Abstract
FMRFamide-Like-Peptides (FLPs) are the largest neuropeptide family in animals, particularly invertebrates. FLPs are characterized by a C-to-N-terminal gradient of decreasing amino acid conservation. NPR-1 is a GPCR (G Protein Coupled Receptor) which has been shown to be a strong regulator of foraging behavior and aggregation responses in Caenorhabditis elegans. Recently, ligands for NPR-1 were identified as neuropeptides coded by the precursor genes flp-18 and flp-21 in C. elegans. The flp-18 gene encodes eight FLPs including DFDGAMPGVLRF-NH2 and EMPGVLRF-NH2. These peptides exhibit considerably different activities on NPR-1, the longer showing lower potency. We have used NMR and biological activity to investigate structural features that may explain these activity differences. Our data demonstrate that long range electrostatic interactions exist between N-terminal aspartates and the C-terminal penultimate arginine as well as N-terminal H-bonding interactions that form transient loops within DFDGAMPGVLRF-NH2. We hypothesize that these loops, along with peptide charge, diminish this peptide's activity on NPR-1 relative to that of EMPGVLRF-NH2. These results provide some insight into the large amino acid diversity in FLPs.
FMRFamide was first discovered in 1977 by Price and Greenberg as a cardioexcitatory peptide from the clam Macrocallista nimbosa (1). FMRFamide-Like-Peptides (FLPs) are the largest family of neuropeptides found in invertebrates (2-5), but mammalian (even human) FLPs have also been identified (6-9). These peptides are characterized by a C- to N-terminal gradient of decreasing sequence conservation, and most end in RF-NH2. This is true when FLP peptide sequences as a whole, from within taxa, within species, or even on specific precursor proteins are compared (2, 4, 5, 10). The peptides, like most neuropeptides and hormones, are synthesized as part of larger precursor proteins and processed in the secretory pathway (11). Peptides on a particular precursor have conserved regions in the mature peptides that are often associated with receptor binding and make up a subfamily (4). For many neuropeptides, including FLPs, this region is the C-terminus. However, other peptide families have different patterns of conservation; for example, in insect orcokinins the N-terminus is the conserved region (12) and in insulin, the cystine framework and other central residues portions are conserved (13). Two examples of FLP precursor proteins, flp-18 from the nematode Caenorhabditis elegans (4) and afp-1 from the nematode Ascaris suum (14) are shown in Table 1.
Table 1.
Sequence comparison of flp-18 and afp-1 precursor proteinsa

|
The first nematode FLP, AF1, was isolated from Ascaris suum (15), and most subsequent early nematode FLP work was done on this species (14, 16-21). FLPs are highly expressed in nematodes, and thus are likely important chemical components of their anatomically simple nervous systems (3, 5, 18). In C. elegans, 28 different genes encoding well over 60 possible FLPs have been identified using bioinformatic approaches (4, 5, 22) and 28 of the putative processed peptides have been detected biochemically (23-27).
FLPs are involved in a wide range of biological processes that have been reviewed previously (10, 28-31). Some of the more prominent functional studies have focused on their role in cardioexcitation (1), muscle contraction (19), modulation of the action of morphine (32), egg laying (33) and feeding behavior (31) in nematodes. Also, disruption of the flp-1 gene in C. elegans resulted in a number of phenotypes (34). Though much work has been done to elucidate the activities of FLPs, the definitive biological functions of the vast majority of FLPs are still unknown.
Two types of receptors for FLPs have been identified: GPCRs (G-Protein Coupled Receptors) (35-40) and a sodium channel gated by FMRF-NH2 (41-43). Other human/mammalian ion channel receptors have been identified whose activities are modulated by FLPs, including the Acid Sensing Ion Channels (ASICs) and Epithelial Na+ Ion Channels (ENaCs) (8, 44). NPR-1, a GPCR that modulates feeding behavior in C. elegans, is activated by two subfamilies of FLPs in C. elegans, including the FLP-18 peptides and FLP-21 peptide (35). All of the FLP-18 peptides occur on the same precursor and are presumably processed and released simultaneously (4). The most active of these peptides is EMPGVLRF-NH2; by comparison DFDGAMPGVLRF-NH2 is significantly less active (35). These observations motivated us to further investigate the structural properties of these peptides relative to their biological activities.
In previous studies, we have suggested that N-terminal hydrogen bonding can influence FLP activity (45). Structural interactions in small peptides such as FLPs are generally invisible to techniques such as X-ray crystallography, because small peptides are dynamic in solution. Also, some NMR parameters such as NOE correlations for distance measurements are of limited value on small peptides due to their dynamic properties in solution (46). However, these structural properties can modulate their activities (45). Although a high resolution X-ray crystallographic structure for a FLP-18 peptide bound to NPR-1 would be extremely useful, it would not provide any information on the unbound state of the peptides. As shown below, we have identified significant structural differences between different unbound FLP-18 peptides, and this work seeks to illuminate the relationship between free ligand conformations and their activities on the NPR-1 receptor.
In the present study we have used NMR for pH titrations, temperature titrations, and chemical shift analyses to identify transient long-range interactions within flp-18 peptides and designed analogues. The sequence and activity diversity among these peptides have motivated us to examine the structural properties of two extreme cases. We have focused on structural features of EMPGVLRF-NH2 and DFDGAMPGVLRF-NH2 that may influence the activity of each peptide on NPR-1. The material presented in this paper examines the hypothesis that local structure in the variable N-terminal regions of flp-18 peptides can modulate their binding to NPR-1.
Experimental Procedures
Peptide Synthesis
Peptides listed in Table 2 were synthesized using standard Fmoc solid phase methods, purified by HPLC, and verified by MALDI-TOF mass spectrometry at the University of Florida Interdisciplinary Center for Biotechnology Research (UF ICBR) protein core facility.
Table 2.
Peptides used in this study
| Name | Sequence a | % Response (n)b |
|---|---|---|
| Peptide 1c | EMPGVLRF-NH2 | 100 |
| Peptide 2d |
|
29.1±5.7 (16) |
| Peptide 3 |
|
0 (3) |
| Peptide 4 |
|
19.0±2.6 (8) |
| Peptide 5 | SGSGAMPGVLRF-NH2 | 118.7±11.0 (4) |
| Peptide 6 | AAAAAMPGVLRF-NH2 | 62.4±3.2 (10) |
| Peptide 7e |
|
49.3±16.5 (4) |
| Peptide 8 |
|
0 (3) |
| Peptide 9f |
|
45.0±10.5 (6) |
| Peptide 10g |
|
104.8±2.8 (8) |
| Peptide 11 | PGVLRF-NH2 | 43.0±5.5 (14) |
| Peptide 12 | PGVLRFPGVLRF-NH2 | 198.1±33.3 (10) |
Naturally occurring sequences are underlined. The conserved PGVLRF-NH2 sequence is in bold. N-terminal “extension” sequences of native long flp-18 peptides are bold and: Red for C. elegans based sequences and Blue for A. suum based sequences.
Peptides (10-6 M) were applied in 2 m pulses to Xenopus oocytes expressing NPR-1 215V. Results expressed as a % of response to 10-6 M Peptide 1 (EMPGVLRF-NH2) +/- SEM
Most active native C. elegans FLP-18 peptide
Longest and least active native FLP-18 peptide
Longest Ascaris AFP-1 peptide
Chimera of long FLP-18 + long AFP-1
Chimera of long AFP-1 + long FLP-1
Peptide Sample Preparation
Lyophilized peptides were weighed and dissolved to ∼1 mM in 95% H2O and 5% D2O, and the pH was adjusted to 5.5. These were aliquoted and frozen at -20 °C until being used for biological assays or NMR experiments. Small aliquots of each sample were submitted for amino acid analysis at the UF ICBR protein core to determine more accurate concentrations. For NMR spectroscopy, the pH-stable chemical shift standard DSS (2,2-dimethyl-2-silapentane-5-sulfonic acid) was added to 600 μL aliquots to a final concentration of 0.17 mM.
Biological Activity Assays - Expression in Xenopus laevis oocytes
Sense cRNA was prepared in vitro using the mCAPTM RNA Capping Kit (Stratagene, La Jolla, CA) from plasmid DNA containing full-length npr-1 215V cDNA cloned in pcDNA3 (Invitrogen Ltd., Paisley UK). RNA transcripts were synthesized using T7 RNA Polymerase (Stratagene, La Jolla, CA) after linearizing the plasmid with Apa I (Promega UK, Southampton, UK) and blunting the 3′ overhangs with T4 DNA Polymerase (Amersham Pharmacia Biotech, Little Chalfont, Bucks, UK). T7 RNA transcripts synthesized in vitro with the mCAPTM RNA Capping Kit are initiated with the 5′ 7MeGpppG 5′ cap analog. Sense cRNA was prepared in a similar manner from the GIRK1 and GIRK2 clones in pBS-MXT (47) (kindly donated by Drs. S.K. Silverman and H.A. Lester, California Institute of Technology, Pasedena, USA) after linearizing the plasmid with Sal I (Promega).
