Abstract
Glutamate transporters (GluTs) are the primary regulators of extracellular concentration of the neurotransmitter glutamate in the central nervous system. In this study, we have investigated the dynamics and coupling of the substrate and Na+ binding sites, and the mechanism of cotransport of Na+ ions, using molecular dynamics simulations of a membrane-embedded model of GluT in its apo (empty form) and various Na+- and/or substrate-bound states. The results shed light on the mechanism of the extracellular gate and on the sequence of binding of the substrate and Na+ ions to GluT during the transport cycle. The results suggest that the helical hairpin HP2 plays the key role of the extracellular gate for the substrate binding site, and that the opening and closure of the gate is controlled by substrate binding. GluT adopts an open conformation in the absence of the substrate exposing the binding sites of the substrate and Na+ ions to the extracellular solution. Based on the calculated trajectories, we propose that Na1 is the first element to bind GluT, as it is found to be important for the completion of the substrate binding site. The subsequent binding of the substrate, in turn, is shown to result in an almost complete closure of the extracellular gate and the formation of the Na2 binding site. Finally, binding of Na2 locks the extracellular gate and completes the formation of the occluded state of GluT.
INTRODUCTION
Glutamate transporters (GluTs) are membrane transporters in neurons and glial cells that remove the neurotransmitter glutamate from the synapse using preexisting ionic gradients as a source of energy (1,2). Glutamate is a major excitatory neurotransmitter and plays critical roles in fundamental processes in the brain, e.g., learning and memory (3). To maintain recurrent and selective signaling, the neurotransmitter must be rapidly removed after its release (4,5). The GluT family, also known as excitatory amino-acid transporters (EAATs), includes five human EAAT subtypes (EAAT1–EAAT5), two neutral amino-acid transporters, and several prokaryotic homologs (6).
By coupling to cotransport of three Na+ and one H+, and countertransport of one K+, GluTs transport their substrates against the concentration gradient (7–10). Hence, glutamate transport by GluT is an electrogenic process resulting in a net translocation of two positive charges to the intracellular side during each transport cycle. In addition to this coupled flux, Na+ and substrate transport by GluT also activates a thermodynamically uncoupled flux of Cl− from the extracellular side to the cytoplasmic side (11,12). The uncoupled Cl− conductance plays an important physiological role by hyperpolarizing the membrane and dissipating the electrical potential generated during the substrate transport (13,14). There is, however, conflicting evidence with regard to the Cl− permeation pathway through GluT. Some studies suggest that individual monomers might provide the pathway for Cl− (15,16), whereas others indicate that Cl− permeates through a pore formed at the center of the trimeric structure of GluT (17,18). Recent S65V mutation experiments in GluT (14) indicate that Na+-driven substrate transport by GluT induces conformational changes of monomers regulating Cl− permeation and that the uncoupled Cl− permeation pathway is different from the substrate transport pathway. However, the problem of the exact permeation pathway of Cl− is still unresolved.
A large number of experimental studies have investigated the structural and functional properties of GluTs (2,6,16,19–49). Based on accessibility measurements, several groups have shown that substrate binding induces conformational changes in GluT (6,19–32). However, the limited spatial resolution of these studies made it difficult to draw specific conclusions about the nature and magnitude of the conformational changes. Earlier models suspected a rocker-switch mechanism with large protein conformational changes for GluT. However, most recent models (28,29) are based on gating mechanisms and propose that localized, small-scale motions (gates) alternate the accessibility of the substrate-binding site to the cytoplasmic (likely through short channels) and the extracellular solution.
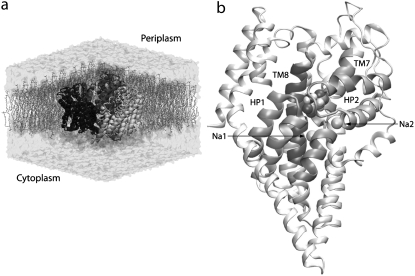
The determination of an x-ray crystal structure of an archaeal homolog of GluT (Gltph) (45) marks the beginning of a new chapter in the study of the structure and mechanism of GluT. Gltph shares ∼37% amino-acid identity with the EAATs, suggesting that it can serve as a structural model for understanding transport in the EAATs. The structure reveals a bowl-shaped trimer, with a solvent-accessible extracellular basin extending halfway across the membrane (Fig. 1 a). Each GluT monomer is composed of eight transmembrane helices (TM1–TM8) and two highly conserved helical hairpins (HP1 and HP2) forming a lumen for binding and permeation of the substrate and Na+ ions (Fig. 1 b). Each substrate binding site is cradled by the two helical hairpins reaching from the opposite sides of the membrane (Fig. 1 b). The functional importance of HP1 and HP2 (45) is supported by biochemical experiments on bacterial (33) and mammalian transporters (21,24,27,34,43). In a recent crystallographic and thermodynamic study of Gltph (31), HP2 was proposed to serve as the extracellular gate that adopts an open conformation exposing the substrate binding site to the extracellular solution in the apo state. The study (31) also showed that substrate binding is coupled to the binding of at least two Na+ ions (Na1 and Na2) and proposed binding sites for them (Fig. 1 b).
FIGURE 1.
Relevant structural features of GluT. (a) The simulation system used in this study is composed of the bowl-shaped trimer of GluT embedded in a lipid bilayer and water. (b) The structure of GluT monomer with bound substrate (shown in VDW) and the two structurally resolved Na+ ions (spheres). Helical hairpins HP1 and HP2, and transmembrane helices TM7 and TM8, which, together, form the substrate binding site, are shown in a darker shade.
Based on the x-ray structure of Gltph (31), Tao al. (35) proposed a structural model for Na+ and glutamate binding to excitatory amino-acid carrier 1 (EAAC1, also named EAAT3) in which one Na+ ion binds first to the empty transporter before the glutamate. They also proposed that conformational changes take place in two glutamate-dependent half-cycles of EAAC1: glutamate-induced closing of an extracellular gate, and the subsequent opening of a cytoplasmic gate that allows glutamate dissociation into the cytoplasm (32).
