Abstract
Respiratory syncytial virus (RSV) is a primary cause of severe lower respiratory tract infection in children worldwide. RSV infects airway epithelial cells, where it activates inflammatory genes via the NF-κB pathway. NF-κB is controlled by two pathways, a canonical pathway that releases sequestered RelA complexes from the IκBα inhibitor, and a second, the noncanonical pathway, that releases RelB from the 100-kDa NF-κB2 complex. Recently we found that the retinoic acid-inducible gene I (RIG-I) is a major intracellular RSV sensor upstream of the canonical pathway. In this study, we surprisingly found that RIG-I silencing also inhibited p100 processing to 52-kDa NF-κB2 (“p52”), suggesting that RIG-I was functionally upstream of the noncanonical regulatory kinase complex composed of NIK·IKKα subunits. Co-immunoprecipitation experiments not only demonstrated that NIK associated with RIG-I and its downstream adaptor, mitochondrial antiviral signaling (MAVS), but also showed the association between IKKα and MAVS. To further understand the role of the NIK·IKKα pathway, we compared RSV-induced NF-κB activation using wild type, Ikkγ-/-, Nik-/-, and Ikkα-/--deficient MEF cells. Interestingly, we found that in canonical pathway-defective Ikkγ-/- cells, RSV induced RelA by liberation from p100 complexes. RSV was still able to activate IP10, Rantes, and Groβ gene expression in Ikkγ-/- cells, and this induction was inhibited by small interfering RNA-mediated RelA knockdown but not RelB silencing. These data suggest that part of the RelA activation in response to RSV infection was induced by a “cross-talk” pathway involving the noncanonical NIK·IKKα complex downstream of RIG-I·MAVS. This pathway may be a potential target for RSV treatment.
In the United States, respiratory syncytial virus (RSV)2 causes about 86,000 hospitalizations for severe lower respiratory tract infections (1). Here, pathologic lesions such as bronchiolar epithelial necrosis, bronchiolar occlusion, parenchymal inflammation, and alveolar exudation are found (2). These features, along with the finding that inhibition of mucosal NF-κB signaling in a mouse model of RSV disease blocks mononuclear infiltration and disease manifestations (3), suggest that the inflammatory response mediates a lot of clinical disease manifestations.
Previous studies have shown that RSV induces expression of 16 cytokines and chemokines (4), many of which are NF-κB dependent (5, 6). Five members of the NF-κB family have been reported, including three subunits with transactivating function, RelA, RelB, c-Rel, and two DNA binding subunits, NF-κB1 (p50) and NF-κB2 (p52) (7). The NF-κB molecules are sequestered in the cytoplasm by interacting with a group of inhibitory proteins including IκBα, IκBβ, IκBε, p100, and p105 (8). Currently, it is understood that NF-κB activation can be controlled by two separate pathways, the canonical and noncanonical pathways, activated by distinct stimuli and under control of distinct IκB kinase (IKK) complexes. The canonical NF-κB pathway is induced by the monokines TNF and interleukin-1, stimulating the IKK complex composed of the catalytic kinases, IKK-α and-β and the regulatory subunit, IKKγ (9). Activated IKK, in turn, induces the phosphorylation-coupled degradation of IκBα, liberating sequestered cytoplasmic RelA allowing RelA to translocate into the nucleus and initiates gene transcription (10). The non-canonical NF-κB pathway is induced by lymphotoxin β (LTβ), the TNF superfamily member, LIGHT, or other lymphokines (11–14). This pathway stimulates a complex of NIK and IKKα, resulting in the phosphorylation-coupled proteolysis of 100-kDa NF-κB2 (“p100”) to form the activated 52-kDa NF-κB2 (“p52”)-RelB complex. Although we found that RSV was able to activate NIK kinase activity and p52 formation (15), the mechanism how RSV activates the NIK·IKKα complex is unknown.
Recently, we found that retinoic acid-inducible gene I (RIG-I), a DEXD/H box RNA helicase, was the initial intracellular sensor for RSV infection and upstream of the canonical NF-κB pathway (16). RIG-I is a central regulator of dsRNA-induced signaling for most of the single-stranded RNA viruses (17, 18). RIG-I is upstream of the mitochondrial antiviral signaling (MAVS) protein, also known as interferon-β promoter stimulator 1, caspase recruitment domain (CARD) adaptor inducing IFN-β (Cardif) or virus-induced signaling adaptor (19–23). Activated RIG-I binds to MAVS through a homologous CARD motif; interestingly this interaction is absolutely required for downstream RIG-I signaling and occurs on the outer mitochondrial membrane (19).
In this study, we discover that siRNA-mediated RIG-I silencing inhibits basal and RSV-inducible p52 formation. Further investigation revealed that NIK associates with the RIG-I·MAVS complex via the RIG-I NH2-terminal CARD domain and the COOH terminus of NIK. Surprisingly, we found RSV induced RelA activation in Ikkγ-/- mouse embryonic fibroblasts (MEFs), cells lacking a functional canonical NF-κB pathway. In nondenaturing co-immunoprecipitation experiments, we demonstrated that the NIK·IKKα complex induced RelA release from cytoplasmic p100 complexes. Together, these findings indicate RIG-I controls RelA activation by two distinct downstream signaling modules, one mediated by canonical pathway activation and the second involving a novel cross-talk pathway dependent on complex formation with NIK·IKKα whose activation liberates RelA from p100 sequestration. Targeted disruption of this pathway may have significant effects in modulating the inflammatory response to RSV without affecting the viral replication.
EXPERIMENTAL PROCEDURES
Cell Cultures—Human A549 pulmonary type II epithelial cells (American Type Culture Collection) were grown in F12K medium (Invitrogen) with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 g/ml) at 37 °C in a 5% CO2 incubator. Wild type, Nik-/- (24), Ikkα-/- (25), and Ikkγ-/- (26) MEFs were cultured in Eagle's minimum essential medium (Invitrogen) with 0.1 mm nonessential amino acids, 1.0 mm sodium pyruvate, and 10% fetal bovine serum. HEK293 cells were cultured in Eagle's minimum essential medium (Invitrogen) with 0.1 mm nonessential amino acids, 1.0 mm sodium pyruvate, and 10% fetal bovine serum.
Virus Preparation and Infection—The human RSV A2 strain was grown in Hep2 cells and prepared as described (27). The viral titer of purified RSV pools was varied from 8 to 9 log plaque forming units/ml, determined by a methylcellulose plaque assay. Viral pools were aliquoted, quick-frozen on dry ice-ethanol, and stored at -70 °C until they were used. For viral adsorption, cells were transferred into culture medium containing 2% (v/v) fetal bovine serum and RSV infected at a multiplicity of infection of 1 for the times as described in the text.
Plasmid Construction—Expression vectors encoding full-length and a series of deletion mutants of NIK were produced by PCR and cloned as BamHI/XbaI sequences into the pEGFP-Myc plasmid (Invitrogen). The sequences of the primers are shown in Table 1. A FLAG-tagged full-length NIK expression vector (FLAG_EGPF_NIK) was constructed as a COOH-terminal fusion in pEGFP plasmid. Expression vectors encoding FLAG epitope-tagged RIG-I and its deletion mutants were under the control of a tetracycline response element in a modified pT1S plasmid as described (16). Expression vectors encoding Myc epitope-tagged MAVS and different deletion mutants were produced by PCR and cloned into HindIII/XbaI of the modified pcDNA3_strawberry plasmid; the primers used are listed in Table 2. PEF6_Flag_IKKα was cloned into PEF6_V5 vector (Invitrogen) using upstream primer: 5′-CTTGGTTATGGACTACAAGGACGACGATGACAAGGGGTCGACCATGGAGCGGCCCCCGGGGCTGCGGCCG-3′, and downstream primer, 5′-TCATTCTGTTAACCAACTCCAATCAAGATTC-3′.
TABLE 1.