All experiments using Xenopus laevis were carried out under a Home Office (UK) Project License. Stage V and VI oocytes from virgin female adult X. laevis were prepared using standard procedures (35, 48-50). Oocytes were then injected with 50 ng of npr-1 receptor sense cRNA, either alone, or together with 0.5 ng each of GIRK 1 and GIRK 2 sense cRNA and incubated at 19oC for 2 - 5 days. Uninjected oocytes were used as controls. Electrophysiological recordings were made from oocytes using a two-microelectrode voltage-clamp technique, (35, 48-50).
NMR Spectroscopy
NMR data were collected at 600 MHz on a Bruker Advance (DRX)-600 console in a 14.1 Tesla magnet equipped with a 5 mm TXI Z-Gradient CryoProbe. Unless otherwise stated, all NMR experiments were collected at 288 K, and spectra were collected with a 6600 Hz spectral width and were referenced by setting the methyl proton resonance peak from DSS protons to 0.0 ppm. The 1H carrier frequency was centered on water which was reduced using a WATERGATE sequence (51) or presaturation. Two-dimensional TOCSY (52) experiments were collected using a DIPSI-2 mixing sequence with a 60 ms mixing time. Two-dimensional ROESY (53) experiments were collected using a 2.27 kHz cw field spinlock applied for 250 ms.
Processing of 1D NMR spectra and creation of stack plots of pH and temperature titrations was done using Bruker XWINNMR and XWINPLOT ver 4.0 software. Two-dimensional NMR datasets were processed with NMRPipe (54) using standard methods: removing residual water by deconvolution, multiplying the data with a squared cosine function, zero-filling, Fourier transformation, and phase correction. Data were analyzed and assigned with NMRView (55) using standard 1H-based methods (56).
One-dimensional pH titration experiments were performed for all peptides in Table 2 that contain aspartate and/or glutamate residue(s), as well as PGVLRF-NH2 and SGSGAMPGVLRF-NH2 as controls. One-dimensional NMR spectra were collected at increments of about 0.2 pH units from 5.5 to 1.9 by adding 1-3 μL of 0.01-0.1 M HCl for each pH value. pKa values and effective populations (c in Equation 1) of pH dependant resonance peaks were calculated using Origin 7.0 software and a modified version of the Henderson-Hasselbach equation below as previously described (57):
| Equation 1 |
where δ(pH) is the experimental chemical shift, δb is the chemical shift at the least acidic condition, δa is the chemical shift at the more acidic condition, pKai is the negative common log of the acid/base equilibrium constant for the ith titration event, and ci is the contribution of the ith titration event to the total pH dependence of chemical shift.
One-dimensional NMR temperature titrations were collected on a standard TXI probe at 5 Kelvin (K) increments from 278 to 328 K, then ramped back to 278 K to check for sample integrity. The temperature for each experiment was calibrated using methanol (for 278.15 – 298.15 K) and ethylene glycol (for 308.15 – 328.15 K) and the corrected temperatures were used for the determination of all temperature coefficients (TC).
Results
Peptide Design Rationale and Physiological Responses
Three major considerations have motivated this study. First, we have been intrigued for some time by the amino acid diversity in FLPs (2, 10, 14-16, 45, 58, 59). In particular, as described above, FLPs display patterns of decreasing amino acid conservation from the C- to the N-termini, and the comparison of the C. elegans flp-18 (4) and A. suum afp-1 (14) genes suggests that the longer peptides produced by these genes are unique (see Table 1). Second, the activity at NPR-1 of the long FLP-18 peptide, DFDGAMPGVLRF-NH2, is significantly lower than the shorter EMPGVLRF-NH2 (35). Finally, in previous work on FLPs from mollusks, we found that different amino acid substitutions significantly changed the conformations of the peptides (45, 58) and that these conformational differences are correlated with their differences in activity (60).
In designing the peptides for this study, we considered several possibilities to explain the difference in NPR-1 activity between two native FLP-18 peptides, EMPGVLRF-NH2 and DFDGAMPGVLRF-NH2: First, the N-terminus could have intrinsic activity or act as a competitive inhibitor; second, a glutamic acid might be required in a position corresponding to the first residue of the more active EMPGVLRF-NH2; third, the added bulk due to the extra amino acids could prevent the active portion of the peptide from efficiently binding to NPR-1; fourth, the N-terminal extension of DFDGAMPGVLRF-NH2 could be involved in structural interactions that cause it to be less potent on NPR-1 than EMPGVLRF-NH2. To address these possibilities, two native FLP-18 peptides, and a range of substituted and derived analogues (Table 2), were tested for their ability to activate the NPR-1 215V receptor expressed as described in the Experimental Methods. In the following, we use the term “activity” to indicate the magnitude of the potassium current evoked by a 10-6 M pulse of peptide as a percentage of the response of the same oocyte to a 10-6 M control pulse of peptide 1 (EMPGVLRF-NH2). We will refer to peptides by their peptide number shown in Table 2.
It was observed that that the long native FLP-18 peptide 2 was much less effective than the shorter FLP-18 peptide 1 at activating the receptor (Table 2), confirming previous observations (35). To test if the N-terminus of peptide 2 could have intrinsic activity or act as a competitive inhibitor, we designed and analyzed the effect of peptide 3. This peptide had no intrinsic activity on the receptor (Table 2) and did not block the effects of 1 μM pulses of peptide 1 (n = 3) (data not shown). To test the possibility that a glutamic acid might be required in a position corresponding to the first residue peptide 1, we examined peptide 4, where a glutamic acid residue is substituted for the alanine at position 5 in peptide 2. However, this substitution did not improve, and in fact weakened, the effectiveness of the long peptide. It also seemed possible that the added bulk of peptide 2 due to the extra amino acids could be preventing access to the NPR-1 binding site. Thus, we analyzed peptides 5 and 6. Peptide 5 was designed to eliminate any potential structure in the N-terminus based on commonly used flexible linker sequences in fusion protein constructs (pET fusion constructs, Novagen Inc.). The N-terminus of peptide 6 was designed to induce a nascent helical structure in the same region (61, 62). It can be seen that peptide 5 completely restored activity compared to peptide 2, whilst peptide 6 only partially restored activity in comparison with the short native peptide.
As shown in Table 1 the longest native peptide from afp-1 in A. suum (19), peptide 7, is two amino acids longer than the corresponding longest peptide from the C. elegans flp-18 gene (4), peptide 2. Thus, we also synthesized and tested peptide 7, as well as its N-terminus, peptide 8. Peptide 7 was slightly more effective than the peptide 2. However, the short N-terminal peptide sequence was again inactive (Table 2) and did not block the effects of 1 μM pulses of peptide 1 (n = 3) (data not shown). In addition, we also made chimeras of the long C. elegans and A. suum sequences, peptides 9 and 10. Peptide 9, in which two extra amino acids (SM) are introduced into the center of peptide 4 to give it the same number of amino acids as peptide 7, showed similar activity to that of the long native Ascaris peptide itself, GFGDEMSMPGVLRF-NH2. However, peptide 10 showed similar activity to that of peptide 1.
As shown later, our results indicate that the conserved C-terminal PGVLRF-NH2 is largely unstructured in solution. Thus, we tested peptide 11 and also peptide 12, in which the conserved sequence was duplicated. The activity of the peptide 11 was less than that of peptide 1 and similar to that of peptides 7 and 9. When compared to peptides 1 and 5, the reduced activity of peptide 11 could indicate that methionine preceding proline is important for activity. However, the C-terminal duplicated peptide with no methionine was approximately twice as active as peptide 1.
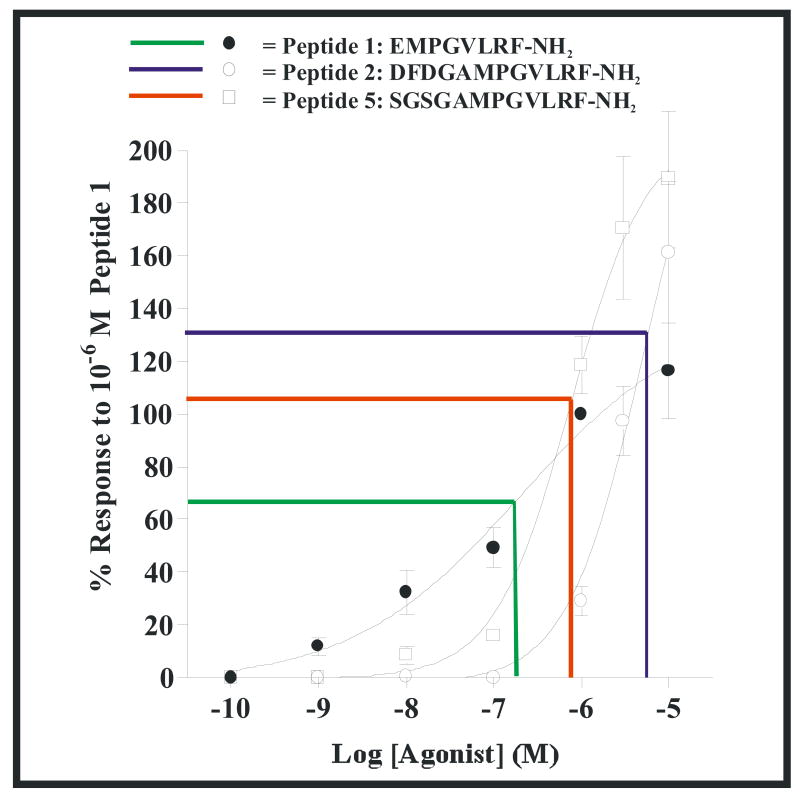
To further investigate the effects of changing the structure of the N-terminal sequence of the C. elegans long FLP-18 peptide, we determined full dose response curves for the peptides 1, 2, and 5 (Fig. 1).
