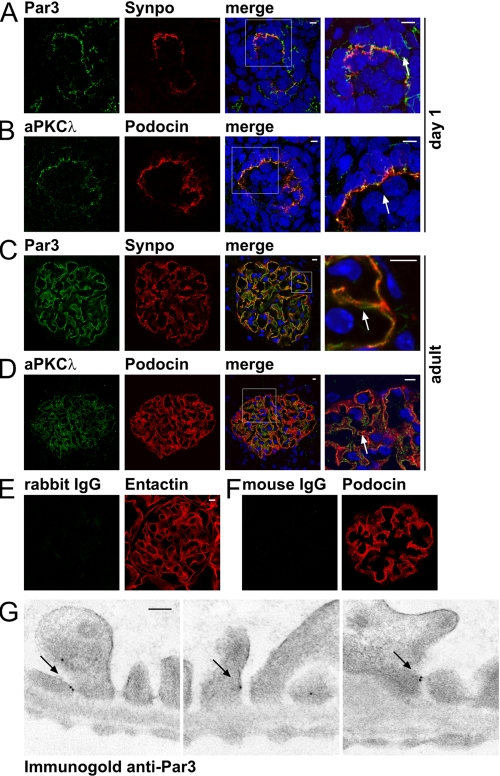
Identification of Par3, Par6, and aPKC as a Novel Slit Diaphragm-associated Protein Complex—Confocal images of immunofluorescence stainings identified a colocalization of the Par3, Par6, and aPKC polarity complex with podocyte foot process markers (Fig. 3, A–D). During glomerular maturation (capillary loop stage), Par3 and aPKC already strongly colocalized with the podocyte foot process markers synaptopodin and podocin, respectively (Fig. 3, A and B). In adult tissue, Par3 and aPKC maintained their foot process expression pattern (Fig. 3, C and D). In immunogold electron microscopic experiments, Par3 mainly localized to the insertion site of the slit diaphragm (Fig. 3G; supplemental Fig. 2), supporting in vivo co-expression with the Neph-Nephrin junctional complex (5, 19).
FIGURE 3.
Identification of Par3, Par6, and aPKC as a novel slit diaphragm-associated protein complex. A, frozen kidney sections of new born Wistar rat (day 1) were stained using antibodies against Par3 and the podocyte foot process marker synaptopodin (Synpo) and subjected to confocal laser microscopy. Par3 signal colocalized to synaptopodin staining (arrow), indicating that Par3 is expressed in podocyte processes during glomerular maturation. All scale bars represent 5 μm. B, dual staining using antibodies against aPKCλ and the podocyte foot process marker podocin showed colocalization of both antigens during glomerular maturation (arrow). C, Par3 and synaptopodin colocalized in podocyte foot processes of the mature glomerulus (arrow). D, aPKCλ and podocin colocalized in podocyte foot processes of the mature glomerulus (arrow). E, secondary antibody (anti-rabbit IgG) only staining served as negative control for anti-Par3 staining. F, secondary antibody (anti-mouse IgG) only staining served as negative control for anti-aPKCλ staining. G, immunogold electron microscopy of rat kidney sections showed specific localization of Par3 at the slit diaphragm in podocyte foot processes (arrows). The scale bar represents 100 nm.