Primers for deletion mutants of NIK
| Name | Span (aa) | Sense | Antisense |
|---|---|---|---|
| NIK_FL | 8–947 | GAATGAGGATCCTGCCCAGGTGCCCCTGGC | GAGGTCTAGATTAGGGCCTGTTCTCCAGCTGGCC |
| NIK-C1 | 380–947 | AACCGGATCCGTCCTGCTCACTGAGAAACTCAAGC | GAGGTCTAGATTAGGGCCTGTTCTCCAGCTGGCC |
| NIK-C2 | 661–947 | GAGGCGGATCCCTGAAGAGCCCTTGGAGGGGAG | GAGGTCTAGATTAGGGCCTGTTCTCCAGCTGGCC |
| NIK-C3 | 796–947 | GAGGCGGATCCCTCTCGTGCCTCAGCATCGAC | GAGGTCTAGATTAGGGCCTGTTCTCCAGCTGGCC |
| NIK-N1 | 8–660 | GAATGAGGATCCTGCCCAGGTGCCCCTGGC | AACGATCTAGATTAACCTCCCACTTGCTGTAGTG CCCGGTTCACC |
| NIK-N2 | 8–387 | GAATGAGGATCCTGCCCAGGTGCCCCTGGC | AACGATCTAGATTACTTGAGTTTCTCAGTGAGCA GGACACCCCTGTTG |
| NIK-N3 | 8–233 | GAATGAGGATCCTGCCCAGGTGCCCCTGGC | CGATCTAGATTAATCGAGGCAGAGCCGGCCGTAGGCCCTCGCCAAGC |
TABLE 2.
Primers for deletion mutants of MAVS
| Name | Sense | Antisense |
|---|---|---|
| MAVS | AAGTATAAGCTTATGCCGTTGCTGAAGAC | TTCATATCTAGACTAGTGCAGACGCCGCCGG |
| MAVS_dN | ATAACTAAGCTTTACCAGCCTCGGACCTCG | TTCATATCTAGACTAGTGCAGACGCCGCCGG |
| MAVS_dC | AAGTATAAGCTTATGCCGTTTGCTGAAGAC | ATACATTCTAGATGAGGGCCTGTGGCATGGC |
siRNA-mediated Gene Silencing—siRNA against human RIG-I (M-012511-00), mouse Relb (M-040784-01), mouse Rela (M-040776-00), and control siRNA (D-001206-13) were commercially obtained from Dharmacon Research, Inc. (Lafayette, CO). The siRNA targeting RIG-I and control siRNA were transfected at 100 nm into A549 cells using a TransIT-siQuest transfection kit (Mirus Bio Corp., Madison, WI) according to the manufacturer's instructions. The control siRNA and the siRNA targeting RelA and RelB were transfected at 50 nm into MEF cells using reverse transfection according to the manufacturer's protocol. Forty-eight hours after transfection, cells were RSV infected at the indicated times. The silencing efficiency of siRNA was evaluated using reverse transcriptase-PCR (RT-PCR) for RIG-I as well as Western immunoblot for RelB and RelA.
RT-PCR and Quantitative Real-time PCR (qRT-PCR)—Total RNA was extracted using acid guanidium phenol extraction (TriReagent; Sigma). One microgram of RNA was reversibly transcribed using Superscript III (Invitrogen) in a 20-μl reaction mixture. One μl of cDNA product was diluted 1:2, and 2 μl was amplified in a 25-μl reaction mixture containing 12.5 μl of SYBR Green supermix (Bio-Rad) and 0.4 μm each of forward and reverse gene-specific primers (Table 3), aliquoted into 96-well, 0.2-mm thin-wall PCR plates, and covered with optical quality sealing tape. The plates were denatured for 90 s at 95 °C and then subjected to 40 cycles of 15 s at 94 °C, 60 s at 60 °C, and 1 min at 72 °C in iCycler (Bio-Rad). After PCR was performed, PCR products were run on 2% agarose gels to assure a single amplification product. Static analysis of gene expression was described before (16).
TABLE 3.
Probes for real-time PCR
| Gene name | Forward sequence | Reverse sequence |
|---|---|---|
| Groß | CACTCTCAAGGGCGGTCAA | TGGTTCTTCCGTTGAGGGAC |
| IP10 | CGATGACGGGCCAGTGA | CGCAGGGATGATTTCAAGCT |
| Rantes | TCCAATCTTGCAGTCGTGTTTG | TCTGGGTTGGCACACACTT |
| GAPDH | CATGGCCTTCCGTGTTCCTA | GCGGCACGTCAGATCCA |
Electrophoretic Mobility Shift Assay—A total of 35 μg of whole cell extracts were incubated in DNA-binding buffer containing 5% glycerol, 12 mm HEPES, 80 mm NaCl, 5 mm dithiothreitol, 5 mm MgCl2, 0.5 mm EDTA, 1 μg of poly(dA-dT), and 100,000 cpm of 32P-labeled double-stranded oligonucleotide containing NF-κB binding sites in a total volume of 25 μl as described (28). Gels were dried and exposed to Bio-Max film (Kodak) for autoradiography.
Co-immunoprecipitation and Western Immunoblot—Whole cell extracts were prepared using modified radioimmunoprecipitation assay (RIPA) buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 0.25% sodium deoxycholate, 1% IGEPAL CA-630, 1 mm phenylmethylsulfonyl fluoride, 1 mm sodium fluoride, 1 mm Na3VO4, and 1 μg/ml each of aprotinin, leupeptin, and pepstatin). Whole cell extracts were pre-cleared with protein A-Sepharose 4B (Sigma) for 10 min at 4 °C and immunoprecipitation was conducted for 2 h at 4 °C with primary Ab. Immune complexes were then precipitated by adding 50 μl of protein A-Sepharose beads (50% slurry) and incubated for 1 h at 4 °C. Beads were washed three times with cold TB buffer (150 mm NaCl, 5 mm EDTA, 50 mm Tris-HCl, pH 7.4, 0.05% IGEPAL CA-630), and immune complexes were fractionated by 10% SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane by electroblotting. Membranes were blocked in 5% nonfat dry milk in Tris-buffered saline, 0.1% Tween and probed with the primary Ab indicated in the figure legends. Membranes were washed and incubated with IRDye 700-conjugated anti-mouse Ab or IRDye 800-conjugated anti-rabbit Ab (Rockland, Inc.). Finally, the membranes were washed three times with TBS-T and scanned by an Odyssey infrared scanner. Sources of primary Ab were: anti-FLAG M2 mAb (Stratagene), anti-Myc mAb (Santa Cruz), rabbit anti-RelA C20 polyclonal Ab (Santa Cruz), anti-p52 polyclonal Ab (Upstate, Charlottesville, VA), and goat anti-RSV antibody (Biodesign). For the experiments requiring the second primary antibody, the membrane was not stripped to remove the first primary antibody, and the second primary antibody was incubated with membrane directly.
Electroporation—Two million freshly isolated MEFs were suspended in 100 μl of MEF2 nucleofactor solution (Amaxa), and transfected (program A023) with 5 μg of plasmid DNA. After transfection, cells were immediately transferred to Dulbecco's modified Eagle's medium and cultured for at least 48 h before treatment.
RESULTS
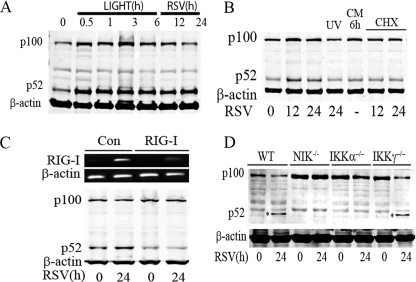
RSV-induced p52 Formation Is RIG-I-, NIK-, and IKKα-dependent—Recently, we have reported that RSV infection activated the noncanonical pathway. To illustrate, we stimulated A549 cells with the LTβ agonist, LIGHT (29), a noncanonical pathway activator, and compared p52 formation with that induced by RSV infection (Fig. 1A). LIGHT rapidly and strongly induced p52 formation within 30 min after treatment, which persisted for more than 6 h. In addition, we noted p100 expression was up-regulated, a consequence of p100 being downstream of the NF-κB pathway (30). Similarly, RSV infection induced p52 formation detectable 12 h after RSV adsorption and persisted for 24 h (Fig. 1A). In contrast to the response to LIGHT, the p100 precursor was transiently induced at the 12-h point and did not persist at 24 h. This is probably due to the transient activation of the noncanonical pathway in RSV infection (15).
FIGURE 1.