Figure 1.
Dose response curves of select FLP-18 peptides. Peptides were applied to Xenopus oocytes expressing NPR-1 215V, and the responses from inward rectifying K+ channels were recorded and normalized to the response of peptide 1 (EMPGVLRF-NH2) at 10-6 M. Filled circles are peptide 1 (EMPGVLRF-NH2) (EC50 = 10-6.80 M), open squares are peptide 5 (SGSGAMPGVLRF-NH2) (EC50 = 10-6.12 M), and open circles are peptide 2 (DFDGAMPGVLRF-NH2) (EC50 = 10-5.28 M). Three measurements at each peptide concentration were obtained and results are shown +/- SEM.
From Figure 1, both peptides 2 and 5 are less potent compared to peptide 1. This suggests that elimination of structure at the N-terminus of peptide 2 can increase its potency on the receptor. Also, peptides 2 and 5 are more efficacious at higher concentrations than peptide 1, suggesting that longer peptides might be more efficacious on NPR-1 than shorter peptides.
NMR Chemical Shifts Reveal Regions of flp-18 Peptides with Significant Structure
Chemical shifts are extremely sensitive to molecular and electronic environments and thus provide unique atomic probes in molecules. Specifically, in peptides and proteins, chemical shifts of many nuclei along the polypeptide backbone have been shown to be dependent on secondary structure (46, 63-68). Thus, the first step in NMR analysis is the assignment of resonance peaks in spectra to atoms in the molecule. For all peptides in Table 2, nearly complete NMR resonance assignments were made using standard two-dimensional 1H-based methods (56) (Supplementary Material S2).
Short, linear peptides are often very dynamic, lack a 3D hydrophobic core, and interconvert rapidly between many different conformations. Despite this inherent flexibility, numerous studies have demonstrated that regions of short peptides can be highly populated in specific types of secondary structure (69-72). A fundamental hypothesis of this study is that differences in local structure of variable N-termini of free FLPs could partially explain differences in their potencies on receptors.
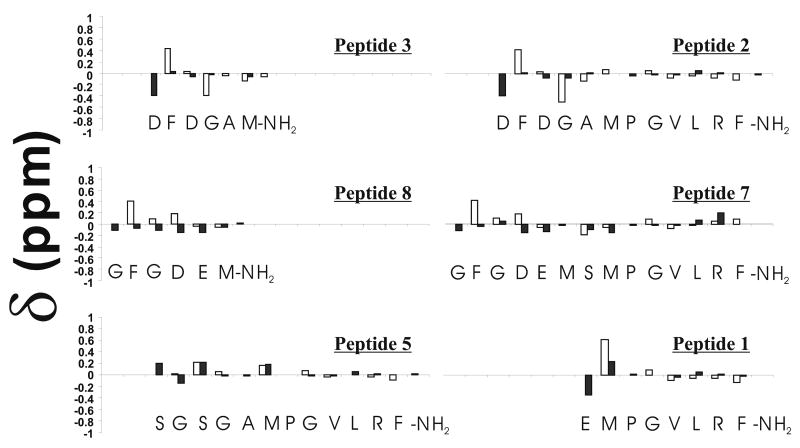
In order to compare chemical shifts from one peptide to another and to identify regions that contain significant populations of secondary structure, it is useful to compare experimental chemical shifts to random-coil values (64-66, 68). Fig. 2 plots the difference between experimental and random-coil values for some of the peptides analyzed in this study. The white and black bars represent deviations from random-coil values for amide and alpha protons, respectively, and the magnitude of the deviations reflects the population of local structure along the backbone of the peptides (64, 68). All data were compared to random coil values at pH 2.3.
Figure 2.
NMR chemical shift deviations from random coil values. Experimental chemical shifts at pH ∼2.3 were subtracted from sequence corrected random coil values (64, 65). Filled and open bars represent alpha and amide protons, respectively.
Several features in Figure 2 are worth noting. First, the chemical shifts of residues in the conserved PGVLRF-NH2 regions of each peptide are close to random-coil values, and rather similar among all the peptides examined. This suggests that this conserved sequence is unstructured in solution and that flexibility is important for binding to NPR-1. This flexibility may help the ligand diffuse/maneuver more effectively into a binding pocket on the receptor. Second, the N-terminal extension of peptide 2 shows significant deviation from random-coil values. In particular, the G4 amide proton has a very large deviation, suggesting significant structure. Third, the chemical shift deviations of amide and alpha protons of peptides 3 and 8 are nearly identical to the corresponding regions of the full-length peptides 2 and 7, respectively. This indicates that these N-terminal extensions are behaving as independent structural units. Finally, peptide 5, designed to lack N-terminal structure, indeed shows a consistent very small deviation from random-coil values in its first five residues.
pH Dependence of Amide Proton Chemical Shifts Reveal Regions of flp-18 Peptides with Significant Structure
The sensitivity of NMR chemical shifts to electronic structure and hydrogen bonding make them ideal probes of longer-range interactions with titratable side-chains. NMR studies of peptides that utilize amide protons often need to be conducted below ∼pH 6 to prevent amide proton exchange (56, 73). By varying the pH from about 5.5 to 2, both aspartic and glutamic acid side-chains will be converted from negatively charged and deprotonated to neutral and protonated. These different charge states of the carboxylate groups will produce changes in the electronic environment in interacting atoms proportional to 1/R3, where R is the distance between the charged group and chemical shift probe. Backbone amide resonances are particularly sensitive to interactions such as H-bonding (57, 66) and thus provide ideal probes of long-range interactions with side-chain carboxylates. This phenomenon provides a powerful mechanism to study long-range hydrogen bonding and salt bridge interactions in small peptides (45, 73, 74).
Many of the FLP-18 peptides contain aspartic or glutamic acids, so we performed 1D NMR pH titration experiments on all peptides in Table 2 containing these residues and, as controls, on peptides 5 and 11, neither of which showed any pH dependence in the proton chemical shifts. Stackplots of the amide region for a representative set of peptides are shown in Fig. 3.
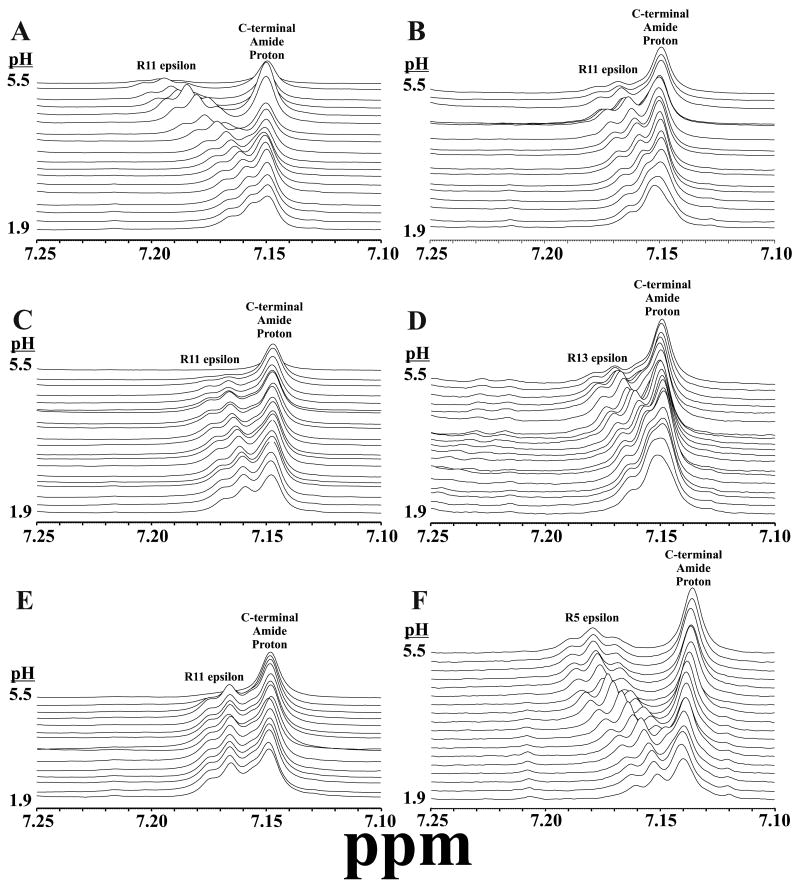
Figure 3.