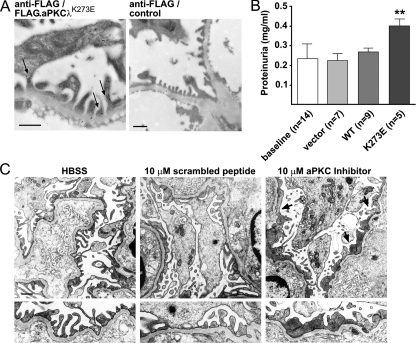
aPKC Is Essential for Foot Process Architecture and Function—We next tested the effect of transient in vivo expression of a dominant-negative aPKC (Flag.aPKCλK273E) construct by intravenous gene delivery in mice (20, 21). Using this approach, we found FLAG.aPKCλK273E in podocyte foot processes partially localizing in close vicinity to the slit diaphragm 16 h after gene delivery by immunogold electron microscopy (Fig. 4A). Mice administered an injection with cDNA encoding for aPKCλK273E displayed rapid-onset proteinuria 16 h after gene injection (Fig. 4B), whereas aPKCλ wild type cDNA or empty vector control had no effect. To further substantiate a critical role of aPKC activity for the integrity of the glomerular filtration barrier, we treated freshly isolated mouse glomeruli with a cell-permeable pseudosubstrate inhibitor of aPKC. Scrambled peptide or Hanks' balanced salt solution served as negative control. Inhibition of aPKC resulted in a phenotype that mimicked Neph1 or Nephrin gene deletion (3, 22) with a loss of the regular foot process architecture and flattened, effaced, and coarse secondary processes (Fig. 4C). Taken together, these data suggest that the Par3-Par6-aPKC complex is fundamental for foot process polarity and architecture.
FIGURE 4.
aPKC is essential for secondary foot process architecture and function. A, a dominant-negative FLAG.aPKCλK273E plasmid was introduced into C57BL/6 mice by hydrodynamic gene delivery. Expression of FLAG.aPKCλK273E in podocyte foot processes was detected using a rabbit polyclonal anti-FLAG antibody (arrows). Anti-FLAG staining of an uninjected mouse kidney section served as negative control for the staining. Scale bars represent 500 nm. B, urine was collected before injection (baseline) and 16 h after injection. Gene delivery of aPKCλK273E (48 μg) resulted in rapid-onset proteinuria. Control mice were injected with empty vector (48 μg) or aPKCλ wild type (WT) cDNA (48 μg) (**, p < 0.01, when compared with all three controls; n, number of mice). C, isolated mouse glomeruli were incubated in vitro with a cell-permeable myristoylated pseudosubstrate inhibitor of atypical PKC or myristoylated scrambled peptide as negative control (10 μm in Hanks' balanced salt solution (HBSS)). Inhibition of aPKC resulted in loss of regular foot process architecture with flattened, effaced, and coarse secondary foot processes (arrows). The scale bar represents 1 μm.
DISCUSSION
To execute their filtration function, podocytes have to develop and maintain an incredible complex and polarized three-dimensional architecture. The signal mechanisms necessary to establish and maintain the orientation and polarization of the foot process network are largely unknown. Mature podocytes lack tight junctions and instead form macromolecular slits to filter large volumes of solutes.
To determine the developmental timing of the expression of the Par3-Par6-aPKC polarity complex and Neph-Nephrin proteins as the core proteins of the slit diaphragm, we assayed mRNA expression in in situ hybridization experiments. Nephrin, Neph, and Par protein expression could be detected as early as embryonic day 14.5, and they all were significantly co-expressed at their highest levels at embryonic day 16.5. In addition, we demonstrated that Par3 colocalized with the known podocyte junctional molecule ZO-1 during all steps of podocyte development. ZO-1 has previously been shown by the Farquhar group (18) to localize to tight junctions of presumptive podocytes during S-shaped body stage and to the slit diaphragm of mature podocytes. These observations indicate that glomerular maturation may depend on polarity signaling provided by the Neph-Nephrin proteins and the Par3-Par6-aPKC complex. We therefore speculated that the Par complex might directly bind to Neph-Nephrin proteins and found that the Neph-Nephrin complex did interact with Par3 and recruited the Par3-Par6-aPKC complex to the slit diaphragm. The interaction between Neph1-Nephrin and Par3 links the specialized Neph-Nephrin-based slit diaphragm to established polarity pathways. Par3 interacts with Par6 that links Rac1/Cdc42 GTPase activity to aPKC by specifically interacting with the active form of Rac1/Cdc42. Par3 also binds directly to aPKC and other binding partners at the cell membrane. Such an interaction provides a platform for the assembly of the polarity complex at the membrane (7). In recent years, several Par3-interacting transmembrane proteins that target the preformed Par3-Par6-aPKC complex to the plasma membrane and cell-cell contacts have been identified including junctional adhesion molecule 1 (JAM1) and the immunoglobulin-like cell-cell adhesion molecules Nectin1 and Nectin3 (23–25). Our findings suggest that glomerular podocytes utilize members of the Neph-Nephrin family to immobilize the Par polarity complex at the plasma membrane. With respect to the glomerular filter, the slit diaphragm is the only cell-cell contact between adjacent podocytes that can provide the information to orient the complex three-dimensional network of podocyte foot processes.