RIG-I, NIK, and IKKα mediate RSV-induced p52 formation. A, A549 cells were treated with LIGHT at 0, 0.5, 1, 3, and 6 h, as well as infected with RSV (multiplicity of infection 1) at 12 and 24 h. Whole cell extracts were collected and Western immunoblot was conducted to detect the expression of p100 and its proteolytic product p52. β-Actin staining was used as a loading control. B, A549 cells were treated with RSV, UV-inactivated RSV, or RSV conditioned medium (CM) for the indicated times (in h, bottom). Right panel, cells were RSV infected and treated with cycloheximide (CHX, 25 μg/ml) for the indicated times. The abundance of p100 and p52 were determined by Western immunoblot. β-Actin staining was used as a loading control. C, A549 cells transfected with control (Con) siRNA and RIG-I siRNA for 48 h were then RSV infected for 0 and 24 h. Total RNA was extracted and assayed by RT-PCR to measure the expression of RIG-I (upper panel). β-Actin is a control. Shown is an ethidium bromide-stained agarose gel. Whole cell lysates were prepared from the same cell treatment and p100/p52 by Western immunoblot using NH2-terminal anti-NF-κB2 Ab (lower panel). The blot was probed with β-actin as a loading control. D, WT, Nik-/-, Ikkα-/-, and Ikkγ-/- MEFs were infected by RSV (multiplicity of infection 1) for 0 and 24 h. Whole cell lysates were assayed by Western immunoblot to detect p100 expression and p52 formation (indicated by asterisk). β-Actin was a loading control. p52 was detected only in RSV-infected WT and Ikkγ-/- cells.
Previous work from our laboratory has shown that RSV-induced NF-κB activation is dependent on active viral replication, with either UV-inactivated RSV or RSV-conditioned medium being unable to induce NF-κB binding upon exposure to naïve cells (5). To determine whether RSV replication is necessary for p100 processing, A549 cells were exposed to RSV, UV-inactivated RSV, or conditioned medium from RSV-infected cells. Although replication competent RSV induced p52 formation, neither UV-inactivated RSV nor CM from RSV-infected cells produced detectable p52 formation (Fig. 1B). Interestingly, p52 formation has previously been shown to be mediated via a co-translational mechanism, where newly synthesized p100 was the preferential target for processing to p52 (31). In this study, it was shown that cycloheximide (CHX) inhibits the formation of both p100 and p52. To determine whether RSV induced p52 formation also via a co-translational mechanism, RSV infection was repeated in the presence of CHX. We noted that CHX inhibited the formation of the p100 precursor and inducible p52 formation was inhibited (Fig. 1B, right panel). These data indicated that RSV replication was required for p52 formation, and that newly synthesized p100 was the primary source of p52 processing.
RIG-I is the major cytoplasmic sensor of RSV infection upstream of the canonical pathway, but the pathway mediating noncanonical activation is unknown (16). We next examined whether RIG-I mediated RSV-induced p52 formation. For this purpose, we silenced RIG-I expression using siRNA transfection. In comparison with control siRNA-transfected cells, where RIG-I mRNA was not detectable in uninfected cells and its expression was strongly up-regulated 24 h after RSV infection, accumulation of RIG-I mRNA was significantly reduced in cells transfected with RIG-I-specific siRNA (Fig. 1C, top). Importantly, both the basal and RSV-induced p52 accumulation was significantly decreased in RIG-I-silenced cells (Fig. 1C, bottom).
To confirm the NIK and IKKα dependence in noncanonical pathway activation, p52 formation was determined in wild type (WT), NIK-deficient (Nik-/-), IKKα-deficient (Ikkα-/-), and IKKγ-deficient (Ikkγ-/-) MEFs. Here, p52 formation was observed in WT and Ikkγ-/- MEFs 24 h after RSV infection, but was completely abolished in Nik-/- and Ikkα-/- cells (Fig. 1D). These data suggested that RSV-induced p52 formation was RIG-I dependent, involving both the NIK and IKKα kinases.
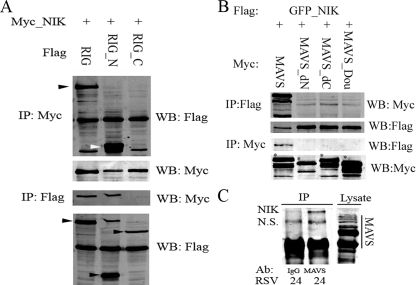
RIG-I and MAVS Associate with NIK and IKKα—To further investigate the novel functional interaction between RIG-I and NIK, and determine the domains involved, co-immunoprecipitation experiments were conducted. First, full-length Myc_NIK and different FLAG_RIG-I constructs were co-transfected into HEK293 cells. The RIG-I constructs included full-length RIG-I, the RIG-I NH2 terminus containing the two CARD domains (RIG_N), and the RIG-I COOH-terminal part containing the helicase domain (RIG_C). NIK was precipitated using the anti-Myc Ab, and RIG-I association was detected by Western immunoblot using anti-FLAG Ab (Fig. 2A, first panel from top). These results indicated that NIK bound to full-length RIG-I and RIG_N, but not RIG_C, suggesting that the two CARD domains of RIG-I are required for RIG-I·NIK complex formation.
FIGURE 2.
RIG-I and MAVS complex with NIK. A, HEK293 cells were transfected with eukaryotic expression vectors encoding Myc-tagged NIK (Myc_ NIK) and different deletion mutants of FLAG epitope-tagged RIG-I, including full-length RIG-I (RIG), the NH2-terminal CARD domains (RIG_N), and the COOH-terminal helicase domain (RIG_C). 36 h after transfection, whole cell lysates were prepared and Myc_NIK immunoprecipitated (IP) using anti-Myc Ab. RIG-I association were detected by Western immunoblot (WB) probing with anti-FLAG Ab. Black arrowhead shows the location of full-length of RIG-I; white arrowhead indicates RIG_N (the first panel from top). To monitor IP recovery, the presence of Myc_NIK in the immunoprecipitate was measured by probing the same membrane with anti-Myc Ab (second panel from top). Conversely, Flag_RIG and its deletion mutants were immunoprecipitated using anti-FLAG Ab, and Western immunoblot was performed using anti-Myc Ab (third panel from top). Note that Myc-NIK is only seen in the IPs from RIG and RIG_N co-transfected cells. To monitor IP recovery, the presence of RIG-I was measured by anti-FLAG Ab; each isoform is indicated by black arrowheads (bottom panel). B, HEK293 cells were transfected with Flag_GFP_NIK and different forms of Myc_MAVS. IP was conducted using anti-FLAG and MAVS detected by anti-Myc Ab in a Western blot (top panel). IP of Flag_GF-P_NIK was confirmed by Western blot (WB) (second panel). Conversely, the MAVS was immunoprecipitated using anti-Myc Ab, and the presence of NIK was determined in Western blot using anti-FLAG Ab (third panel). IP of Myc-MAVS was confirmed by reprobing the membrane with anti-Myc (bottom panel). Note that only full-length MAVS associates with NIK. C, nondenaturing coimmunoprecipitation of RSV-infected cell extracts using anti-IgG (lane 1) or anti-MAVS Ab (lane 2). Immune complexes were washed, and the presence of NIK determined by Western immunoblot. Reactivity of anti-MAVS Ab was determined by probing cellular lysates (lane 3), where full-length MAVS and its characteristic alternative translational initiation forms are seen (compare B, lane 1). A specific band of 150 kDa was observed only in the MAVS immunoprecipitates. NS, nonspecific.
To confirm this finding, the reverse experiment was performed using the anti-FLAG Ab to precipitate RIG-I and NIK association determined using anti-Myc Ab in the Western immunoblot. The same result was observed in this experiment (Fig. 2A, third panel from top).
We next investigated whether the downstream RIG-I adapter, MAVS, complexed with NIK. Co-immunoprecipitation experiments were performed using full-length FLAG- and GFP-tagged NIK and different deletion mutations of MAVS. For this experiment, 4 forms of Myc and Strawberry-tagged MAVS plasmids were used including wild type MAVS (MAVS), MAVS deleted in its NH2-terminal CARD domain (MAVS_dN), MAVS deleted in its COOH-terminal transmembrane domain (MAVS_dC), and MAVS without both its CARD and transmembrane domains (MAVS_Dou). First, full-length NIK was precipitated using anti-FLAG Ab and MAVS association detected by anti-Myc Ab in Western immunoblot (Fig. 2B, top panel). The converse experiment was conducted to precipitate MAVS using anti-Myc Ab, and NIK detected using anti-FLAG in Western blot (Fig. 2B, third panel from top). Both experiments produced the same result, only the full-length MAVS associated with NIK.