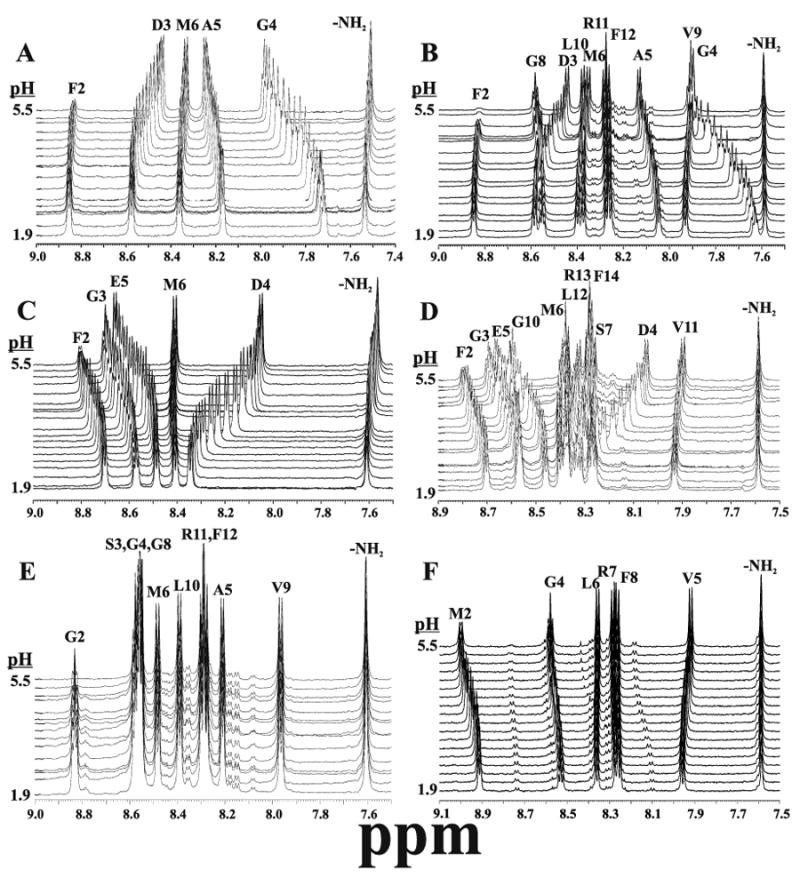
Amide region of one-dimensional NMR data, collected as a function of pH from about 1.9 to 5.5. Peaks are labeled with their assigned amino acids and the panels correspond to the following peptides: (ADD peptide sequences here as well) A. Peptide 3 = DFDGAM-NH2, B. Peptide 2 = DFDGAMPGVLRF-NH2, C. Peptide 8 = GFGDEM-NH2, D. Peptide 7 = GFGDEMSMPGVLRF-NH2, E. Peptide 5 = SGSGAMPGVLRF-NH2, F. Peptide 1 = EMPGVLRF-NH2. pH dependent interactions are summarized in Figure 5, and complete pKa analyses are provided in supplementary material.
No chemical shifts in peptide 5 (Figs 3E and 4E) or peptide 11 (data not shown) have any pH dependence, demonstrating that backbone amide proton chemical shifts are not intrinsically pH-dependent in this pH range. Second, several resonances in other peptides have large pH-dependent shifts. To our knowledge no systematic study has been undertaken to identify the maximum change in chemical shift of backbone amide protons as a function of pH, but Wüthrich and coworkers showed that in a small protein with a well-defined (∼70-90% populated) hydrogen-bond between an aspartic acid side chain and a backbone amide proton led to a change of 1.45 ppm over the titratable range of the aspartic acid (75). Thus, in Figure 3, some amide proton resonances of non-titratable amino acids have pH-dependent shifts that are characteristic of significant H-bond interactions. Others have smaller pH-dependent changes, suggesting either more transient dynamic interactions or much longer and weaker H-bonds. In contrast, several resonances in peptides with titratable groups show little or no pH dependence, showing that these effects are relatively specific. Next, the spectra from the N-terminal truncated peptides 3 and 8 are highly dependent on pH and are nearly perfect subsets of the same regions in their full length counterparts. Consistent with chemical shift data in Figure 2, this demonstrates that the N-terminal extensions of peptides 2 and 7 behave as independent structural units. The extensions also do not interact significantly with the more C-terminal backbone atoms, which show relatively little pH dependence, indicating that the conserved C-termini are less structured than the N-terminal extensions.
Figure 4.
Arg Hε region of one-dimensional NMR data, collected as a function of pH from 1.9 to 5.5. These long-range interactions on the penultimate C-terminal Arg result from the titratable carboxylate groups on the N-termini. pH dependent interactions are summarized in Figure 5, and complete pKa analyses are provided in supplementary material. Legend: A. Peptide 4 = DFDGEMPGVLRF-NH2, B. Peptide 2 = DFDGAMPGVLRF-NH2, C. Peptide 10 = GFGDAMPGVLRF-NH2, D. Peptide 7 = GFGDEMSMPGVLRF-NH2, E. Peptide 5 = SGSGAMPVLRF-NH2, F. Peptide 1 = EMPGVLRF-NH2
pH Dependence of Arginine Side-Chains Reveal Long-Range Interactions
The penultimate arginine residue is highly conserved and found in the same position in all FLPs. This arginine is at least 7 residues away from any carboxyl groups, so we were surprised to find in several FLP-18 peptides that its epsilon proton (Arg Hε) is pH dependant (Fig. 4).
Peptide 5 demonstrates that there is no intrinsic pH dependence Arg Hε over the pH range investigated, and we conclude that in other peptides there are long-range interactions between the Arg and the N-terminal carboxylates. Such an interaction would indicate a non-covalent ring structure. These interactions show up in most of the FLP-18 analogues having N-terminal carboxyl sidechains and, at first glance, do not appear to relate to the activity of the peptides (Table 2). For example, peptide 1 (one of the more active peptides) has nearly the same Arg Hε pH dependence as peptide 4 (the least active PGVLRF-NH2 containing examined). Moreover, peptide 10, with similar activity on to peptide 1, has nearly no Arg Hε pH dependence. Thus, Arg interaction with acidic residues alone is not sufficient to explain the difference in activity among the FLP-18 analogues tested.
Quantitative Determination of pKa Reveals Multiple Interactions
Several of the peptides in Table 2 have more than one carboxylate, so it is not always obvious which is responsible for the pH dependence of a particular resonance. If the titrating groups have distinct pKa values, then it should be possible to determine the contribution of each carboxylate on each titrating resonance using Equation 1. Every peak that exhibited pH-dependent chemical shifts was fitted using first one, then two, then three pKa values. In all cases we used the minimum number of interacting pKa values to get a good quality of fit and maximum linear regression coefficient (R2) to the experimental data. In the peptides with three titrating groups, inclusion of three interacting groups in the calculation did not improve the fits more so than including only two.
The complete table of relative pKa contributions (c from Eq 1) and pKa values are provided as supplementary material (Table S1), and the interactions are represented graphically in Fig. 5. As we discuss below, the interactions between titrating groups and resonances in these peptides is rather complicated and dynamic. The data presented here illustrate that, though there is a heterogeneous ensemble of H-bonding interactions between various backbone amide protons, certain ones are prominent.
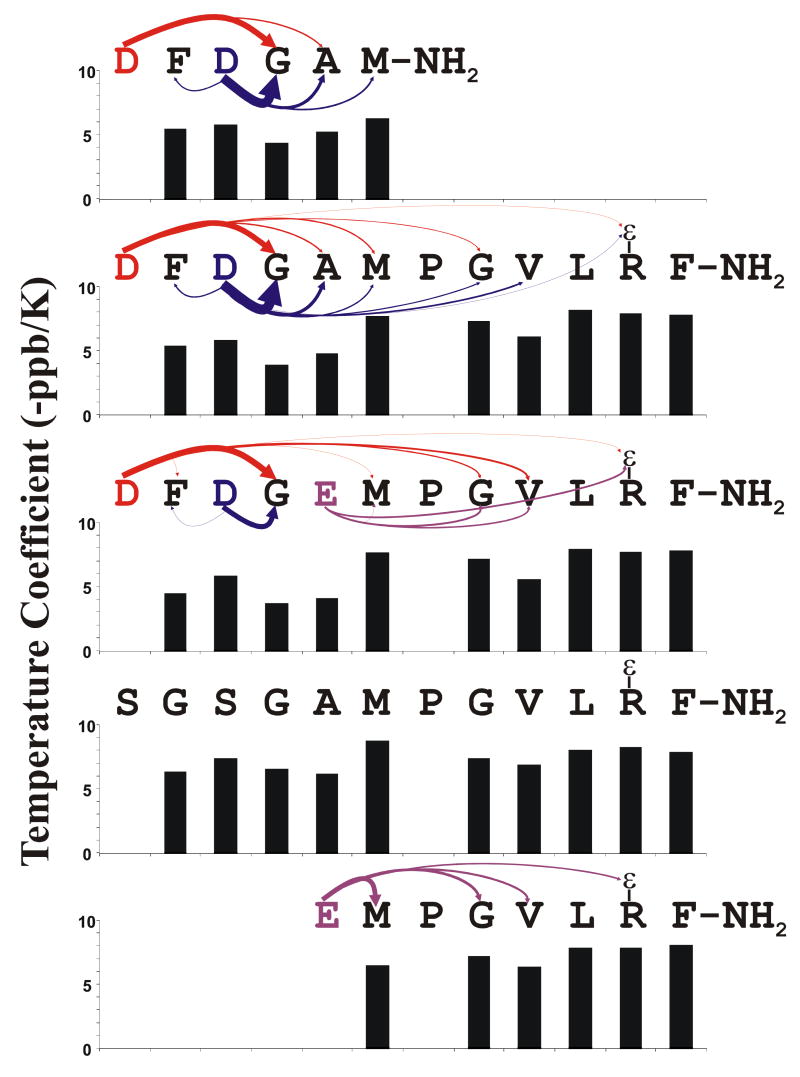
Figure 5.