Although these experiments have provided key information regarding the transport cycle of GluT, the details of the mechanism of extracellular gating, the sequence of binding of the substrate and cotransported Na+ ions, and the role of these ions in transport remain elusive. Here, using molecular dynamics (MD) simulations of a membrane-embedded model of GluT, in its apo and different bound states, we have investigated the dynamics and energetics of the binding sites, and the coupling of the substrate and Na+ ions in GluT. We note that in this study we are not simulating the actual transition between the studied bound states, which might take place on a much longer timescale than those achieved by our simulations. Rather, by placing GluT at various bound states and comparing the dynamics of those states, we try to gather information about the molecular events that might be involved during the transition between these states.
Our simulations show that the helical hairpin HP2 undergoes large-scale motions in the apo state exposing the substrate binding site to the extracellular solution, providing direct evidence for dynamical role of this loop in the gating of the substrate binding site in GluT, which was proposed based on structural and biochemical measurements (31,35). Further analysis of the trajectories shows that substrate binding only results in a partial closure of the extracellular gate, but is necessary for the formation of the Na2 binding site, and that binding of Na2 to its newly formed binding site locks the extracellular gate and completes the formation of the occluded state of the transporter. We also suggest that Na1 binding precedes that of the substrate, as Na1 is found to be necessary for maturation of the substrate binding site. Based on the results, a hypothetical mechanism for the binding of the substrate and the two Na+ binding ions during the extracellular half of the transport cycle is proposed.
MATERIALS AND METHODS
Simulation setup
The simulation system was constructed by embedding the trimeric GluT, taken from the x-ray structure (accession code 2NWX from the Protein Data Bank) (31), in a POPE membrane containing 437 lipids surrounded by water molecules with a system size of 143 × 141 × 107 Å3 (Fig. 1 a). Na+ and Cl− ions with a physiological concentration of 0.1 M were added to the bulk water to neutralize the total charge of the system. The x-ray structure (31,45) shows a bowl-shaped trimer with a concave aqueous basin facing the extracellular side. In the PDB file, each monomer includes the protein residues 12–416, a substrate aspartate, two Na+ ions, three detergent molecules, and one water molecule. The detergent molecules were removed. Missing side chains and hydrogen atoms were added by the psfgen plug-in in VMD (50) employing the CHARMM27 topology files.
Simulation protocol
All simulations used TIP3P model for explicit water (51), and the CHARMM27 force field (52) for proteins, lipids, and ions including the CMAP corrections (53). All simulations were performed under periodic boundary conditions with a time step of 1 fs using NAMD 2.6 (54). First, lipids were melted in a 500-ps NVT (constant volume/constant temperature) simulation at 303 K in which all atoms except the lipid tails were fixed. The system was then equilibrated in an NPT (constant pressure/constant temperature) ensemble with a constant pressure of 1 atm for 2 ns during which the heavy atoms of the protein were constrained by harmonic potentials (k = 2 kcal/mol/Å2) to allow for packing of the lipid molecules around the protein. Next, all the constraints were removed, and the equilibration was continued for additional 2 ns. Finally, the production runs were performed under constant area of the lipid bilayer in the NPnT ensemble with a constant pressure of 1 atm. Constant temperature was maintained by employing Langevin dynamics with a damping coefficient of 0.5 ps−1. The Langevin piston method (55,56) was employed to maintain a constant pressure of 1.0 atm with a piston period of 100 fs. Short-range nonbonded interactions were calculated using a cutoff distance of 12 Å, and long-range electrostatic interactions were calculated using the particle mesh Ewald method (57). All simulations were performed under fully equilibrium conditions, and no membrane potential was used.
The simulation system includes the trimer of GluT, embedding POPE bilayer and water, and is composed of ∼225,000 atoms. As the substrate binds independently to each monomer (16,31), we were able to cover, when desired, up to three different bound states of GluT in each simulation system by including different combinations of the substrate and two Na+ ions in various monomers. A summary of the simulation systems (S1–S3) investigated in this study is given in Table 1. The transport cycle involves three Na+ ions, a K+ ion, and a proton. Two of the Na+ binding sites have been resolved crystallographically (31). The precise location of the third Na+ (Na3) binding site is unknown, even though mutagenesis experiments on EAAC1 (35) suggest that D367 (corresponding to D312 in the Gltph) is involved in coordination of the bound Na+ (Na3) in the substrate-free form. The binding site for H+ is also unknown. K+ is transported during a different transport hemicycle from the transport of the substrate, the three Na+ ions, and H+ (10,29). In this study, we have concentrated only on the coupling of the binding of the substrate and the two Na+ ions (Na1 and Na2) for which binding sites have been suggested in the crystal structure of GluT (31).
TABLE 1.
The summary of GluT simulation systems reported in this study
| System | Monomer | Substrate | Na1 | Na2 |
|---|---|---|---|---|
| S1 | S1A | + | + | + |
| S1B | + | + | − | |
| S1C | + | − | + | |
| S2 | S2A | + | − | − |
| S2B | − | − | − | |
| S2C* | − | − | − | |
| S3 | S3A | − | + | + |
| S3B | − | + | − | |
| S3C | − | − | + |
The simulation time for each system is 20 ns.
In this monomer two serine residues (S277 and S279) on HP1 were mutated to alanines.
For clarity we refer to the protein-only system, i.e., GluT without the substrate and the Na+ ions, as the apo state. Systems S1–S3 were set up to study equilibrium dynamics of GluT in the presence and absence of the substrate and the Na+ ions. In S1, each monomer of GluT is substrate-bound but includes different combinations of Na+ ions: one monomer with two Na+ ions (S1A), one monomer with only Na1 (S1B), and the third with only Na2 (S1C). To investigate the structural dynamics of GluT in the presence and absence of the substrate and the role of the three conserved serine residues (S277, S278, and S279) of HP1, we set up the simulation system S2, in which one monomer is only bound to the substrate (S2A), one monomer is in the apo state of wild-type GluT (S2B), and the third monomer is the apo form of the S277A/S279A double mutant (S2C). To further investigate the role of the bound Na+ ions in the conformational changes of the binding sites, simulation system S3 was set up in which the three monomers are void of the substrate, but bound with Na1 (S3B), Na2 (S3C), or both Na1 and Na2 (S3A), respectively. Note that the initial protein structure in all simulations was the occluded state, i.e., the structure reported in the crystal structure (31).
RESULTS AND DISCUSSION
In this study we investigate the dynamics of the binding sites of the substrate and the two structurally resolved Na+ ions and the coupling between their binding. Based on the results, we propose a mechanism for the extracellular gating of GluT, and a sequence of binding events for the substrate and the two Na+ ions.