The Par polarity complex has emerged as a central player in regulating cell polarity in different cell types (26–29). In epithelial cells, Par3, Par6, and aPKC localize to tight junctions (30). We now identified the Par3-Par6-aPKC complex as a novel component of the molecular network at the slit diaphragm. Par3 and aPKC colocalized to the developing podocyte structures as well as to terminally developed foot processes, indicating a role for the Par complex for glomerular development and maintenance. The importance of the Par3-Par6-aPKC complex for the integrity of the glomerular filter of the kidney was confirmed by the effect of transient in vivo expression of a dominant-negative aPKC (aPKCλK273E) construct by intravenous gene delivery in mice. The method of in vivo gene delivery has been shown to be effective in targeting glomerular podocytes (16, 20, 21). Mice that were administered an injection of aPKCλK273E displayed mild but significant proteinuria. The relative mild phenotype is most likely explained by the relative low level of expression of the dominant-negative aPKC construct by this method. Therefore, to further evaluate the role of Par3, Par6, and aPKC activity for the maintenance of the foot process architecture, we incubated freshly isolated mouse glomeruli with a specific pseudosubstrate inhibitor of aPKC, which resulted in loss of regular foot process architecture with effaced foot processes in comparison with control conditions, indicating the importance of the Par3-Par6-aPKC complex for the maintenance of the filtration barrier.
The molecular mechanisms underlying Neph-Nephrin-mediated processes are complex and probably involve the integration of numerous signaling pathways that impinge on the cytoskeleton. We have previously shown that the Nephrin-CD2AP protein complex induces phosphatidylinositol 3-kinase signaling at the slit diaphragm of podocytes (12). Interestingly, phosphatidylinositol 3-kinase activity has been reported to also be required for proper localization of the Par complex at the tips of growing axons (29), indicating that Neph and Nephrin proteins supply both phosphatidylinositol 3-kinase activity and transmembrane molecules specifically interacting with the Par complex to ensure efficient polarity signaling. Another hallmark of the activation of the Par3-Par6-aPKC complex is the activation of Cdc42, which in turn binds and activates aPKC scaffolded on Par6 (7). Recently, IQGAP1, which binds actin and directly activates Cdc42, has been identified as a binding partner of the Neph-Nephrin protein complex at the slit diaphragm (31) and is therefore likely to provide GTP loaded Cdc42 to the Par3-Par6-aPKC complex.
In summary, this study identified Par3-Par6-aPKC as a novel slit diaphragm-associated protein complex that interacts with Neph-Nephrin proteins. Our results suggest that the Neph-Nephrin-Par multiprotein complex is essential for the maintenance of the filtration barrier. In general, we propose that Neph-Nephrin proteins via recruitment of the Par3-Par6-aPKC protein complex control subcellular polarization cues that are required for podocyte process development.
Supplementary Material
Acknowledgments
We thank Shigeo Ohno (Yokohama City University School of Medicine, Yokohama, Japan) for providing full-length cDNA of rat Par3 and dominant-negative mouse aPKCλK273E. We thank Peter Mundel (Department of Medicine, Mount Sinai School of Medicine, New York, NY) for providing anti-synaptopodin mouse monoclonal antibody. We thank Dörte Thiel and Charlotte Meyer (Renal Division, University Hospital Freiburg, Freiburg, Germany) for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant DK073495 (to J. R). This work was also supported by a grant from the Kristyna M. Driehaus foundation (to J. R.), by Deutsche Forschungsgemeinschaft Grants BE 2212 (to T. B.) and HU 1016/2-1) (to T. B. H.), and by Deutsche Forschungsgemeinschaft Sonderforschungsbereich 592 (to T. B. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains two supplemental figures.