Proposed H-bonding Interactions Between Backbone Amide Protons and Carboxyl Side-chains: DFDGAM-NH2 = Peptide 3, DFDGAMPGVLRF-NH2 = Peptide 2, DFDGEMPGVLRF-NH2 = Peptide 4, SGSGAMPGVLRF-NH2 = Peptide 5, EMPGVLRF-NH2 = Peptide 2. Each H-bond acceptor residue is color coded to match the arrows leading from it to its H-bond donors. The arrow widths are proportional to the relative extent to which that particular interaction affects the chemical shift of the amide proton at the point end of the arrow. The bar plots show the temperature coefficient of the backbone amide proton resonances
Using the pKas calculated for the pH dependent resonances for the peptides in this study we can assign most H-bonding interactions between titrating carboxyl side-chains and either backbone amide or Arg Hε protons. Figure 5 also illustrates the relative strength of these interactions. The most significant interaction (the largest shift from a long range interaction) is from a hydrogen bond between the D1 carboxylate and G4 amide in all peptides containing the N-terminal DFDG sequence. It contributes 40% to the observed titration of the G4 amide in peptides 2 and 3, and 55% to that of peptide 4 (supplementary data). The calculated pKa of D1 (∼3.0) is significantly lower than that of D3 (∼4.0), indicating that D1 is likely interacting with the positively charged amino terminus and stabilizing its negative charge. This is also seen in peptide 1, as E1 also has an unusually low pKa (∼3.5). Additional support for this pKa assignment comes from the pKa of the alpha protons of D1 and D3 of peptide 3 and D1 of peptide 2, which are 3.23, 4.09, and 2.97, respectively (supplementary data).
The interactions observed from pH titrations of peptides beginning in GFGD are different from and less substantial than those beginning in DFDG. For example, the largest pH dependant chemical shift change of the amide proton of a non-acidic amino acid for peptide 7 is ∼0.12 ppm (G3), whereas G4 in peptide 2 is ∼0.28 ppm. Also, the arginine sidechain of peptides 7 and 9 show a rather small chemical shift change in the pH titration (Fig. 4D) compared the same resonance for peptides 2 and 4. Additionally, the D4 sidechain of GFGD containing peptides seems to interact primarily with backbone amides N-terminal to it (Supplemental Material S1). This is a different conformation entirely than that observed in DFDG containing peptides, where D1 has a substantial interaction with G4.
Temperature Dependence of Amide Chemical Shifts Corroborates Regions with H-Bonding
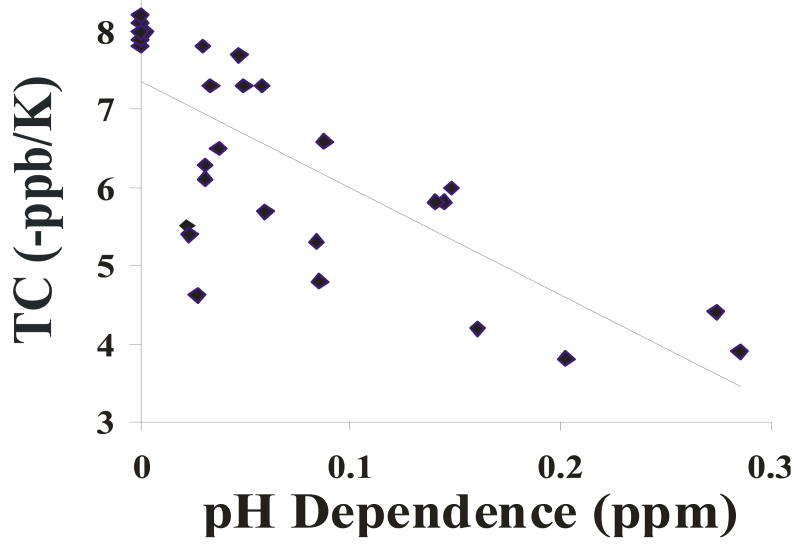
Although complicated and often over-interpreted, the temperature dependence of amide proton chemical shifts in polypeptides can be associated with hydrogen bonding (46, 76). Additionally, some peptides analyzed here lacked carboxyl side-chains, so pH titration results were not valid in determining possible structural interactions in these peptides. We therefore measured temperature coefficients (TCs) for several relevant peptides (Fig. 5). A rough guideline to interpreting TCs is that an absolute value less than 4 indicates an internal hydrogen bond, values between 4 and 6 indicate weak hydrogen bonding, and values greater than 6 are not involved in hydrogen bonding (46, 76, 77). The magnitudes of the temperature coefficients for all amide protons in this study are inversely correlated with the magnitude of chemical shift pH dependence for those resonances (Fig. 6), which is consistent with H-bonding interactions as described above.
Figure 6.
Relationship between Temperature Coefficients and pH dependence of chemical shift among Backbone Amide Protons: Plotted here is the chemical shift change with pH vs with Temperature for backbone amide resonances. R2 for the linear fit is 0.58. All data from peptides 1-4 are represented.
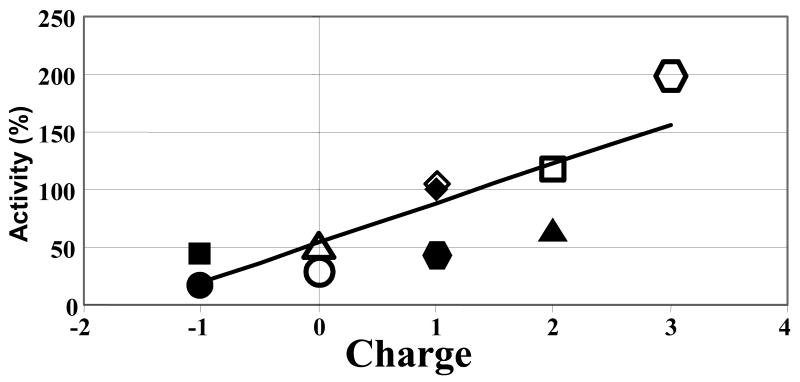
Overall Peptide Charge is Correlated With Activity on NPR-1
The experimental data presented above demonstrate that acidic residues in the N-terminal regions of FLP-18 peptides can interact with numerous amide protons and the conserved penultimate Arg. Although there are many additional factors influencing activity as addressed below, there appears to be a qualitative relationship between their charge properties (particularly of the N-terminus) and activities on NPR-1. This relationship is demonstrated in Figure 7 where the overall net charge at pH 7 of the entire peptide is plotted against its activity on NPR-1.
Figure 7.
Relationship between overall peptide charge and activity on the NPR-1 receptor. The overall peptide charge at neutral pH is plotted against activity for all of the peptides analyzed in this study. For the linear fit, R2 = 0.67. Legend: Filled circle = 4, Open Circle = 2, Filled Square = 9, Open Square = 5, Filled Triangle = 6, Open Triangle = 7, Filled Diamond = 1, Open Diamond = 10, Filled Hexagon = 11, Open Hexagon = 12 (numbers correspond to peptide numbers in Table 2)
Discussion
The goal of this work has been to determine the conformational properties of unbound FLP-18 neuropeptides from C. elegans and how these may affect their potencies on NPR-1. The starting point for this study was the knowledge that two of the peptides encoded by the flp-18 gene have significantly different potencies on NPR-1 (35). The major findings reported above can be summarized as follows:
The backbone of the conserved PGVLRF-NH2 is predominantly unstructured.
DFDG forms a structural loop stabilized by H-bonding.
Another loop forms when N-terminal acidic residue(s) interact with the conserved C-terminal penultimate arginine side-chain.
The DFDG loop interacts with the second loop to form a dynamic bicyclic structure that might influence binding to NPR-1.
Charge also affects the activity of FLP-18 peptides on NPR-1.