Molecular nature of the extracellular gate
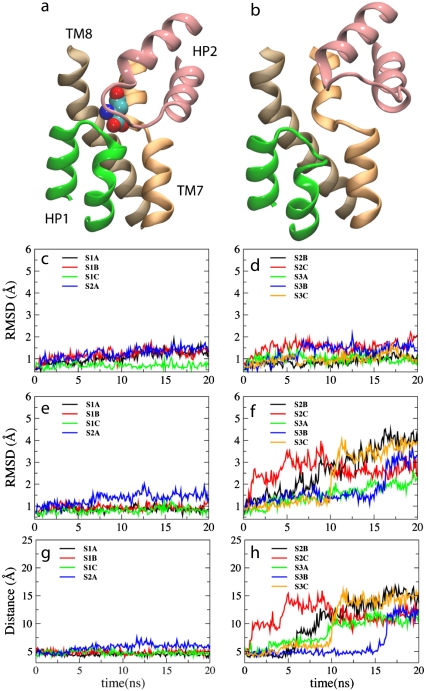
The conformational change of HP2 in the absence of the substrate was one of the most significant events observed in our simulations. Table 2 lists average Cα-RMSD values of each monomer, as well as individual helical hairpins HP1 and HP2, and the two studied Na+ ions during the last 5 ns of the simulations. The RMSD of HP1 in all the simulations is very small. In the presence of the substrate (simulations S1A–S1C and S2A), the RMSD of HP2 is also small. However, in the absence of the substrate (simulations S2B, S2C, and S3A–S3C) HP2 shows large deviations from the original structure. The time evolution of RMSD of HP1 and HP2 (Fig. 2, c–f) clearly shows that HP2 is stable in the presence of the substrate (Fig. 2 e), but experiences large amplitude motions once the substrate is removed, regardless of the number of Na+ ions bound to the transporter (Fig. 2 f). For instance, in the apo state, the RMSD of HP2 exhibits a sharp increase to ∼2.5 Å within the first 6 ns of the equilibration, and approaches 4.0 Å at t = 20 ns. HP1, on the other hand, exhibits only minor fluctuations in all the simulations regardless of the bound state of the substrate and the Na+ ions (Fig. 2, c and d; Table 2). Clearly, HP2 is conformationally more flexible than HP1; however, substrate binding confines its flexible character. In contrast, despite the apparent structural symmetry to HP2, HP1 exhibits a high degree of conformational stability regardless of the substrate. This observation, which can be attributed to the shorter length of the loop region in HP1 (when compared to HP2), suggests that at least during the extracellular half of the transport cycle, HP1 does not play a dynamical role.
TABLE 2.
Conformational fluctuation of each monomer, of the helical hairpins HP1 and HP2, and of the two Na+ ions
| S1A | S1B | S1C | S2A | S2B | S2C | |
|---|---|---|---|---|---|---|
| Monomer | 1.6 (0.1) | 1.5 (0.1) | 1.6 (0.2) | 1.8 (0.1) | 2.6 (0.1) | 1.9 (0.1) |
| HP1 | 0.8 (0.3) | 1.3 (0.1) | 1.2 (0.1) | 1.5 (0.2) | 1.1 (0.1) | 1.8 (0.2) |
| HP2 | 0.9 (0.1) | 0.9 (0.1) | 1.0 (0.1) | 1.5 (0.2) | 4.0 (0.3) | 2.6 (0.3) |
| Na1 | 0.6 (0.2) | 0.8 (0.2) | ||||
| Na2 | 0.8 (0.2) | 0.7 (0.2) | ||||
| S3A | S3B | S3C | ||||
| Monomer | 1.8 (0.1) | 2.3 (0.1) | 2.7 (0.2) | |||
| HP1 | 0.9 (0.1) | 1.5 (0.2) | 1.2 (0.2) | |||
| HP2 | 2.1 (0.2) | 2.8 (0.6) | 3.6 (0.3) | |||
| Na1 | 1.0 (0.2) | 2.4 (0.2) | ||||
| Na2 | 1.3 (0.4) | 2.7 (1.0) |
Average Cα-RMSD values (Å) with the standard deviation are calculated using snapshots taken every 0.1 ns from the last 5 ns of the equilibration simulations, with reference to the initial structure. Large RMSD values are highlighted in bold.
FIGURE 2.
Dynamics of the extracellular gate in GluT. Left and right panels show the results of the simulations performed in the presence and in the absence of the substrate, respectively. See Table 1 for the description of the systems. (a) The last frame of simulation S1A showing the occluded state of GluT. (b) Opening of the substrate binding site upon removal of the substrate is mediated through a large displacement of HP2 and its separation from HP1, as captured, e.g., in one of the last frames of simulation S2B. (c–f) Time evolution of the RMSDs of the helical hairpins HP1 and HP2. Panels c and d show the RMSDs of HP1, and panels e and f show the RMSDs of HP2. (g and h) Time evolution of the distance between HP1 and HP2, measured as the distance between S277(Cα) and G355(Cα), located respectively at the tips of HP1 and HP2. A clear dependence of the dynamics of HP2 on the presence of the substrate is evident.
Comparison of the simulated apo state and the crystal structure of GluT in complex with an inhibitor (TBOA, PDB code: 2NWW, (31)), we notice a high degree of similarity in the opening motion of HP2. The extent of opening, however, appears to be larger in the simulated apo state, probably due to the presence of the inhibitor in the crystal structure which could prevent HP2 from achieving a complete open conformation (Fig. 3 c). The RMSD of HP2 in the simulated apo state (averaged over the last 5 ns of the simulation S2B) with reference to the crystal structures of substrate-bound GluT and TBOA-bound GluT is 4.0 and 3.3 Å, respectively.
FIGURE 3.