To confirm that this observation was not the result of artifactual overexpression, we sought to co-immunoprecipitate the endogenous proteins. For this experiment, RSV-infected cells were immunoprecipitated with IgG, or with anti-MAVS Abs. The presence of NIK was then assayed in the washed immune complexes by Western immunoblot. A specific NIK band was observed only in the MAVS immunoprecipitate (Fig. 2C). These data confirmed that the endogenous proteins formed a complex in cellulo.
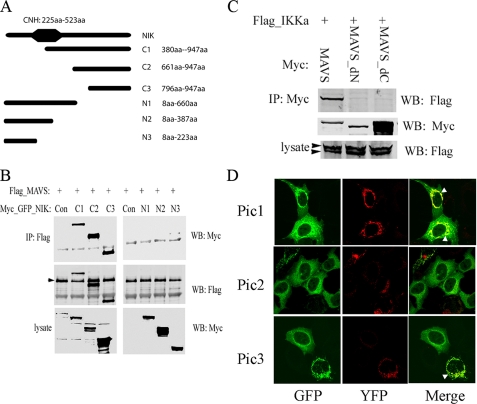
To identify which NIK domains were required for MAVS complex formation, co-immunoprecipitation experiments were conducted using different NIK deletion mutants. A series of expression vectors encoding Myc epitope-tagged NH2 and COOH-terminal domain deletions were tested (Fig. 3A). These constructs were co-transfected with full-length FLAG-tagged MAVS, and MAVS-associated complexes precipitated using anti-FLAG Ab. Our results suggested that the N-terminal deletion mutants of NIK (C1, C2, and C3) associate with MAVS, but the COOH-terminal deletions (N1, N2, and N3) did not (Fig. 3B). This result suggested that the COOH terminus of NIK spanning amino acids 660–947 was required for complexing with MAVS.
FIGURE 3.
NIK and IKKα interact with MAVS. A, schematic diagram of NIK and deletion mutants. The Ser-Thr kinase domain is indicated by a black octagon. NH2-terminal deletion mutants C1, C2, and C3, and COOH-terminal deletion mutants N1, N2, and N3 are indicated. At right is the span (in amino acids) of the various deletions. B, full-length FLAG-MAVS was transfected in the absence (Con) or presence of different deletion forms of Myc-tagged NIK into HEK293 cells. Whole cell extracts were prepared and subjected to co-IP experiment using anti-FLAG Ab. Association with NIK C1, C2, C3, N1, N2, and N3 deletions was detected in Western blot (WB) using anti-Myc Ab (upper panel). The presence of Flag_MAVS in the IP was detected by Western blot. Its location is indicated by the black arrowhead (middle panel). The expression levels of different deletion forms of NIK were measured in Western blot in the lysates used for IP (lower panel). Note that only the COOH-terminal fragments (C1, C2, and C3) bind MAVS. C, IKKα association with full-length MAVS. Expression vectors encoding full-length FLAG-tagged IKKα (Flag_IKKα) and different deletion forms of Myc_MAVS were co-transfected into HEK293 cells. Myc_MAVS was immunoprecipitated (IP) using anti-Myc Ab and the presence of Flag_IKKα was detected using anti-FLAG Ab (upper panel). On the same membrane, IP of Myc_MAVS mutants were measured in Western blot using anti-Myc Ab (middle panel). Flag_IKKα expression was also measured in lysates using anti-IKKα Ab (bottom panel). The upper black arrowhead showed the ectopic Flag_IKKα and the lower black arrowhead indicates the endogenous IKKα. Only full-length MAVS associates with IKKα. D, GFP-tagged IKKα was co-transfected with YFP-tagged MAVS into A549 cells. 36 h after transfection, cells were fixed by 4% paraformaldehyde and analyzed by confocal microscopy (Zeiss LSM510 META ×40). Three different individual views are shown (Pic1, Pic2, and Pic3). Green color shows the GFP-tagged IKKα, red color demonstrates YFP tagged MAVS, and the colocalization of these two molecules is shown by the yellow merge color, highlighted by white arrowheads.
Because of the central role of IKKα in mediating the noncanonical pathway, and its functional requirement for RSV-induced p52 formation (shown in Fig. 1D), the association of IKKα and MAVS was also investigated. Full-length FLAG-tagged IKKα was co-expressed with full-length or deletion mutants of MAVS. The complex was precipitated using anti-Myc Ab and IKKα association detected using anti-FLAG Ab in the Western immunoblot. Our result suggested that only full-length MAVS binds IKKα (Fig. 3C). To confirm this finding, GFP-tagged IKKα and YFP-tagged MAVS were cotransfected into A549 cells. 36 h after transfection, colocalization of IKKα and MAVS were studied by confocal microscopy. Three images are shown in Fig. 3D. The first image shows two cells expressing both GFP_IKKα and YFP_MAVS. The colocalization of these two molecules is demonstrated by the yellow color in the merged figure (at right). In the second image, most cells express only GFP_IKKα; here the IKKα distribution is evenly dispersed through the cytoplasm. In the third image, one cell is expressing GFP_IKKα only and the fluorescence distribution is even throughout the cytoplasm. The other cell expressing both YFP_MAVS and GFP_IKKα, the redistribution of green fluorescence to a mitochondrial pattern is again demonstrated. Together these data indicate that IKKα is recruited to colocalize with MAVS on the mitochondrion.
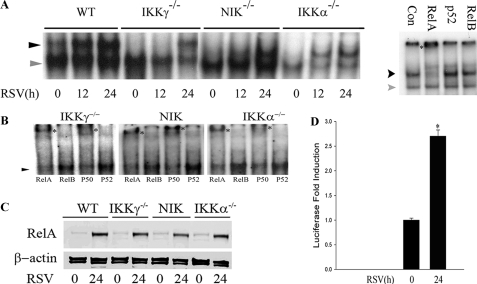
RSV Activates RelA Translocation in Ikkγ-/- MEFs—To further understand the function of the NIK·IKKα complex in RSV-induced signaling, we infected WT, Ikkγ-/-, Nik-/-, and Ikkα-/- MEFs with RSV and assayed nuclear extracts (NE) for canonical NF-κB DNA binding activity using a RelA/NF-κB1-selective probe (28). In WT cells, RSV induces the presence of a RelA-containing DNA binding complex 12 and 24 h after viral adsorption. Surprisingly, in Ikkγ-/- cells, although RelA DNA binding activity was not detected 12 h after RSV infection, RelA binding was strongly induced 24 h after infection (Fig. 4A, second panel from left). Similar complexes were observed in Nik-/- and Ikkα-/- MEFs. To confirm which member of the NF-κB family forms this DNA protein complex, NE from RSV-infected Ikkγ-/- cells were incubated with anti-RelA, anti-RelB, or anti-p52 Abs and a supershift assay was performed. Only the sample incubated with RelA produced a retarded band, leading us to conclude that RelA is contained in the DNA binding complex in RSV-infected Ikkγ-/- cells.
FIGURE 4.