The backbone structure of the conserved PGVLRF-NH2 is predominantly unstructured
All NMR structural parameters measured in this study for the PGVLRF-NH2 region of FLP-18 peptides indicate that the peptide backbone of this conserved sequence is predominantly unstructured. The only significant evidence for any kind of structural motif is the interaction between the conserved penultimate arginine side-chain and acidic residues in the N-termini. These results suggest that the primary receptor-binding region of FLP-18 peptides is highly flexible before interacting with NPR-1.
DFDG forms a structural loop stabilized by H-bonding
We observe transient H-bonding and ionic interactions within FLP-18 peptides beginning in the sequence DFDG. Specifically, acidic residues in the variable N-termini form substantial H-bonds to backbone amides N-terminal to the conserved proline (Figures 5 and 8).
Figure 8.
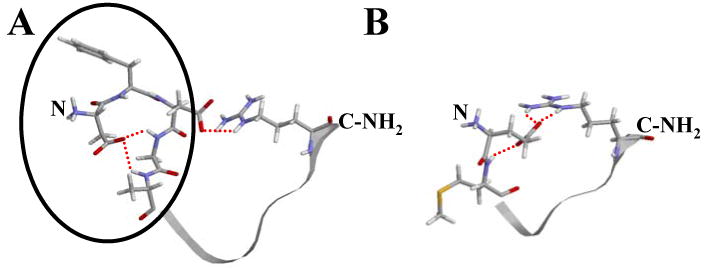
Model of interactions thought occurring within native FLP-18 peptides. This figure shows the most significant H-bonding interactions supported by NMR data. A: For DFDGAMPGVLRF-NH2, H-bonds between the D1 side-chain carboxylate and G4/A5 backbone amide protons as well as H-bonding/ionic interaction between the D3 side-chain by red dashed lines. The N-terminal loop structure implicated in inhibiting binding to NPR-1 is circled in black. B: EMPGVLRF-NH2 is shown with the most significant H-bonding and ionic interactions for which we have evidence. Notice that it has no N-terminal loop, in contrast to DFDGAMPGVLRF-NH2. Also, the same unstructured region for both peptides is shown in ribbon view. The N- and C- termini are also labeled on both peptides.
In the DFDG containing peptides, G4 has the smallest temperature coefficient of all amides in the study; this is characteristic of involvement in a significant H-bonding interaction (46, 76, 78). This phenomenon is particularly prominent in peptide 4, where the D1 pKa rather than that of D3 is the most significant contributor to the G4 amide proton titration. It is also the least active PGVLRF-NH2 containing peptide tested. Also, weak ROESY peaks were observed between D1 beta protons and G4 alpha protons in both peptides 2 and 3 (data not shown). This further corroborates the pH titration results that indicate significant long-range H-bonding between the D1 sidechain and G4 backbone amide proton of peptides beginning with DFDG. In contrast, the N-terminal SGSG region of peptide 5 is unstructured based on our NMR results, and is one of the most active peptides analyzed.
The DFDG loop may interact with the second loop to form a dynamic bicyclic structure which reduces binding to NPR-1
There is no direct or simple correlation between the activity data and any one set of NMR data. However, the two carboxylate residues in peptides 2 and 4 allow both the N-terminal loops as well as the ionic interaction between the conserved arginine and the aspartates (Fig. 8A). The increased activities of peptides 7 and 9, along with their apparent weaker interaction between the penultimate arginine and acidic residues relative to peptides 2 and 4, illustrate that the residues SM inserted in the middle of these peptides can interfere with loop formation between the N- and C- termini. FLP-18 peptides are short and flexible, and both loop interactions are likely dynamic. However, there is a possibility that the bulk of the N-terminal loop in DFDG containing peptides is brought into proximity of the conserved receptor-binding region by the action of the second loop involving the penultimate arginine. We propose that this bicyclic structure reduces binding to NPR-1.
Charge is also important in determining the activity of flp-18 peptides on NPR-1
There is a significant correlation between charge and activity such that more positively charged peptides tend to activate NPR-1 better than more negatively charged ones. Interestingly, the vast majority of predicted FLPs in C. elegans tend to be positively charged (4), including the peptide encoded by flp-21, which has an overall charge of +3 and is active on both naturally occurring isoforms of NPR-1 (215F and 215 V). However, peptides 4 and 9 have the same charge but different activities on NPR-1. The acidic residues of peptides 4 and 9 differ substantially in their interaction with the C-terminal arginine. This is likely due to the insertion of the residues SM in the middle of peptide 9. Thus, the N-terminal DFDG loop in peptide 9 does not interact well with the penultimate arginine, whereas that of peptide 4 does. This further supports the bicyclic model and the affect of a two-loop conformation on the activities of DFDG containing peptides.
Peptides 6 and 12 were often outliers in our attempts to correlate specific NMR data parameters to activity results. Peptide 6 was designed to possess a helix in the N-terminus, and we predicted reduced binding to NPR-1 resulting in activity similar to that of peptide 2. This prediction was incorrect, and peptide 6 had more activity than peptide 2. However, with no carboxylates, peptide 6 lacks the ability to form sidechain mediated H-bonding loops, which our model suggests should give it an activity more like that of peptides 1 and 5. Thus, the activity of peptide 6 (intermediate between peptides 1 and 2) suggests that other properties of its structure modulate its potency.
Peptide 12 unexpectedly had nearly exactly twice the activity of peptide 1. It is composed of two copies of the conserved PGVLRF sequence that is responsible for FLP-18 activity on NPR-1. Previous studies on FLP receptors show that the C-terminal amide group is necessary for activity (40), so it is extremely unlikely that the C-terminal PGVLRF in peptide 12 can interact with the active site of NPR-1. However, this peptide is also the most positively charged of all among those tested. This is consistent with our observation that a peptide's charge influences its activity on NPR-1.
Both native FLP-18 peptides in this study, DFDGAMPGVLRF-NH2 and EMPGVLRF-NH2, differ in both potency and efficacy. We have shown that N-terminal structure, peptide charge, loop formation and backbone flexibility in PGVLRF-NH2 all modulate the activity of FLP-18 peptides on NPR-1. One interesting feature of the dose response curves in Figure 1 is that the two longer peptides have a larger maximal response and a steeper linear portion than the shorter peptide. This suggests that the native peptides could induce different configurations of the NPR-1 receptor with different abilities to couple the G-protein pathway under study and perhaps to other additional second messenger pathways as yet untested (79-82). Both the A. suum and C. elegans long peptides have been isolated (19, 23), demonstrating that these exist in vivo. However, other studies (83, 84) have shown that many peptide degradation products can also be found in cells. Perhaps multiple forms of FLP-18 peptides could shape the behavioral response to NPR-1 activity in a way that could not be achieved by any one peptide alone. It is possible that the ensemble of peptides functions as a bouquet to achieve a unique, beneficial, fine tuned response (2, 45).
Supplementary Material
S1: Calculated pKas for all pH dependent resonances in all peptides examined by NMR pH titration and calculated proportion contributions for each pKa to each pH dependant resonance.
S2: NMR chemical shift assignments for all peptides examined by NMR
Acknowledgments
Peptides were synthesized and purified by Alfred Chung and amino acid analyses were done by Scott McClung in the ICBR Protein Core facility at the University of Florida. We thank Jim Rocca and Dan Plant in the McKnight Brain Institute AMRIS facility for excellent NMR support. We thank Omjoy Ganesh for writing a program used to implement published sequence dependant corrections to the random coil chemical shifts. A.S. Edison thanks Tony Stretton for many helpful and pleasant discussions.
This work was supported by a NSF CAREER grant to A.S. Edison, NIH 5P41RR016105, and the Human Frontier Science Program.
The abbreviations used are
- FMRFamide
Phe-Met-Arg-Phe-NH2
- FLP
FMRFamide-Like-Peptide
- NMR
nuclear magnetic resonance
- AAA
Amino Acid Analysis
- TOCSY
total correlation spectroscopy
- NOESY
nuclear Overhauser effect spectroscopy
- ROESY
rotating frame Overhauser effect spectroscopy
- D2O
deuterium oxide
- GPCR
G-Protein Coupled Receptor
- C. elegans
Caenorhabditis elegans
- TC
Temperature Coefficient
- K
Kelvin
- NPR-1
neuropeptide resemblance receptor-1
- flp-n
gene name
- flp-n
precursor protein
- FLP-N
peptide name
Footnotes
Author Contributions – A.T.D and C.Z. did the NMR data analysis. V.R. did electrophysiological activity measurements. H.C. prepared cDNA and cRNA for the NPR-1 receptor, which was supplied by M.deB. A.T.D., P.D.E., and A.S.E. formulated the hypotheses, interpreted the results, and developed the conclusions.