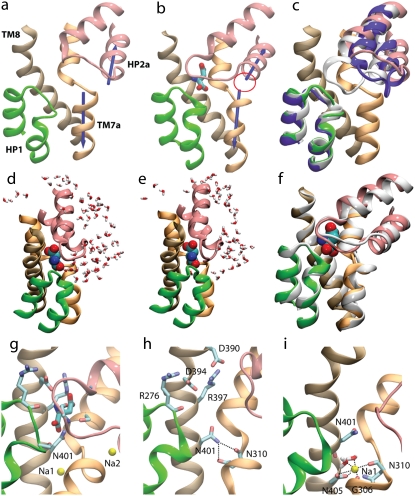
Coupling of binding of the two Na+ ions and the substrate. (a and b) Substrate-induced formation of Na2 binding site. Formation of the Na2 binding site upon substrate binding is shown by comparing the structure of the binding site in the absence (a, simulation S2B) and in the presence (b, simulation S2A) of the substrate. Blue arrows represent the dipole moments of the half-helices TM7a and HP2a. The dipole moments are misaligned in the absence of the substrate, but exhibit a significant alignment upon substrate binding, resulting in the formation of the Na2 binding site (marked with a circle in b). (c) Comparison of the binding site of the fully open state from the simulations (last snapshot of the simulation of the apo form, S2B, shown in the same colors as in other panels) with the crystal structure of the occluded state (in the presence of the substrate and the two Na+ ions, shown in white) and the crystal structure of the open state (bound with TBOA, shown in violet). (d–f) Na2-induced formation of the occluded state. Binding of Na2 to GluT results in complete closure of the binding site to the extracellular solution. In the absence of Na2 (d, simulation S2A), the binding pocket is accessible to water, whereas, upon Na2 binding (e, simulation S1C), the binding site is completely sealed. An overlay of the two states (colored, before Na2 binding; white, after Na2 binding) is shown in panel f highlighting the small change in the conformation of HP2 upon Na2 binding. (g–i) Indirect effect of Na1 on the substrate binding site. (g) The substrate binding site in the occluded state of GluT showing all residues that directly interact with the substrate. (h) In the absence of Na1, the side chain of N401 leaves the substrate binding site due to direct interactions with N310. (i) Na1 binding to GluT results in the upward shift of N401 and the recovery of the substrate binding site.
Each GluT monomer also displays a slight fluctuation in the presence of the substrate, with the average RMSDs of ∼1.6 Å. However, in the absence of the substrate, the RMSD of the monomers shows a large increase, approaching, e.g., 2.7 Å in the simulation S3C (see Table 2). Comparing the RMSDs of the monomers with those of HP2s, the fluctuation of each monomer seems to be primarily due to the motion of HP2. For instance, in S2B and S3C, the monomers exhibit the largest fluctuation among the simulation systems, and this corresponds to the largest RMSD values observed for HP2 among the simulations. The RMSDs of the TMs in S3C, which is one of the monomers displaying a large monomeric RMSD, are 2.3 ± 0.2 (TM1), 2.1 ± 0.2 (TM2), 1.0 ± 0.1 (TM3), 3.3 ± 0.3 (TM4a), 1.6 ± 0.3 (TM4b), 0.8 ± 0.2 (TM4c), 1.6 ± 0.2 (TM5), 1.4 ± 0.2 (TM6), 1.3 ± 0.1 (TM7), and 1.2 ± 0.1 Å (TM8), respectively. Except TM4a, which is located on the surface of the protein and is largely exposed to the solution, TMs of protein display only slight deviations from their original structures. TM7 and TM8, which form the binding pocket for the substrate along with HP1 and HP2, are found to be particularly stable.
Fig. 2, g and h, compares the displacement of HP2 away from HP1 toward the aqueous basin during all the simulations. In the presence of the substrate, the distance between HP1 and HP2 (Fig. 2 g) shows minimal changes also suggesting a stabilizing effect of the substrate on HP2 (Fig. 2 a). In the absence of the substrate, however, a large movement of HP2 is evident from the change of its distance from HP1 (Fig. 2 h). In the apo state (simulation S2B), for instance, HP2 starts to shift away from HP1 at t = 6 ns, and by the end of the simulation it has moved up toward the water basin by ∼12 Å from its original position in the closed conformation (Fig. 2 b). Once the original tight packing of the two hairpins is disrupted, HP2 tends to remain open and highly disordered as indicated by the large amplitude fluctuations in the distance between HP1 and HP2 (Fig. 2 h). In the substrate-free GluT, HP2 undergoes a large opening motion resulting in the complete opening and exposure of the substrate binding site to the extracellular solution (Fig. 2 b). Opening of the substrate binding site is accompanied by its full hydration. Consequently, the substrate can diffuse into its binding site by gravitating toward the charged and polar residues of the binding site.
Comparison of the dynamics of the substrate-bound and the substrate-free states of GluT in these simulations suggests that HP2 plays the key role of the extracellular gate that adopts an open conformation only in the absence of the substrate. The opening and closure of the gate is primarily controlled by substrate binding, and seems to be irrespective of Na+ binding, in agreement with the measurements of temperature-dependent kinetics of substrate transport suggesting that glutamate binding induces closing of an extracellular gate (32). A gating role of HP2 has been also suggested based on the structure of GluT in the presence of an inhibitor (31), as well as the results of biochemical measurements (35). Our simulations provide a dynamical mechanistic view for the extracellular gating of GluT and clearly show the substrate dependence of the process.
Structural inspection of the trajectories shows that fluctuations of HP2 are confined mainly to its loop region (by loop region we refer to the polypeptide stretch of G351–G359 that connects the two helical regions of the hairpin structure), which contains four highly conserved glycines (45), G351, G354, G357, and G359, as well as five hydrophobic residues Y352, A353, V355, P356, and A358. The loop region has a great flexibility, as described above. In the absence of the substrate, the loop region of HP2 has only one attractive interaction with the transporter protein provided by the loop region of HP1 in the closed state. The attraction between the inconsecutive hydrophobic residues seems to be the main drive to disorder the conformation of the flexible loop region of HP2. Therefore, after water molecules disrupt the interaction between the tips of HP1 and HP2, the hydrophobic interactions disorder the conformation of the loop of HP2 resulting in the opening of the substrate binding site.
HP1 includes three highly conserved serine residues (S277–S279) in its loop region (45). These residues collectively are involved in hydrogen bonds between HP1 and HP2. During the simulation S2C (in which two of these serine side chains were mutated to alanines, S277A/S279A), the fluctuation of HP1 becomes significantly larger (Table 2, Fig. 2 e) indicating that these residues are important for structural stabilization of HP1. Moreover, during this simulation, HP2 rapidly dissociates from HP1, suggesting that HP1 plays a role in stabilizing the structure of HP2 in the occluded state, which forms upon substrate binding.