RelA is activated in Ikkγ-/- MEFs. A, WT, Ikkγ-/-, Nik-/-, and Ikkα-/- MEFs were infected with RSV (multiplicity of infection = 1) for the indicated time periods, and whole cell lyses were assayed by electrophoretic mobility shift assay using a DNA probe specifically targeting RelA (left panel). The location of inducible RelA·NF-κB1 complex is shown by the arrowhead; the NF-κB1 homodimer is indicated by a gray arrowhead. On the right, proteins from RSV-infected Ikkγ-/- cells were incubated with anti-RelA, anti-RelB, and anti-p52 Abs for 1 h and supershift experiments were conducted. The asterisk marks the supershifted band in the RelA complex; note the significant attenuation of inducible complex binding in the extracts incubated with anti-RelA Ab. B, supershift experiment using RSV-infected NE from Ikkγ-/-, Nik-/-, and Ikkα-/- MEFs. NE were preincubated in the presence of anti-RelA, anti-RelB, anti-p50, and anti-p52 Abs as indicated prior to nondenaturing fractionation. For each extract, RelA and p50 Abs produced a reduction in the RSV inducible complex and the appearance of a supershifted band indicated by an asterisk. The location of the RSV-inducible NF-κB complex is indicated by a black arrowhead. C, NE from Ikkγ-/-, Nik-/-, and Ikkα-/- MEF were prepared from uninfected (0 h) or RSV-infected cells (24 h). Shown is a Western immunoblot using anti-RelA Ab (upper panel). β-Actin was used as a loading control. Note the RelA nuclear translocation in response to RSV infection in all NE. D, Ikkγ-/- MEFs were transfected with an NF-κB-dependent PRD II reporter gene, subsequently RSV was infected for 24 h. Luciferase reporter assay was measured. Shown is -fold change in normalized luciferase reporter activity. The asterisk indicates a significant increase of reporter gene expression at 24 h of RSV infection compared with 0 h of infection (p < 0.05, t test).
To further examine the complexes in Ikkγ-/-, Nik-/-, and Ikkα-/- MEFs, RSV-infected NEs were subjected to supershift assay for RelA, RelB, NF-κB1/p50, and NF-κB2/p52 complexes (Fig. 4B). With all three cell types, the predominant members of the inducible DNA binding complex were RelA and NF-κB1/p50 as indicated by the ability of these Abs to reduce the RSV inducible complex and form a supershifted band (* in Fig. 4B).
To more convincingly demonstrate RelA translocation, sucrose cushion purified NE from RSV-infected WT, Ikkγ-/-, Nik-/-, and Ikkα-/- MEFs were assayed for changes in RelA abundance by Western immunoblot. Here, RSV infection strongly induced the nuclear RelA signal in all cell types. Importantly, RSV infection increased the nuclear accumulation of RelA in Ikkγ-/- MEFs (Fig. 4C), leading us to conclude that RSV induces RelA translocation independently of the canonical pathway.
To determine whether that RelA was transcriptionally competent, a NF-κB luciferase reporter gene (the IFNβ PRD II domain (32)) was transfected into Ikkγ-/- MEFs and exposed to the absence or presence of RSV. A 3-fold increase in normalized luciferase activity was observed in the RSV-infected cells (Fig. 4D).
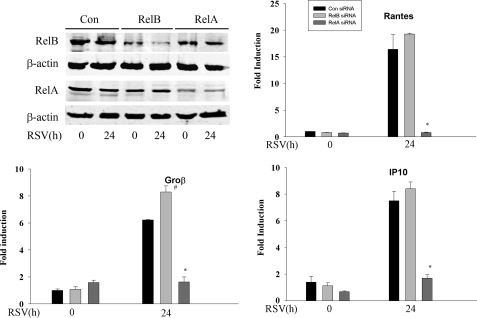
We further tested the requirement of RelA in RSV-induced gene expression in Ikkγ-/- cells. To isolate its function, we separately silenced RelA or RelB by siRNA transfection. The efficiency of siRNA knockdown was evaluated using Western immunoblot to detect RelA and RelB expression. Here, a >50% inhibition of RelA or RelB expression was produced by the cognate siRNA transfection (Fig. 5). In the Control, RelA-, or RelB-silenced IKKγ-/- cells, the RSV-induced expression of IP10, Rantes, and Groβ were measured by qRT-PCR. In cells transfected with control siRNA, Ip10, Rantes, and Groβ increased about 8-, 15-, and 7-fold, respectively. A similar level of induction was observed in the cells transfected with siRNA targeting RelB, whereas the expression of all the three genes decreased significantly after RelA was silenced in IKKγ-/- cells (Fig. 5). These data suggested the existence of an RSV-inducible IKKγ-independent signaling pathway that activates RelA translocation whose presence is required for transcriptional activity of endogenous target genes.
FIGURE 5.
RelA is the major transactivator in Ikkγ-/- MEFs. Top left, Ikkγ-/- MEFs were transfected with control, RelB-specific, or RelA-specific siRNA for 48 h. Cells were either uninfected or RSV infected for 24 h as indicated. Western immunoblot was performed to evaluate silencing efficiency. Top panel, anti-RelB, third panel, anti-RelA. Each blot was reprobed with anti-β-actin as loading controls. Ikkγ-/- MEFs were transfected by control, RelA-, and RelB-specific siRNA. 48 h after transfection, cells were infected with RSV for another 24 h. RNAs were extracted and qRT-PCR was conducted using probes specific for IP10, Rantes, and Groβ. Shown is -fold induction of mRNA. Note that RSV inducible gene expression is significantly inhibited only in the Rela silenced cells. Asterisk indicates a significant decrease of gene expression in RSV-infected cells transfected by Rela siRNA compared with RSV-infected cells transfected by Relb siRNA or Control siRNA (p < 0.001, t test). # indicates a increase of Groβ expression in RSV-infected cells transfected by Relb siRNA compared with RSV-infected cells transfected by Control siRNA (p < 0.05, t test).
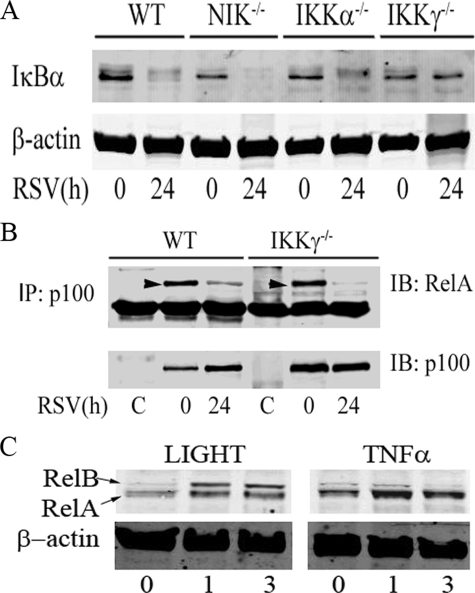
An IKKγ-independent Component of RelA Activation Involves Liberation from p100 Complexes—To determine canonical pathway activation, whole cell extracts were prepared from RSV-infected WT, Nik-/-, Ikkα-/-, and Ikkγ-/- MEFs, and Western immunoblot was performed using anti-IκBα Ab. RSV-induced IκBα proteolysis was observed in WT, Nik-/-, and Ikkα-/- cells; however, no detectable IκBα degradation was observed in Ikkγ-/- MEFs (Fig. 6A). This data not only demonstrated that RSV was able to activate the canonical NF-κB pathway, but also indicated that RSV-induced RelA activation in Ikkγ-/- cells was independent of IκBα proteolysis.
FIGURE 6.
RelA is released from p100 associated complexes. A, IκBα degradation is IKKγ-dependent. WT, Nik-/-, Ikkα-/-, and Ikkγ-/- MEFs were mock or RSV infected for 24 h. IκBα was measured using Western immunoblot (top panel); β-actin was used as an internal control (bottom panel). Note that IκBα is degraded in WT, Nik-/-, and Ikkα-/- MEFs, but not significantly in Ikkγ-/- MEFs. B, RelA is released from p100 complexes. WT and Ikkγ-/- MEFs were infected by RSV for 0 and 24 h. p100 was subjected to nondenaturing co-immunoprecipitation (IP) using anti-p100 Ab; nonimmune rabbit IgG was used as nonspecific binding control. RelA association was detected by Western immunoblot (IB) using anti-RelA Ab (top). Specific RelA staining is indicated by a black arrowhead. To monitor p100 recovery, the membrane was re-probed with anti-NF-κB2 Ab (bottom). Note that RelA associates with p100 in uninfected cells, but this binding is lost in response to RSV infection. C, RelA release upon noncanonical pathway activation. A549 cells were stimulated with LIGHT or TNF for the indicated times (in h, bottom). NE were prepared and RelB or RelA translocation determined by Western immunoblot. β-Actin staining is used as an internal control. Note that LIGHT induces both RelA and RelB translocation, whereas TNF only induces RelA translocation.