References
- 1.Price DA, Greenberg MJ. Structure of a Molluscan Cardioexcitatory Neuropeptide. Science. 1977;197:670–671. doi: 10.1126/science.877582. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg MJ, Price DA. Relationships among the FMRFamide-like peptides. Prog Brain Res. 1992;92:25–37. doi: 10.1016/s0079-6123(08)61162-0. [DOI] [PubMed] [Google Scholar]
- 3.Li C, Nelson LS, Kim K, Nathoo A, Hart AC. Neuropeptide gene families in the nematode Caenorhabditis elegans. Ann N Y Acad Sci. 1999;897:239–52. doi: 10.1111/j.1749-6632.1999.tb07895.x. [DOI] [PubMed] [Google Scholar]
- 4.Li C, Kim K, Nelson LS. FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. Brain Res. 1999;848:26–34. doi: 10.1016/s0006-8993(99)01972-1. [DOI] [PubMed] [Google Scholar]
- 5.McVeigh P, Leech S, Mair GR, Marks NJ, Geary TG, Maule AG. Analysis of FMRFamide-like peptide (FLP) diversity in phylum Nematoda. Int J Parasitol. 2005;35:1043–60. doi: 10.1016/j.ijpara.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Vilim FS, Aarnisalo AA, Nieminen ML, Lintunen M, Karlstedt K, Kontinen VK, Kalso E, States B, Panula P, Ziff E. Gene for pain modulatory neuropeptide NPFF: induction in spinal cord by noxious stimuli. Mol Pharmacol. 1999;55:804–11. [PubMed] [Google Scholar]
- 7.Perry SJ, Yi-Kung Huang E, Cronk D, Bagust J, Sharma R, Walker RJ, Wilson S, Burke JF. A human gene encoding morphine modulating peptides related to NPFF and FMRFamide. FEBS Lett. 1997;409:426–30. doi: 10.1016/s0014-5793(97)00557-7. [DOI] [PubMed] [Google Scholar]
- 8.Perry SJ, Straub VA, Schofield MG, Burke JF, Benjamin PR. Neuronal expression of an FMRFamide-gated Na+ channel and its modulation by acid pH. J Neurosci. 2001;21:5559–67. doi: 10.1523/JNEUROSCI.21-15-05559.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kivipelto L, Panula P. Comparative Distribution of Neurons Containing FLFQPQRFamide-like (morphine-modulating) Peptide and Related Neuropeptides in the Rat Brain. Eur J Neurosci. 1991;3:175–185. doi: 10.1111/j.1460-9568.1991.tb00078.x. [DOI] [PubMed] [Google Scholar]
- 10.Espinoza E, Carrigan M, Thomas SG, Shaw G, Edison AS. A statistical view of FMRFamide neuropeptide diversity. Mol Neurobiol. 2000;21:35–56. doi: 10.1385/MN:21:1-2:035. [DOI] [PubMed] [Google Scholar]
- 11.Fisher JM, Scheller RH. Prohormone processing and the secretory pathway. J Biol Chem. 1988;263:16515–8. [PubMed] [Google Scholar]
- 12.Pascual N, Castresana J, Valero ML, Andreu D, Belles X. Orcokinins in insects and other invertebrates. Insect Biochem Mol Biol. 2004;34:1141–6. doi: 10.1016/j.ibmb.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Conlon JM. Evolution of the insulin molecule: insights into structure-activity and phylogenetic relationships. Peptides. 2001;22:1183–93. doi: 10.1016/s0196-9781(01)00423-5. [DOI] [PubMed] [Google Scholar]
- 14.Edison AS, Messinger LA, Stretton AO. afp-1: a gene encoding multiple transcripts of a new class of FMRFamide-like neuropeptides in the nematode Ascaris suum. Peptides. 1997;18:929–35. doi: 10.1016/s0196-9781(97)00047-8. [DOI] [PubMed] [Google Scholar]
- 15.Cowden C, Stretton AO, Davis RE. AF1, a sequenced bioactive neuropeptide isolated from the nematode Ascaris suum. Neuron. 1989;2:1465–73. doi: 10.1016/0896-6273(89)90192-x. [DOI] [PubMed] [Google Scholar]
- 16.Cowden C, Stretton AO. AF2, an Ascaris neuropeptide: isolation, sequence, and bioactivity. Peptides. 1993;14:423–30. doi: 10.1016/0196-9781(93)90127-3. [DOI] [PubMed] [Google Scholar]
- 17.Sithigorngul P, Cowden C, Guastella J, Stretton AO. Generation of monoclonal antibodies against a nematode peptide extract: another approach for identifying unknown neuropeptides. J Comp Neurol. 1989;284:389–97. doi: 10.1002/cne.902840306. [DOI] [PubMed] [Google Scholar]
- 18.Cowden C, Sithigorngul P, Brackley P, Guastella J, Stretton AO. Localization and differential expression of FMRFamide-like immunoreactivity in the nematode Ascaris suum. J Comp Neurol. 1993;333:455–68. doi: 10.1002/cne.903330311. [DOI] [PubMed] [Google Scholar]
- 19.Cowden C, Stretton AO. Eight novel FMRFamide-like neuropeptides isolated from the nematode Ascaris suum. Peptides. 1995;16:491–500. doi: 10.1016/0196-9781(94)00211-n. [DOI] [PubMed] [Google Scholar]
- 20.Bowman JW, Friedman AR, Thompson DP, Ichhpurani AK, Kellman MF, Marks N, Maule AG, Geary TG. Structure-activity relationships of KNEFIRFamide (AF1), a nematode FMRFamide-related peptide, on Ascaris suum muscle. Peptides. 1996;17:381–7. doi: 10.1016/0196-9781(96)00007-1. [DOI] [PubMed] [Google Scholar]
- 21.Bowman JW, Winterrowd CA, Friedman AR, Thompson DP, Klein RD, Davis JP, Maule AG, Blair KL, Geary TG. Nitric oxide mediates the inhibitory effects of SDPNFLRFamide, a nematode FMRFamide-related neuropeptide, in Ascaris suum. J Neurophysiol. 1995;74:1880–8. doi: 10.1152/jn.1995.74.5.1880. [DOI] [PubMed] [Google Scholar]
- 22.Nathoo AN, Moeller RA, Westlund BA, Hart AC. Identification of neuropeptide-like protein gene families in Caenorhabditiselegans and other species. Proc Natl Acad Sci U S A. 2001;98:14000–5. doi: 10.1073/pnas.241231298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Husson SJ, Clynen E, Baggerman G, De Loof A, Schoofs L. Discovering neuropeptides in Caenorhabditis elegans by two dimensional liquid chromatography and mass spectrometry. Biochem Biophys Res Commun. 2005;335:76–86. doi: 10.1016/j.bbrc.2005.07.044. [DOI] [PubMed] [Google Scholar]
- 24.Rosoff ML, Doble KE, Price DA, Li C. The flp-1 propeptide is processed into multiple, highly similar FMRFamide-like peptides in Caenorhabditis elegans. Peptides. 1993;14:331–8. doi: 10.1016/0196-9781(93)90049-m. [DOI] [PubMed] [Google Scholar]
- 25.Marks NJ, Shaw C, Maule AG, Davis JP, Halton DW, Verhaert P, Geary TG, Thompson DP. Isolation of AF2 (KHEYLRFamide) from Caenorhabditis elegans: evidence for the presence of more than one FMRFamide-related peptide-encoding gene. Biochem Biophys Res Commun. 1995;217:845–51. doi: 10.1006/bbrc.1995.2849. [DOI] [PubMed] [Google Scholar]
- 26.Marks NJ, Maule AG, Geary TG, Thompson DP, Davis JP, Halton DW, Verhaert P, Shaw C. APEASPFIRFamide, a novel FMRFamide-related decapeptide from Caenorhabditis elegans: structure and myoactivity. Biochem Biophys Res Commun. 1997;231:591–5. doi: 10.1006/bbrc.1997.6155. [DOI] [PubMed] [Google Scholar]
- 27.Marks NJ, Maule AG, Geary TG, Thompson DP, Li C, Halton DW, Shaw C. KSAYMRFamide (PF3/AF8) is present in the free-living nematode, Caenorhabditis elegans. Biochem Biophys Res Commun. 1998;248:422–5. doi: 10.1006/bbrc.1998.8982. [DOI] [PubMed] [Google Scholar]
- 28.Mercier AJ, Friedrich R, Boldt M. Physiological functions of FMRFamide-like peptides (FLPs) in crustaceans. Microsc Res Tech. 2003;60:313–24. doi: 10.1002/jemt.10270. [DOI] [PubMed] [Google Scholar]
- 29.Rastogi RK, D'Aniello B, Pinelli C, Fiorentino M, Di Fiore MM, Di Meglio M, Iela L. FMRFamide in the amphibian brain: a comprehensive survey. Microsc Res Tech. 2001;54:158–72. doi: 10.1002/jemt.1130. [DOI] [PubMed] [Google Scholar]
- 30.Brownlee DJ, Fairweather I. Exploring the neurotransmitter labyrinth in nematodes. Trends Neurosci. 1999;22:16–24. doi: 10.1016/s0166-2236(98)01281-8. [DOI] [PubMed] [Google Scholar]
- 31.Dockray GJ. The expanding family of -RFamide peptides and their effects on feeding behaviour. Exp Physiol. 2004;89:229–35. doi: 10.1113/expphysiol.2004.027169. [DOI] [PubMed] [Google Scholar]