Substrate binding to GluT brings the HP2 and HP1 loops close together, through establishing direct interactions between the charged groups of the substrate and the backbone groups of HP2. Specifically, in the substrate-bound form of GluT, HP2 is stabilized by hydrogen bonds between the β-carboxylate group of the substrate and the amino group of A358, and between the amino group of the substrate and the carbonyl group of A355. It has to be noted that, upon substrate binding, only one half of HP2 (the G359 side) is completely sealed, a state that might be best characterized as a partially occluded state (Fig. 3 b). In this state, although the binding site is largely shielded from the extracellular region, water molecules can still move in and out of the binding pocket since the other half of HP2 (the G351 side) is not fully sealed (Fig. 3, d–f). Therefore, a complete occlusion of the binding site requires additional factors, likely binding of Na+ ion(s), that will bring the extracellular gate to a completely closed state during the transport process (Fig. 3, d–f).
Substrate-induced formation of Na2 binding site
In the crystal structure (31), Na2 is bound to a binding site formed between two half-helical structures (HP2a and TM7a, see Fig. 1 b), and coordinated by main-chain carbonyl oxygens of TM7a and HP2a. The dipole moments of these opposing half-helices point toward the Na2 binding site and stabilize the bound Na+ ion.
In the apo state (simulation S2B), the two half-helices of TM7a and HP2a (and their dipole moments) are found to be completely misaligned (Fig. 3 a), indicating that the Na2 binding site likely does not exist in this state, in good agreement with the experimental results indicating that inhibitor-induced opening of HP2 destroys Na2 binding site (31). Upon substrate binding (e.g., in simulation S2A), the two opposing half-helices align such that their dipole moments converge on the same point resulting in the formation of the Na2 binding site (Fig. 3 b). In other words, Na2 binding can only take place after the binding of the substrate. This is supported by the experimental finding (58,59) that substrate binding enables the binding of one of the Na+ ions. When Na2 is bound to a substrate-bound GluT (S1C), it is coordinated by five carbonyl oxygens of the two half-helices TM7a and HP2a. Interestingly, the binding of Na2 further stabilizes HP2, resulting in a completely occluded state of GluT, in which water molecules can no longer access the binding site from the extracellular side (Fig. 3 d). This is in close agreement with the proposed role of Na2 as a lock on the extracellular gate (31). These results suggest that Na2 is most likely the last cofactor that binds GluT and induces the complete closure of the extracellular gate, and, thus, the formation of the occluded state (Fig. 3 d).
One might imagine that due to direct interactions with both half helices, the binding of Na2 would have a similar effect to the binding of the substrate in terms of inducing a closing motion in HP2. However, our simulations clearly show that the binding of Na2 alone is not sufficient to induce the closure of HP2; in both simulations S3A and S3C, despite the presence of Na2, HP2 was observed to show a very similar opening motion to what was described above. As both these simulation systems lack the substrate, it appears that the main determinant of the closure of the binding site by HP2 is the presence of the substrate. During the equilibration of these two systems, Na2 is coordinated by two carbonyl oxygens of TM7a and HP2a and three water molecules.
Na+-dependence of substrate binding
In the apo state, the binding sites of the substrate and the Na+ ions are completely exposed to the extracellular solution (Fig. 3 h), allowing them to enter their binding sites from the extracellular side of GluT. Based on the crystal structure of GluT (31), the binding site for the substrate is formed by residues R276 on HP1, V355 and G359 on HP2, D394, R397, and N401 on TM8, and T314 on TM7 (Fig. 3 g). According to our simulations, in the substrate-free state, the binding site is filled with water.
One of the key residues that directly interacts with the substrate in the binding site is N401. In the apo state (i.e., the protein-only system), however, the side chain of N401 flips away from the substrate binding site as it engages in an interaction with N310, a side chain which would be involved in Na+ coordination once Na1 is in its binding pocket (Fig. 3 h). Therefore, the substrate binding site is not complete in the apo state. When Na1 binds to the transporter (simulation S3B; Fig. 3 i), however, the side chain of N401 moves back to the substrate binding site, since N310 is now involved in the coordination of Na1. These results suggest that Na1 needs to be bound to GluT before the substrate can bind. Dependence of substrate binding on Na1 appears to provide a basis that explains why the binding of the first Na+ ion increases the affinity of GluT for the substrate (35). However, we note that Na1 is coordinated by D405, a residue whose neutralization has been shown not to affect the binding of the first Na+ ion to GluT (35).
In the substrate-bound state (simulation S1A), Na1 is coordinated by the carboxylate group of D405 and three carbonyl oxygens of residues G306, N310, and N401 (Fig. 3 g). After removal of the substrate (simulation S3B), the binding mode of Na1 is slightly affected; Na1 is now coordinated by the carboxylate group of D405, two carbonyl oxygens of residues G306 and N310, and two water oxygens (Fig. 3 i). One of the water molecules appears to replace the carbonyl oxygen of N401 to coordinate the Na1 ion.
While the substrate is bound to GluT, no major conformational changes of HP1 and HP2 were observed throughout the simulations of S1 and S2A within the timescale of the simulations (Fig. 2, c and e). During the time span of the simulations (20 ns), the substrate stays bound to the binding site with key interactions between its α-amino group and residues R276 and D394, its α-carboxylate group and residues S278, V355, and N401, as well as the β-carboxylate group and residues T314, A358, G359, and R397 preserved. Although the limited timescale of the simulations does not allow us to rule out the possibility of eventual unbinding of the substrate in the absence of the Na+ ions, the observed stability of the substrate in its binding pocket in the absence of Na1 suggests that, after the substrate is bound, Na1 does not play a role in its stabilization in the binding site.
The two Na+ ions are always bound in their binding sites during all the Na+-bound simulations. The calculated fluctuations for the Na+ ions in the presence of the substrate are within 0.6–0.8 Å (see Table 2). However, in the absence of the substrate, the RMSDs of the Na+ ions increase, suggesting that these two Na+ binding sites are less stable and that substrate binding has an effect on the stability of the Na+ binding sites, even though the Na+ ions do not directly interact with the substrate (31). Interestingly, it seems that in the absence of the substrate, the two Na+ ions have a stabilizing effect on each other; when the two Na+ ions are simultaneously bound to the transporter protein, their RMSDs are lower than the corresponding values in systems where only one Na+ is bound.