Previous studies have reported that p100 forms a heterodimer with RelA and acted as an inhibitor to sequester RelA in the cytoplasm (14, 33). However, the pathways controlling RelA release from p100 are not known. Because our findings showed that p52 formation was IKKγ-independent (Fig. 1D), we investigated whether RSV induced RelA release from p100-associated complexes in WT and Ikkγ-/- MEFs. To quantitate the p100·RelA complex, nondenaturing co-immunoprecipitation was performed. Here, p100 was precipitated in cytoplasmic extracts from RSV-infected WT and Ikkγ-/- MEFs and Western immunoblot was performed using anti-NF-κB2 and anti-RelA Abs. In uninfected WT MEFs, p100 was strongly associated with RelA; 24 h after RSV adsorption, p100-associated RelA was significantly decreased. Importantly, the same phenomenon occurred in the Ikkγ-/- MEFs (Fig. 6B). These data suggested that RSV-induced RelA translocation is, in part, mediated by an IKKγ-independent proteolysis of p100.
These data indicate that RSV-induced activation of the noncanonical pathway results in RelA translocation. To determine whether a similar phenomenon occurred in response to a prototypical activator of the noncanonical pathway, we examined whether LIGHT induced RelA translocation. In this experiment, A549 cells were stimulated with LIGHT or TNFα for 0, 1, or 3 h. NE extracts were then assayed for RelA and RelB translocation by Western immunoblot. We observed that LIGHT induced both RelA and RelB translocation within 1 h of stimulation, whereas TNFα only induced RelA translocation (Fig. 6C).
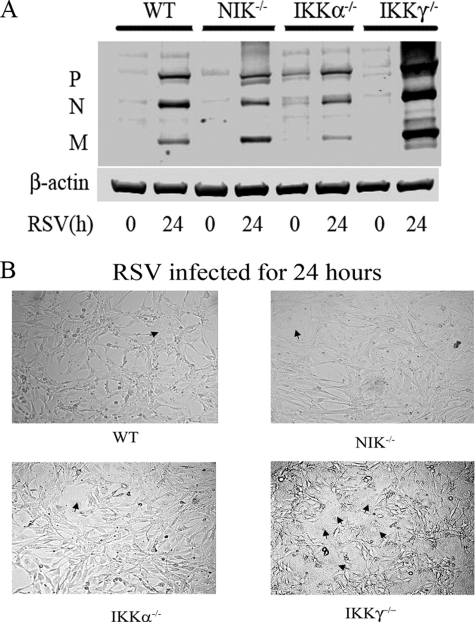
RSV Replication Was Increased in Ikkγ-/-, but Not in Nik-/- and Ikkα-/- MEFs—To determine the role of the canonical and noncanonical/cross-talk pathways in anti-viral response, WT, Nik-/-, Ikkα-/-, and Ikkγ-/- MEFs were RSV infected. Expression of viral proteins was then determined using Western immunoblot using anti-RSV P, N, and M protein Ab. Although the level of viral protein expression was similar in WT, Nik-/-, and Ikkα-/- MEFS, a significant increase in viral protein expression was observed in the Ikkγ-/- MEFs (Fig. 7A). In addition, in contrast to the modest cytopathic effect and cell fusion observed in WT, Nik-/-, and Ikkα-/- MEFs, a significant increase in cytopathic effect and cell fusion was observed in the Ikkγ-/- MEFs (Fig. 7B). These data suggested that IKKγ mediates anti-viral signaling but the noncanonical/cross-talk pathways do not. By extension, this finding suggests that inhibition of the NIK-IKKα signaling pathway may reduce inflammatory chemokine expression, but will not increase viral replication.
FIGURE 7.
RSV replication in Nik-/- and Ikkα-/- MEFs. A, WT, Nik-/-, Ikkα-/-, and Ikkγ-/- MEFs were RSV infected for 24 h. Whole cell extracts were prepared and the expression of RSV P, N, and M proteins were detected by Western blot (upper panel). β-Actin was used as a loading control (lower panel). The expression of RSV proteins are significantly increased in Ikkγ-/- cells. B, cytopathic effect and cell fusion in response to RSV. WT, Nik-/-, Ikkα-/-, and Ikkγ-/- MEFs were cultured on coverslips and RSV infected for 24 h. Shown is a representative field from light microscopy (×10 magnification). Note multinucleated and fused cells in Ikkγ-/- MEFs (black arrowheads).
DISCUSSION
RIG-I is a major initial intracellular sensor that detects RSV infection and activates the downstream NF-κB and IRF3 pathways by complexing with the MAVS adapter (16). In this study, we focused on the mechanistic details for how RSV induces NF-κB pathways. Currently it is thought that NF-κB is regulated by two separate mechanisms, termed the canonical and noncanonical pathways. The canonical NF-κB pathway is IKKγ-dependent and liberates RelA from cytoplasmic IκBα complexes. By contrast, the noncanonical pathway is both NIK- and IKKα-dependent and results in RelB release from cytoplasmic p100 complexes. Although most stimuli activate either the canonical pathway (TNF/IL-1) or the noncanonical pathway (LIGHT/LTβ), RSV efficiently activates both (5, 15, 34). The mechanism by which RIG-I couples to the canonical pathway is largely understood, but the mechanism how RSV activates the noncanonical pathways via the NIK·IKKα kinase complex is unclear. Here we make the surprising findings that RIG-I activates RelA translocation from p100 complexes by the noncanonical NIK·IKKα subunits in Ikkγ-/- cells. The findings that noncanonical pathway activation is dependent on active RSV replication, an event that results in production of vRNAs, and that this pathway is inhibited upon RIG-I knockdown indicate that RIG-I activation is required and upstream of the noncanonical pathway. These data indicate the existence of an additional RSV-inducible “cross-talk” pathway that mediates translocation of the potent transcriptional RelA transactivator.
To our knowledge, this data is the first to demonstrate that NIK associates with the RIG-I-MAVS signaling complex. NIK is a serine-threonine kinase of the mitogen-activated MAP kinase family known to associate with the TNF receptor-associated factors (TRAFs)-2, and 3, the TRAF- and NIK-associated factor (TNAP), and the IKKα kinase (35–39). TRAF association allows NIK to couple with activated receptors in the TNF superfamily. Additionally, previous work shows that NIK serves as a scaffolding molecule, binding IKKα, an event that permits IKKα to complex with p100 to phosphorylate and initiate p52 formation (11). These multiprotein interactions have been partially mapped to the NIK molecule. The NH2 terminus of NIK binds TRAF3 and TNAP (38, 39), whereas the COOH terminus is known to interact with TRAF2 (amino acids 624–947 of NIK) and IKKα (amino acids 735–947 of NIK) (35–37). Our deletion experiments indicate that the NIK COOH terminus (amino acids 660–947) is also required for MAVS interaction. Because of the number of protein interactions, it is highly likely that a macromolecular complex is being formed between NIK, IKKα, RIG-I, and MAVS to produce a functional signaling complex. More detailed work will be required to identify which proteins directly interact.
MAVS is an essential signal transducer for mediating activated RIG-I signaling. MAVS does not have known enzymatic activity and apparently serves as a site for signaling complex assembly. In this regard, the RIG-I·MAVS complex has been shown to be uniquely localized to the surface of mitochondria via a short COOH-terminal transmembrane domain on the MAVS protein (19). Previous work has shown that in the absence of MAVS, cells are unable to activate the canonical NF-κB- or the IRF3 signaling pathways in response to dsRNA or viral infections (19–23). Interestingly, in the absence of mitochondrial targeting, MAVS is unable to associate with RIG-I or mediate its signaling. Our data indicate that mitochondrial localization is required for NIK interaction, because NIK being unable to bind to the COOH terminally deleted MAVS, which aberrantly targets to the cytoplasm (Fig. 2B and Ref. (19)). A similar finding is made for the IKKα-MAVS interaction (Fig. 3C). The explanation for a mitochondrial requirement for signaling is currently unknown. Our data is the first to demonstrate that NIK association with the RIG-I·MAVS complex mediates a third signaling pathway, a cross-talk pathway involved in RelA release from p100 complexes.