Footnotes
The abbreviations used are: aPKC, atypical protein kinase C; GST, glutathione S-transferase; MBP, maltose-binding protein; mAb, monoclonal antibody; pAb, polyclonal antibody; HEK, human embryonic kidney; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
References
- 1.Dressler, G. R. (2006) Annu. Rev. Cell Dev. Biol. 22 509–529 [DOI] [PubMed] [Google Scholar]
- 2.Huber, T. B., and Benzing, T. (2005) Curr. Opin. Nephrol. Hypertens. 14 211–216 [DOI] [PubMed] [Google Scholar]
- 3.Donoviel, D. B., Freed, D. D., Vogel, H., Potter, D. G., Hawkins, E., Barrish, J. P., Mathur, B. N., Turner, C. A., Geske, R., Montgomery, C. A., Starbuck, M., Brandt, M., Gupta, A., Ramirez-Solis, R., Zambrowicz, B. P., and Powell, D. R. (2001) Mol. Cell Biol. 21 4829–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerke, P., Huber, T. B., Sellin, L., Benzing, T., and Walz, G. (2003) J. Am. Soc. Nephrol. 14 918–926 [DOI] [PubMed] [Google Scholar]
- 5.Gerke, P., Sellin, L., Kretz, O., Petraschka, D., Zentgraf, H., Benzing, T., and Walz, G. (2005) J. Am. Soc. Nephrol. 16 1693–1702 [DOI] [PubMed] [Google Scholar]
- 6.Kestila, M., Lenkkeri, U., Mannikko, M., Lamerdin, J., McCready, P., Putaala, H., Ruotsalainen, V., Morita, T., Nissinen, M., Herva, R., Kashtan, C. E., Peltonen, L., Holmberg, C., Olsen, A., and Tryggvason, K. (1998) Mol. Cell 1 575–582 [DOI] [PubMed] [Google Scholar]
- 7.Suzuki, A., and Ohno, S. (2006) J. Cell Sci. 119 979–987 [DOI] [PubMed] [Google Scholar]
- 8.Sellin, L., Huber, T. B., Gerke, P., Quack, I., Pavenstadt, H., and Walz, G. (2003) FASEB. J. 17 115–117 [DOI] [PubMed] [Google Scholar]
- 9.Huber, T. B., Kottgen, M., Schilling, B., Walz, G., and Benzing, T. (2001) J. Biol. Chem. 276 41543–41546 [DOI] [PubMed] [Google Scholar]
- 10.Tsiokas, L., Kim, E., Arnould, T., Sukhatme, V. P., and Walz, G. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 6965–6970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schermer, B., Ghenoiu, C., Bartram, M., Muller, R. U., Kotsis, F., Hohne, M., Kuhn, W., Rapka, M., Nitschke, R., Zentgraf, H., Fliegauf, M., Omran, H., Walz, G., and Benzing, T. (2006) J. Cell Biol. 175 547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber, T. B., Hartleben, B., Kim, J., Schmidts, M., Schermer, B., Keil, A., Egger, L., Lecha, R. L., Borner, C., Pavenstadt, H., Shaw, A. S., Walz, G., and Benzing, T. (2003) Mol. Cell Biol. 23 4917–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takemoto, M., Asker, N., Gerhardt, H., Lundkvist, A., Johansson, B. R., Saito, Y., and Betsholtz, C. (2002) Am. J. Pathol. 161 799–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horvat, R., Hovorka, A., Dekan, G., Poczewski, H., and Kerjaschki, D. (1986) J. Cell Biol. 102 484–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Standaert, M. L., Bandyopadhyay, G., Sajan, M. P., Cong, L., Quon, M. J., and Farese, R. V. (1999) J. Biol. Chem. 274 14074–14078 [DOI] [PubMed] [Google Scholar]