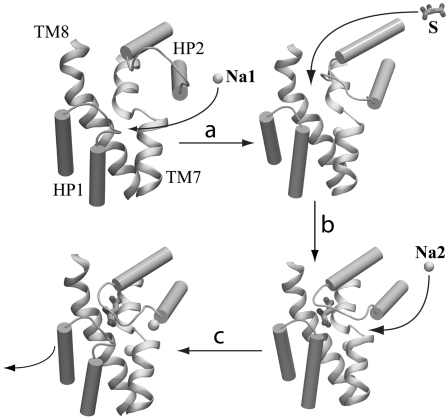
Based on these results, we might propose a sequence for binding of the substrate and the two Na+ ions and a mechanism for the closure of the extracellular gate that consists of the following steps:
(Fig. 4 a) Na1 binds first to extracellular-open GluT, resulting in the completion of the substrate binding site.
(Fig. 4 b) The substrate binds next, and induces a partial closure of the extracellular gate and the formation of the Na2 binding site.
(Fig. 4 c) Na2 then enters its binding site from the extracellular side, and locks the extracellular gate in its fully closed form, thus resulting in an occluded state of the transporter.
FIGURE 4.
Hypothetical mechanism for binding of the substrate and the two structurally resolved Na+ ions during the extracellular half of the transport cycle in GluT. Substrate is shown in licorice. Na+ ions are represented by spheres. (a) Na1 binding to GluT results in the complete formation of the substrate binding site. (b) The substrate binds to GluT and partially closes the extracellular gate HP2, resulting in the formation of the Na2 binding site. (c) Finally, Na2 enters its newly formed binding site from the extracellular side, and locks the extracellular gate in its fully closed form, resulting in the formation of the occluded state. The molecular images are made using the last snapshots of the simulations S2B (apo GluT), S3B (GLuT + Na1), S1B (GluT + Na1 + substrate), and S1A (GluT + Na1 + Na2 + substrate), respectively.
We note, however, that in this study, we have not investigated the roles of the third Na+ ion (Na3) and a proton in the transport cycle. Additional simulations taking into account the roles of these ions will be required to provide a more complete picture of the events involved in the function of GluT.
CONCLUSIONS
The mechanism of transport in the Glutamate transporter (GluT) consists of a large number of steps whose molecular details and sequence are largely unknown. These include the binding, stepwise translocation, and unbinding of both the substrate and the ions that provide the energy required for the active transport, as well as protein conformational changes that are coupled to these events. In this study, starting with an experimental structure of GluT, which is resolved with the substrate and two Na+ ions bound, we employed molecular dynamics simulations of a membrane-bound model of GluT at all possible combinations of the transporter, the bound substrate, and the two Na+ ions. Equilibrium simulations of the apo and the bound states revealed relevant features based on which a detailed mechanism for the extracellular gating and a hypothetical sequence of binding of the substrate and the two Na+ ions are proposed.
The simulations clearly show that one of the helical hairpin structures, HP2, plays the role of the extracellular gate in GluT. HP2 undergoes a large conformational change in the apo GluT resulting in the complete opening of the substrate binding site to the extracellular solution. Conformational flexibility of HP2 is key to this process. Interestingly, despite its apparent symmetry to HP2, HP1 is found to be very rigid and does not appear to directly participate in the extracellular half of the transport cycle. Further investigation is required to determine whether HP1 might play a role during the later stages of the transport cycle. The binding of the substrate results in a partial closure of HP2 and the formation of the Na2 binding site through alignment of two half-helical elements in GluT. The binding of Na2, in turn, results in the complete closure of the substrate binding site and the formation of the occluded state. Furthermore, it appears that the substrate binding site residues are in their most optimal configuration only when Na1 is bound to its binding site, as its binding affects the conformation of a key side chain that directly interacts with the substrate. Based on these results, a sequence for the binding of the substrate and the two Na+ ions during the extracellular half of the transport cycle in GluT is proposed.
Further investigations of the mechanism of transport in GluT would study the role of the other ions that are known to be involved in the transport cycle (7–10). In this study, we have only studied the coupling of binding of the substrate and the two Na+ ions for which binding sites are suggested in the crystal structure of GluT (31). Cotransport of a proton and a third Na+ ion, countertransport of a K+ ion, and uncoupled flux of Cl− ions are involved in the function of GluT. However, the binding site and pathway for conduction of these ions are largely unknown. Theoretical studies, which have successfully predicted putative binding sites for cations and provided useful information regarding the selectivity of these sites (60,61), combined with mutagenesis studies investigating the role of different amino acids in ion binding, will shed more light on the binding site of these ions, thus, allowing one to study the coupling of these ions and the substrate in a more complete model of GluT.
Acknowledgments
The simulations have been performed using the TERAGRID resources (grant No. MCA06N060), the Big Red cluster at Indiana University, and the National Center for Supercomputer Applications Abe cluster, as well as on the Computational Science and Engineering Turing cluster of the University of Illinois at Urbana-Champaign.
Editor: Edward H. Egelman.