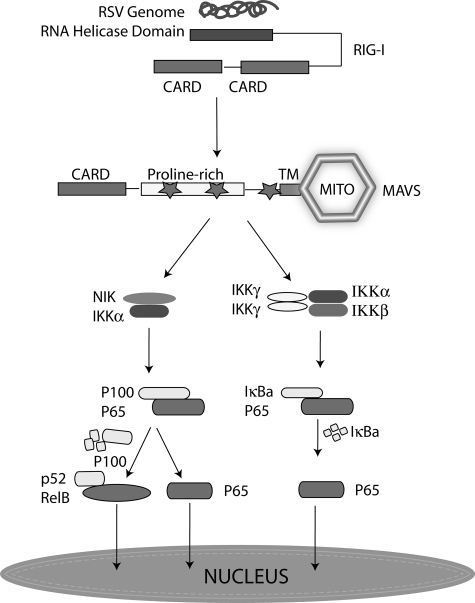
RelA is sequestered in the cytoplasm through association with discrete IκB-like proteins, including IκBα, IκBβ, IκBε, BCL-3, and p100, which serve to function as reservoirs for NF-κB (14, 33, 40–42). The canonical pathway primarily involves stimulus-induced RelA liberation from IκBα-, IκBβ-, and IκBε-sequestered complexes. In this study, we find that RSV induces the degradation of IκBα in WT, Nik-/-, and Ikkα-/- MEFs, but not in Ikkγ-/- MEFs. This result suggests that RSV-induced IκBα proteolysis is IKKγ dependent. However, surprisingly, RSV is still able to activate RelA translocation and transcriptional activation in Ikkγ-/- MEFs. We note that Lin and co-workers (43) also reported a similar 3-fold increase of NF-κB-dependent reporter gene expression in Ikkγ-/- MEFs infected with vesicular stomatitis virus. Although the identity of the transactivator was not investigated, they did note that reporter gene activity was inhibited by dominant negative IκBα, suggesting RelA involvement (43). Our study provides a mechanistic link for the viral inducible activation of RelA via forming a complex with RIG-I-MAVS and the noncanonical NIK-IKKα kinases. Our working model of the viral-induced cross-talk pathway is shown in Fig. 8.
FIGURE 8.
Cross-talk pathway in RSV infection. Shown is a schematic model that shows the signaling cascade downstream of RIG-I that diverges to activate the canonical, noncanonical, and cross-talk pathways. The presence of RSV dsRNA is sensed by the cytosolic RIG-I helicase, which associates with the mitochondrial bound MAVS via heterotypic CARD domain interactions. This complex mediates activation of the typical IKK resulting in RelA release from IκBα-sequestered complexes. In parallel, the NIK·IKKα complex is activated, resulting in activation of the noncanonical and cross-talk pathway by stimulating the 100-kDa NF-κB2 proteolysis. Mito, mitochondria; p65, RelA.
RelA liberated as a result of p100 processing appears to be under a separate stimulus-specific control than the canonical pathway. For example, this cross-talk pathway was recently described as being downstream of LTβ, a TNF superfamily ligand that also induces IκBα independent RelA release from cytoplasmic p100 complexes (44). We demonstrate here that LIGHT also induces RelA translocation. Because LTβ and LIGHT signaling are independent of RIG-I-MAVS, we conclude that several signaling cascades can converge on the cytoplasmic p100·RelA complexes including those activated by TNF superfamily of receptors and cytoplasmic RIG-I-like helicases. Finally, we emphasize that although the existence of this cross-talk pathway was initially indicated by the ability of RSV to induce RelA activation in Ikkγ-/- MEFs, this pathway is activated in wild type MEFs and A549 epithelial cells. In both cell types, cytoplasmic RelA is associated with p100, and that p100 processing is induced (as exemplified by p52 accumulation) in response to RSV infection.
Our study indicates that the activation of the noncanonical NIK·IKKα complexes induces the release of p100-associated RelA as well as the noncanonical RelB·NF-κB2 (p52) complex (Fig. 8). The prototypical NF-κB DNA binding complex is composed of a heterodimer of a transactivating subunit (RelA, RelB) with a DNA binding subunit (NF-κB1, NF-κB2). Currently we understand the canonical RelA·NF-κB1 heterodimer is the predominant DNA binding complex in A549 cells detected in gel binding studies, confirmed by our supershift studies (Fig. 3, A and B). In this complex, RelA is responsible for most of the transcriptional activating properties (Fig. 5 and Refs. 5 and 28). Discussed earlier, RSV-induced RelA is coming from two separate cytoplasmic pools, IκBα/IκBβ and p100. Although p100 processing releases both RelA and RelB in Ikkγ-/- MEFs, we find here by side-by-side siRNA knockdown that RelA, and not RelB, is the major mediator of RSV-induced Rantes, Ip10, and Groβ expression.
Our findings that p52 formation is sensitive to the protein synthesis inhibitor cytoheximide (CHX) is similar to those of Mordmuller et al. (31), who demonstrated that p52 processing is coupled to p100 translation in response to LPS and LT. We interpret these findings to indicate that RNA virus infection also activates RelA release from sequestered p100 via a co-translational mechanism. The exact role of the noncanonical RelB·NF-κB2 (p52) complex in epithelial cells is less clear. Previous work using siRNA-mediated knockdown of p52 indicates that p52 complexes only contribute to a minor degree RSV-inducible cytokine expression (15). In lymphocytes, the RelB·NF-κB2 complex activates a distinct spectrum of genes, including BAFF, SDF, and BLC-3 (11). Because these genes are not expressed by epithelial cells the role of the noncanonical pathway is presently unclear. Presently the only suggestive data is that the RelB·NF-κB2 complex modifies the rate of canonical pathway activation in RSV infection (15).
Previously our group showed that inhibition of the canonical NF-κB pathway using a peptide that disrupted IKKγ-IKKβ interaction (the NEMO binding peptide) significantly down-regulated the inflammatory reaction in RSV-infected mice, despite robust, increased, RSV replication (3). Recently, IKKγ was also shown to mediate viral-induced activation of the IRF3 pathway downstream of the MAVS complex; inhibition of IKKγ was permissive for increased viral replication (43). Together, these findings indicate that inhibiting IKKγ function will affect both the NF-κB-dependent inflammatory response as well as the IRF-3-mediated anti-viral response. In our study of the cross-talk pathway, we have made the intriguing finding that a portion of RSV-induced inflammatory cytokine production is mediated by the noncanonical NIK·IKKα complex. Inhibiting NIK and IKKα signaling does not result in increased RSV replication. These characteristics of the NIK·IKKα-mediated cross-talk pathway makes NIK and IKKα potential targets for therapeutics targeting acute broncholitis caused by RSV infection.
Acknowledgments
We appreciate the gift of PRDII luciferase plasmid from Michael Gale (University of Washington) and AmGen for the gift of NIK-deficient cells.
This work was supported, in whole or in part, by National Institutes of Health Grant AI062885 (to A. R. B.), NHLBI Contract BAA-HL-02-04 (to A. Kurosky), and NIEHS Grant P30 ES06676 (to J. Ward, University of Texas Medical Branch). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: RSV, respiratory syncytial virus; IKK, IκB kinase; MAVS, mitochondrial antiviral signaling; NIK, NFκB inducing kinase; RIG-I, retinoic acid-inducible gene I; WT, wild type; CARD, caspase recruitment domain; TNF, tumor necrosis factor; LTβ, lymphotoxin β; dsRNA, double-stranded RNA; siRNA, small interfering RNA; MEF, mouse embryonic fibroblast; RT, reverse transcriptase; mAb, monoclonal antibody; CHX, cycloheximide; GFP, green fluorescent protein; YFP, yellow fluorescent protein; NE, nuclear extract; TRAF, tumor necrosis factor receptor-associated factor.