- 16.Moller, C. C., Wei, C., Altintas, M. M., Li, J., Greka, A., Ohse, T., Pippin, J. W., Rastaldi, M. P., Wawersik, S., Schiavi, S., Henger, A., Kretzler, M., Shankland, S. J., and Reiser, J. (2007) J. Am. Soc. Nephrol. 18 29–36 [DOI] [PubMed] [Google Scholar]
- 17.Reiser, J., Polu, K. R., Moller, C. C., Kenlan, P., Altintas, M. M., Wei, C., Faul, C., Herbert, S., Villegas, I., Avila-Casado, C., McGee, M., Sugimoto, H., Brown, D., Kalluri, R., Mundel, P., Smith, P. L., Clapham, D. E., and Pollak, M. R. (2005) Nat. Genet. 37 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnabel, E., Anderson, J. M., and Farquhar, M. G. (1990) J. Cell Biol. 111 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, G., Kaw, B., Kurfis, J., Rahmanuddin, S., Kanwar, Y. S., and Chugh, S. S. (2003) J. Clin. Investig. 112 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sever, S., Altintas, M. M., Nankoe, S. R., Moller, C. C., Ko, D., Wei, C., Henderson, J., del Re, E. C., Hsing, L., Erickson, A., Cohen, C. D., Kretzler, M., Kerjaschki, D., Rudensky, A., Nikolic, B., and Reiser, J. (2007) J. Clin. Investig. 117 2095–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei, C., Moller, C. C., Altintas, M. M., Li, J., Schwarz, K., Zacchigna, S., Xie, L., Henger, A., Schmid, H., Rastaldi, M. P., Cowan, P., Kretzler, M., Parrilla, R., Bendayan, M., Gupta, V., Nikolic, B., Kalluri, R., Carmeliet, P., Mundel, P., and Reiser, J. (2008) Nat. Med. 14 55–63 [DOI] [PubMed] [Google Scholar]
- 22.Putaala, H., Soininen, R., Kilpelainen, P., Wartiovaara, J., and Tryggvason, K. (2001) Hum. Mol. Genet. 10 1–8 [DOI] [PubMed] [Google Scholar]
- 23.Ebnet, K., Suzuki, A., Horikoshi, Y., Hirose, T., Meyer Zu Brickwedde, M. K., Ohno, S., and Vestweber, D. (2001) EMBO J. 20 3738–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh, M., Sasaki, H., Furuse, M., Ozaki, H., Kita, T., and Tsukita, S. (2001) J. Cell Biol. 154 491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takekuni, K., Ikeda, W., Fujito, T., Morimoto, K., Takeuchi, M., Monden, M., and Takai, Y. (2003) J. Biol. Chem. 278 5497–5500 [DOI] [PubMed] [Google Scholar]
- 26.Ohno, S. (2001) Curr. Opin. Cell Biol. 13 641–648 [DOI] [PubMed] [Google Scholar]
- 27.Nishimura, T., Kato, K., Yamaguchi, T., Fukata, Y., Ohno, S., and Kaibuchi, K. (2004) Nat. Cell Biol. 6 328–334 [DOI] [PubMed] [Google Scholar]
- 28.Macara, I. G. (2004) Curr. Biol. 14 R160–R162 [PubMed] [Google Scholar]
- 29.Shi, S. H., Jan, L. Y., and Jan, Y. N. (2003) Cell 112 63–75 [DOI] [PubMed] [Google Scholar]
- 30.Hurd, T. W., Gao, L., Roh, M. H., Macara, I. G., and Margolis, B. (2003) Nat. Cell Biol. 5 137–142 [DOI] [PubMed] [Google Scholar]
- 31.Lehtonen, S., Ryan, J. J., Kudlicka, K., Iino, N., Zhou, H., and Farquhar, M. G. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 9814–9819 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.