References
- 1.Chen, N. H., M. E. Reith, and M. W. Quick. 2004. Synaptic uptake and beyond: the sodium- and chloride-dependent neurotransmitter transporter family SLC6. Pflugers Arch. 447:519–531. [DOI] [PubMed] [Google Scholar]
- 2.Grewer, C., and T. Rauen. 2005. Electrogenic glutamate transporters in the CNS: molecular mechanism, pre-steady-state kinetics, and their impact on synaptic signaling. J. Membr. Biol. 203:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dingledine, R., K. Borges, D. Bowie, and S. F. Traynelis. 1999. The glutamate receptor ion channels. Pharmacol. Rev. 51:7–61. [PubMed] [Google Scholar]
- 4.Clements, J. D. 1996. Transmitter timecourse in the synaptic cleft: its role in central synaptic function. Trends Neurosci. 19:163–171. [DOI] [PubMed] [Google Scholar]
- 5.Bergles, D. E., J. S. Diamond, and C. E. Jahr. 1999. Clearance of glutamate inside the synapse and beyond. Curr. Opin. Neurobiol. 9:293–298. [DOI] [PubMed] [Google Scholar]
- 6.Slotboom, D. J., W. N. Konings, and J. S. Lolkema. 1999. Structural features of the glutamate transporter family. Microbiol. Mol. Biol. Rev. 63:293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zerangue, N., and M. P. Kavanaugh. 1996. Flux coupling in a neuronal glutamate transporter. Nature. 383:634–637. [DOI] [PubMed] [Google Scholar]
- 8.Levy, L. M., O. Warr, and D. Attwell. 1998. Stoichiometry of the glial glutamate transporter GLT-1 expressed inducibly in a Chinese hamster ovary cell line selected for low endogenous Na+-dependent glutamate uptake. J. Neurosci. 18:9620–9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pines, G., and B. I. Kanner. 1990. Counterflow of l-glutamate in plasma membrane vesicles and reconstituted preparations from rat brain. Biochemistry. 29:11209–11214. [DOI] [PubMed] [Google Scholar]
- 10.Kavanaugh, M. P., A. Bendahan, N. Zerangue, Y. Zhang, and B. I. Kanner. 1997. Mutation of an amino acid residue influencing potassium coupling in the glutamate transporter GLT-1 induces obligate exchange. J. Biol. Chem. 272:1703–1707. [DOI] [PubMed] [Google Scholar]
- 11.Billups, B., D. Rossi, and D. Attwell. 1996. Anion conductance behavior of the glutamate uptake carrier in salamander retinal glial cells. J. Neurosci. 16:6722–6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eliasof, S., and C. E. Jahr. 1996. Retinal glial cell glutamate transporter is coupled to an anionic conductance. Proc. Natl. Acad. Sci. USA. 93:4153–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veruki, M. L., S. H. Morkve, and E. Hartveit. 2006. Activation of a presynaptic glutamate transporter regulates synaptic transmission through electrical signaling. Nat. Neurosci. 9:1388–1396. [DOI] [PubMed] [Google Scholar]
- 14.Ryan, R. M., and J. A. Mindell. 2007. The uncoupled chloride conductance of a bacterial glutamate transporter homolog. Nat. Struct. Mol. Biol. 14:365–371. [DOI] [PubMed] [Google Scholar]
- 15.Ryan, R. M., A. D. Mitrovic, and R. J. Vandenberg. 2004. The chloride permeation pathway of a glutamate transporter and its proximity to the glutamate translocation pathway. J. Biol. Chem. 279:20742–20751. [DOI] [PubMed] [Google Scholar]
- 16.Grewer, C., P. Balani, C. Weidenfeller, T. Bartusel, Z. Tao, and T. Tauen. 2005. Individual subunits of the glutamate transporter EAAC1 homotrimer function independently of each other. Biochemistry. 44:11913–11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eskandari, S., M. Kreman, and M. P. Kavanaugh. 2000. Pentameric assembly of a neuronal glutamate transporter. Proc. Natl. Acad. Sci. USA. 97:8641–8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres-Salazar, D., and C. Fahlke. 2006. Intersubunit interactions in EAAT4 glutamate transporters. J. Neurosci. 26:7513–7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grunewald, M., and B. I. Kanner. 1995. Conformational changes monitored on the glutamate transporter GLT-1 indicated the existence of two neurotransmitter-bound state. J. Biol. Chem. 270:17017–17024. [DOI] [PubMed] [Google Scholar]
- 20.Grunewald, M., A. Bendahan, and B. I. Kanner. 1998. Biotinylation of single cysteine mutants of the glutamate transporter GLT-1 from rat brain reveals its unusual topology. Neuron. 21:623–632. [DOI] [PubMed] [Google Scholar]
- 21.Grunewald, M., and B. I. Kanner. 2000. The accessibility of a novel reentrant loop of the glutamate transporter GLT-1 is restricted by is substrate. J. Biol. Chem. 275:9684–9689. [DOI] [PubMed] [Google Scholar]
- 22.Lolkema, D. J. S. J. S., and W. N. Konings. 1996. Membrane topology of the C-terminal half of the neuronal, glial, and bacterial glutamate transporter family. J. Biol. Chem. 271:31317–31321. [DOI] [PubMed] [Google Scholar]
- 23.Slotboom, D. J., W. N. Konings, and J. S. Lolkema. 2001. Cysteine-scanning mutagenesis reveals a highly amphipathic, pore-lining membrane-spanning helix in the glutamate transporter GltT. J. Biol. Chem. 276:10775–10781. [DOI] [PubMed] [Google Scholar]
- 24.Seal, R. P., and S. G. Amara. 1998. A reentrant loop domain in the glutamate carrier EAAT1 participates in substrate binding and translocation. Neuron. 21:1487–1498. [DOI] [PubMed] [Google Scholar]
- 25.Seal, R. P., B. H. Leighton, and S. G. Amara. 2000. A model for the topology of excitatory amino acid transporters determined by the extracellular accessibility of substituted cysteines. Neuron. 25:695–706. [DOI] [PubMed] [Google Scholar]
- 26.Zarbiv, R., M. Grunewald, M. P. Kavanaugh, and B. I. Kanner. 1998. Cysteine scanning of the surroundings of an alkali-ion binding site of the glutamate transporter GLT-1 reveals a conformationally sensitive residue. J. Biol. Chem. 273:14231–14237. [DOI] [PubMed] [Google Scholar]
- 27.Borre, L., M. P. Kavanaugh, and B. I. Kanner. 2002. Dynamic equilibrium between coupled and uncoupled modes of a neuronal glutamate transporter. J. Biol. Chem. 277:13501–13507. [DOI] [PubMed] [Google Scholar]
- 28.Koch, H. P., and P. H. Larsson. 2005. Small-scale molecular motions accomplish glutamate uptake in human glutamate transporters. J. Neurosci. 25:1730–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsson, P. H., A. V. Tzingounis, H. P. Koch, and M. P. Kavanaugh. 2004. Fluorometric measurements of conformational changes in glutamate transporters. Proc. Natl. Acad. Sci. USA. 101:3951–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leighton, B. H., R. P. Seal, S. D. Watts, M. O. Skyba, and S. G. Amara. 2006. Structural rearrangements at the translocation pore of the human glutamate transporter EAAT1. J. Biol. Chem. 281:29788–29796. [DOI] [PubMed] [Google Scholar]