References
- 1.Paramore, L. C., Ciuryla, V., Ciesla, G., and Liu, L. (2004) Pharmacoeconomics 22 275-284 [DOI] [PubMed] [Google Scholar]
- 2.Aherne, W., Bird, T., Court, S. D., Gardner, P. S., and McQuillin, J. (1970) J. Clin. Pathol. (Lond.) 23 7-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haeberle, H., Casola, A., Gatalica, Z., Petronella, S., Dieterich, H.-J., Ernst, P. B., Brasier, A. R., and Garofalo, R. P. (2004) J. Virol. 78 2232-2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang, Y., Luxon, B. A., Casola, A., Garofalo, R. P., Jamaluddin, M., and Brasier, A. R. (2001) J. Virol. 75 9044-9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jamaluddin, M., Casola, A., Garofalo, R. P., Han, Y., Elliott, T., Ogra, P. L., and Brasier, A. R. (1998) J. Virol. 72 4849-4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu, T., Castro, S., Brasier, A. R., Jamaluddin, M., Garofalo, R. P., and Casola, A. (2004) J. Biol. Chem. 279 2461-2469 [DOI] [PubMed] [Google Scholar]
- 7.Siebenlist, U., Franzoso, G., and Brown, K. (1994) Annu. Rev. Cell Biol. 10 405-455 [DOI] [PubMed] [Google Scholar]
- 8.Baldwin, A. S., Jr. (1996) Annu. Rev. Immunol. 14 649-683 [DOI] [PubMed] [Google Scholar]
- 9.Zandi, E., and Karin, M. (1999) Mol. Cell. Biol. 19 4547-4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh, S., and Baltimore, D. (1990) Nature 344 678-682 [DOI] [PubMed] [Google Scholar]
- 11.Dejardin, E., Droin, N. M., Delhase, M., Haas, E., Cao, Y., Makris, C., Li, Z. W., Karin, M., Ware, C. F., and Green, D. R. (2002) Immunity 17 525-535 [DOI] [PubMed] [Google Scholar]
- 12.Xiao, G., Harhaj, E. W., and Sun, S. C. (2001) Mol. Cell 7 401-409 [DOI] [PubMed] [Google Scholar]
- 13.Xiao, G., Fong, A., and Sun, S. C. (2004) J. Biol. Chem. 279 30099-30105 [DOI] [PubMed] [Google Scholar]
- 14.Kanno, T., Franzoso, G., and Siebenlist, U. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 12634-12638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choudhary, S., Boldogh, S., Garofalo, R. P., Jamaluddin, M., and Brasier, A. R. (2005) J. Virol. 79 8948-8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, P., Jamaluddin, M., Li, K., Garofalo, R. P., Casola, A., and Brasier, A. R. (2007) J. Virol. 81 1401-1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato, H., Sato, S., Yoneyama, M., Yamamoto, M., Uematsu, S., Matsui, K., Tsujimura, T., Takeda, K., Fujita, T., Takeuchi, O., and Akira, S. (2005) Immunity 23 19-28 [DOI] [PubMed] [Google Scholar]
- 18.Kato, H., Takeuchi, O., Sato, S., Yoneyama, M., Yamamoto, M., Matsui, K., Uematsu, S., Jung, A., Kawai, T., Ishii, K. J., Yamaguchi, O., Otsu, K., Tsujimura, T., Koh, C. S., Reis e Sousa, C., Matsuura, Y., Fujita, T., and Akira, S. (2006) Nature 441 101-105 [DOI] [PubMed] [Google Scholar]
- 19.Seth, R. B., Sun, L., Ea, C. K., and Chen, Z. J. (2005) Cell 122 669-682 [DOI] [PubMed] [Google Scholar]
- 20.Kawai, T., Takahashi, K., Sato, S., Coban, C., Kumar, H., Kato, H., Ishii, K. J., Takeuchi, O., and Akira, S. (2005) Nat. Immunol. 6 981-988 [DOI] [PubMed] [Google Scholar]
- 21.Kumar, H., Kawai, T., Kato, H., Sato, S., Takahashi, K., Coban, C., Yamamoto, M., Uematsu, S., Ishii, K. J., Takeuchi, O., and Akira, S. (2006) J. Exp. Med. 203 1795-1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meylan, E., Curran, J., Hofmann, K., Moradpour, D., Binder, M., Bartenschlager, R., and Tschopp, R. (2005) Nature 437 1167-1172 [DOI] [PubMed] [Google Scholar]
- 23.Opitz, B., Vinzing, M., van Laak, V., Schmeck, B., Heine, G., Gunther, S., Preissner, R., Slevogt, H., N′Guessan, P. D., Eitel, J., Goldmann, T., Flieger, A., Suttorp, N., and Hippenstiel, S. (2006) J. Biol. Chem. 281 36173-36179 [DOI] [PubMed] [Google Scholar]
- 24.Yin, L., Wu, L., Wesche, H., Arthur, C. D., White, J. M., Goeddel, D. V., and Schreiber, R. D. (2001) Science 291 2162-2165 [DOI] [PubMed] [Google Scholar]
- 25.DiDonato, J. A., Hayakawa, M., Rothwarf, D. M., Zandl, E., and Karin, M. (1997) Nature 388 548-554 [DOI] [PubMed] [Google Scholar]
- 26.Yamaoka, S., Courtois, G., Bessia, C., Whiteside, S. T., Weil, R., Agou, F., Kirk, H. E., Kay, R. J., and Israel, A. (1998) Cell 93 1231-1240 [DOI] [PubMed] [Google Scholar]
- 27.Ueba, O. (1978) Acta Med. Okayama 32 265-272 [PubMed] [Google Scholar]
- 28.Tian, B., Zhang, Y., Luxon, B. A., Garofalo, R. P., Casola, A., Sinha, M., and Brasier, A. R. (2002) J. Virol. 76 6800-6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, Y. S., Nedospasov, S. A., and Liu, Z. G. (2005) Mol. Cell. Biol. 25 2130-2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian, B., and Brasier, A. R. (2003) Recent Prog. Horm. Res. 58 95-130 [DOI] [PubMed] [Google Scholar]
- 31.Mordmuller, B., Krappmann, D., Esen, M., Wegener, M., and Scheidereit, C. (2003) EMBO Rep. 4 82-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garoufalis, E., Kwan, I., Lin, R., Mustafa, A., Pepin, N., Roulston, A., Lacoste, J., and Hiscott, J. (1994) J. Virol. 68 4707-4715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun, S. C., Ganchi, P. A., Beraud, C., Ballard, D. W., and Greene, W. C. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 1346-1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garofalo, R., Sabry, M., Jamaluddin, M., Yu, R. K., Casola, A., Ogra, P. L., and Brasier, A. R. (1996) J. Virol. 70 8773-8781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malinin, N. L., Boldin, M. P., Kovalenko, A. V., and Wallach, D. (1997) Nature 385 540-544 [DOI] [PubMed] [Google Scholar]
- 36.Xiao, G., and Sun, S. C. (2000) J. Biol. Chem. 275 21081-21085 [DOI] [PubMed] [Google Scholar]
- 37.Luftig, M. A., Cahir-McFarland, E., Mosialos, G., and Kieff, E. (2001) J. Biol. Chem. 276 14602-14606 [DOI] [PubMed] [Google Scholar]
- 38.Liao, G., Zhang, M., Harhaj, E. W., and Sun, S. C. (2004) J. Biol. Chem. 279 26243-26250 [DOI] [PubMed] [Google Scholar]
- 39.Hu, W. H., Mo, X. M., Walters, W. M., Brambilla, R., and Bethea, J. R. (2004) J. Biol. Chem. 279 35975-35983 [DOI] [PubMed] [Google Scholar]
- 40.Mercurio, F., DiDonato, J. A., Rosette, C., and Karin, M. (1993) Genes Dev. 7 705-718 [DOI] [PubMed] [Google Scholar]
- 41.Naumann, M., Nieters, A., Hatada, E. N., and Scheidereit, C. (1993) Oncogene 8 2275-2281 [PubMed] [Google Scholar]
- 42.Scheinman, R. I., Beg, A. A., and Baldwin, A. S., Jr. (1993) Mol. Cell Biol. 13 6089-6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao, T., Yang, L., Sun, Q., Arguello, M., Ballard, D. W., Hiscott, J., and Lin, R. (2007) Nat. Immunol. 8 592-600 [DOI] [PubMed] [Google Scholar]
- 44.Basak, S., Kim, H., Kearns, J. D., Tergaonkar, V., O'Dea, E., Werner, S. L., Benedict, C. A., Ware, C. F., Ghosh, G., Verma, I. M., and Hoffmann, A. (2007) Cell 128 369-381 [DOI] [PMC free article] [PubMed] [Google Scholar]