- 31.Boudker, O., R. M. Ryan, D. Yernool, K. Shimamoto, and E. Gouaux. 2007. Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature. 445:387–393. [DOI] [PubMed] [Google Scholar]
- 32.Mim, C., Z. Tao, and C. Grewer. 2007. Two conformational changes are associated with glutamate translocation by the glutamate transporter EAACl. Biochemistry. 46:9007–9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slotboom, D. J., I. Sobczak, W. N. Konings, and J. S. Lolkema. 1999. A conserved serine-rich stretch in the glutamate transporter family forms a substrate-sensitive reentrant loop. Proc. Natl. Acad. Sci. USA. 96:14282–14287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leighton, B. H., R. P. Seal, K. Shimamoto, and S. G. Amara. 2002. A hydrophobic domain in glutamate transporters forms an extracellular helix associated with the permeation pathway for substrates. J. Biol. Chem. 277:29847–29855. [DOI] [PubMed] [Google Scholar]
- 35.Tao, Z., Z. Zhang, and C. Grewer. 2006. Neutralization of the aspartic acid residue Asp-367, but not Asp-454, inhibits binding of Na+ to the glutamate-free form and cycling of the glutamate transporter EAAC1. J. Biol. Chem. 281:10263–10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, Z., Z. Tao, A. Gameiro, S. Barcelona, S. Braams, and T. Rauen. 2007. Transport direction determines the kinetics of substrate transport by the glutamate transporter EAAC1. Proc. Natl. Acad. Sci. USA. 104:18025–18030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haugeto, O., K. Ullensvang, L. M. Levy, F. A. Chaudhry, T. Honore, M. Mielsen, K. P. Lehre, and N. C. Danbolt. 1996. Brain glutamate transporter proteins form homomultimers. J. Biol. Chem. 271:27715–27722. [DOI] [PubMed] [Google Scholar]
- 38.Kanai, Y., M. Stelzner, S. Nussberger, S. Khawaja, S. C. Hebert, C. P. Smith, and M. A. Hediger. 1994. The neuronal and epithelial human high affinity glutamate transporter. Insights into structure and mechanism of transport. J. Biol. Chem. 269:20599–20606. [PubMed] [Google Scholar]
- 39.Kanner, B. I., and L. Borre. 2002. The dual-function glutamate transporters: structure and molecular characterization of the substrate-binding sites. Biochim. Biophys. Acta. 1555:92–95. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, Y., and B. Kanner. 1999. Two serine residues of the glutamate transporter GLT-1 are crucial for coupling the fluxes of sodium and the neurotransmitter. Proc. Natl. Acad. Sci. USA. 96:1710–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bendahan, A., A. Armon, N. Madani, M. P. Kavanaugh, and B. I. Kanner. 2000. Arginine 447 plays a pivotal role in substrate interactions in a neuronal glutamate transporter. J. Biol. Chem. 275:37436–37442. [DOI] [PubMed] [Google Scholar]
- 42.Reference deleted in proof.
- 43.Ryan, R. M., and R. J. Vandenberg. 2002. Distinct conformational states mediated the transport and anion channel properties of the glutamate transporter EAAT-1. J. Biol. Chem. 277:13494–13500. [DOI] [PubMed] [Google Scholar]
- 44.Amara, S. G., and A. C. K. Fontana. 2002. Excitatory amino acid transporters: keeping up with glutamate. Neurochem. Int. 41:313–318. [DOI] [PubMed] [Google Scholar]
- 45.Yernool, D., O. Boudker, Y. Jin, and E. Gouaux. 2004. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 431:811–818. [DOI] [PubMed] [Google Scholar]
- 46.Watzke, N., T. Rauen, E. Bamberg, and C. Grewer. 2000. On the mechanism of proton transport by the neuronal excitatory amino acid carrier 1. J. Gen. Physiol. 11:609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grewer, C., N. Watzke, M. Wiessner, and T. Rauen. 2000. Glutamate translocation of the neuronal glutamate transporter EAAC1 occurs within milliseconds. Proc. Natl. Acad. Sci. USA. 97:9706–9711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grewer, C., N. Watzke, T. Rauen, and A. Bicho. 2003. Is the glutamate residues Glu-373 the proton acceptor of the excitatory amino acid carrier 1? J. Biol. Chem. 278:2585–2592. [DOI] [PubMed] [Google Scholar]
- 49.Rosental, N., A. Bendahan, and B. I. Kanner. 2006. Multiple consequences of mutation two conserved β-bridge forming residues in the translocation cycle of a neuronal glutamate transporter. J. Biol. Chem. 281:27905–27915. [DOI] [PubMed] [Google Scholar]
- 50.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD—visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- 51.Jorgensen, W. L., J. Chandrasekhar, J. D. Madura, R. W. Impey, and M. L. Klein. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935. [Google Scholar]
- 52.MacKerell, A., Jr., D. Bashford, M. Bellott, R. L. Dunbrack, Jr., J. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, I. W. E. Reiher, B. Roux, M. Schlenkrich, J. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiorkiewicz-Kuczera, D. Yin, and M. Karplus. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102:3586–3616. [DOI] [PubMed] [Google Scholar]
- 53.MacKerrell, A. D., Jr., M. Feig, and C. L. Brooks III. 2004. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25:1400–1415. [DOI] [PubMed] [Google Scholar]
- 54.Phillips, J. C., R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kale, and K. Schulten. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martyna, G. J., D. J. Tobias, and M. L. Klein. 1994. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101:4177–4189. [Google Scholar]
- 56.Feller, S. E., Y. H. Zhang, R. W. Pastor, and B. R. Brooks. 1995. Constant pressure molecular dynamics simulation—the Langevin piston method. J. Chem. Phys. 103:4613–4621. [Google Scholar]
- 57.Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald. An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- 58.Watzke, N., E. Bamberg, and C. Grewer. 2001. Early intermediates in the transport cycle of the neuronal excitatory amino acid carrier EAAC1. J. Gen. Physiol. 117:547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bergles, D. E., A. V. Tzingounis, and C. E. Jahr. 2002. Comparison of coupled and uncoupled currents during glutamate uptake by GLTi-1 transporters. J. Neurosci. 22:10153–10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noskov, S. Y., and B. Roux. 2007. Importance of hydration and dynamics on the selectivity of the KcsA and NaK channels. J. Gen. Physiol. 129:135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noskov, S. Y., and B. Roux. 2008. Control of ion selectivity in LeuT: two Na+ binding sites with two different mechanisms. J. Mol. Biol. 377:804–818. [DOI] [PMC free article] [PubMed] [Google Scholar]