- 32.Yang HY, Fratta W, Majane EA, Costa E. Isolation, sequencing, synthesis, and pharmacological characterization of two brain neuropeptides that modulate the action of morphine. Proc Natl Acad Sci U S A. 1985;82:7757–61. doi: 10.1073/pnas.82.22.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waggoner LE, Hardaker LA, Golik S, Schafer WR. Effect of a neuropeptide gene on behavioral states in Caenorhabditis elegans egg-laying. Genetics. 2000;154:1181–92. doi: 10.1093/genetics/154.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson LS, Rosoff ML, Li C. Disruption of a neuropeptide gene, flp-1, causes multiple behavioral defects in Caenorhabditis elegans. Science. 1998;281:1686–90. doi: 10.1126/science.281.5383.1686. [DOI] [PubMed] [Google Scholar]
- 35.Rogers C, Reale V, Kim K, Chatwin H, Li C, Evans P, de Bono M. Inhibition of Caenorhabditis elegans social feeding by FMRFamide-related peptide activation of NPR-1. Nat Neurosci. 2003;6:1178–85. doi: 10.1038/nn1140. [DOI] [PubMed] [Google Scholar]
- 36.Kubiak TM, Larsen MJ, Nulf SC, Zantello MR, Burton KJ, Bowman JW, Modric T, Lowery DE. Differential activation of “social” and “solitary” variants of the Caenorhabditis elegans G protein-coupled receptor NPR-1 by its cognate ligand AF9. J Biol Chem. 2003;278:33724–9. doi: 10.1074/jbc.M304861200. [DOI] [PubMed] [Google Scholar]
- 37.Mertens I, Vandingenen A, Meeusen T, Janssen T, Luyten W, Nachman RJ, De Loof A, Schoofs L. Functional characterization of the putative orphan neuropeptide G-protein coupled receptor C26F1.6 in Caenorhabditis elegans. FEBS Lett. 2004;573:55–60. doi: 10.1016/j.febslet.2004.07.058. [DOI] [PubMed] [Google Scholar]
- 38.Mertens I, Meeusen T, Janssen T, Nachman R, Schoofs L. Molecular characterization of two G protein-coupled receptor splice variants as FLP2 receptors in Caenorhabditis elegans. Biochem Biophys Res Commun. 2005;330:967–74. doi: 10.1016/j.bbrc.2005.03.071. [DOI] [PubMed] [Google Scholar]
- 39.Kubiak TM, Larsen MJ, Zantello MR, Bowman JW, Nulf SC, Lowery DE. Functional annotation of the putative orphan Caenorhabditis elegans G-protein-coupled receptor C10C6.2 as a FLP15 peptide receptor. J Biol Chem. 2003;278:42115–20. doi: 10.1074/jbc.M304056200. [DOI] [PubMed] [Google Scholar]
- 40.Johnson EC, Bohn LM, Barak LS, Birse RT, Nassel DR, Caron MG, Taghert PH. Identification of Drosophila neuropeptide receptors by G protein-coupled receptors-beta-arrestin2 interactions. J Biol Chem. 2003;278:52172–8. doi: 10.1074/jbc.M306756200. [DOI] [PubMed] [Google Scholar]
- 41.Lingueglia E, Champigny G, Lazdunski M, Barbry P. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature. 1995;378:730–3. doi: 10.1038/378730a0. [DOI] [PubMed] [Google Scholar]
- 42.Cottrell GA. The first peptide-gated ion channel. J Exp Biol. 1997;200:2377–86. doi: 10.1242/jeb.200.18.2377. [DOI] [PubMed] [Google Scholar]
- 43.Furukawa Y, Miyawaki Y, Abe G. Molecular cloning and functional characterization of the Aplysia FMRFamide-gated Na(+) channel. Pflugers Arch. 2005 doi: 10.1007/s00424-005-1498-z. [DOI] [PubMed] [Google Scholar]
- 44.Xie J, Price MP, Wemmie JA, Askwith CC, Welsh MJ. ASIC3 and ASIC1 mediate FMRFamide-related peptide enhancement of H+-gated currents in cultured dorsal root ganglion neurons. J Neurophysiol. 2003;89:2459–65. doi: 10.1152/jn.00707.2002. [DOI] [PubMed] [Google Scholar]
- 45.Edison AS, Espinoza E, Zachariah C. Conformational ensembles: the role of neuropeptide structures in receptor binding. J Neurosci. 1999;19:6318–26. doi: 10.1523/JNEUROSCI.19-15-06318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andersen NH, Neidigh JW, Harris SM, Lee GM, Liu Z, Tong H. Extracting Information from the Temperature Gradients of Polypeptide NH Chemical Shifts. 1. The Importance of Conformational Averaging. J Am Chem Soc. 1997;119:8547–8561. [Google Scholar]
- 47.Silverman SK, Lester HA, Dougherty DA. Subunit stoichiometry of a heteromultimeric G protein-coupled inward-rectifier K+ channel. J Biol Chem. 1996;271:30524–8. doi: 10.1074/jbc.271.48.30524. [DOI] [PubMed] [Google Scholar]
- 48.Van Renterghem C, Bilbe G, Moss S, Smart TG, Constanti A, Brown DA, Barnard EA. GABA receptors induced in Xenopus oocytes by chick brain mRNA: evaluation of TBPS as a use-dependent channel-blocker. Brain Res. 1987;388:21–31. doi: 10.1016/0169-328x(87)90017-9. [DOI] [PubMed] [Google Scholar]
- 49.Brown NA, McAllister G, Weinberg D, Milligan G, Seabrook GR. Involvement of G-protein alpha il subunits in activation of G-protein gated inward rectifying K+ channels (GIRK1) by human NPY1 receptors. Br J Pharmacol. 1995;116:2346–8. doi: 10.1111/j.1476-5381.1995.tb15077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reale V, Hannan F, Hall LM, Evans PD. Agonist-specific coupling of a cloned Drosophila melanogaster D1-like dopamine receptor to multiple second messenger pathways by synthetic agonists. J Neurosci. 1997;17:6545–53. doi: 10.1523/JNEUROSCI.17-17-06545.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–5. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 52.Schweiger A, Braunschweiler L, Fauth J, Ernst RR. Coherent and incoherent echo spectroscopy with extended-time excitation. Physical Review Letters. 1985;54:1241–1244. doi: 10.1103/PhysRevLett.54.1241. [DOI] [PubMed] [Google Scholar]
- 53.Bothner-By AA, Stephens RL, Lee J, Warren CD, Jeanloz RW. Structure determination of a tetrasaccharide: transient nuclear Overhauser effects in the rotating frame. J Am Chem Soc. 1984;106:811–813. [Google Scholar]
- 54.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 55.Johnson BA, B R. a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 56.Wuthrich K. NMR of Proteins and Nucleic Acids. New York, NY: 1986. [Google Scholar]
- 57.Betz M, Lohr F, Wienk H, Ruterjans H. Long-range nature of the interactions between titratable groups in Bacillus agaradhaerens family 11 xylanase: pH titration of B. agaradhaerens xylanase. Biochemistry. 2004;43:5820–31. doi: 10.1021/bi049948m. [DOI] [PubMed] [Google Scholar]
- 58.Carlacci L, Edison AS. Computational analysis of two similar neuropeptides yields distinct conformational ensembles. Proteins. 2000;40:367–77. doi: 10.1002/1097-0134(20000815)40:3<367::aid-prot30>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 59.Stretton AO, Cowden C, Sithigorngul P, Davis RE. Neuropeptides in the nematode Ascaris suum. Parasitology. 1991;102 Suppl:S107–16. doi: 10.1017/s0031182000073339. [DOI] [PubMed] [Google Scholar]
- 60.Payza K, Greenberg MJ, Price DA. Further characterization of Helix FMRFamide receptors: kinetics, tissue distribution, and interactions with the endogenous heptapeptides. Peptides. 1989;10:657–61. doi: 10.1016/0196-9781(89)90156-3. [DOI] [PubMed] [Google Scholar]
- 61.Marqusee S, Robbins VH, Baldwin RL. Unusually stable helix formation in short alanine-based peptides. Proc Natl Acad Sci U S A. 1989;86:5286–90. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chakrabartty A, Kortemme T, Baldwin RL. Helix propensities of the amino acids measured in alanine-based peptides without helix-stabilizing side-chain interactions. Protein Sci. 1994;3:843–52. doi: 10.1002/pro.5560030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol. 1991;222:311–33. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 64.Schwarzinger S, Kroon GJ, Foss TR, Wright PE, Dyson HJ. Random coil chemical shifts in acidic 8 M urea: implementation of random coil shift data in NMRView. J Biomol NMR. 2000;18:43–8. doi: 10.1023/a:1008386816521. [DOI] [PubMed] [Google Scholar]
- 65.Schwarzinger S, Kroon GJ, Foss TR, Chung J, Wright PE, Dyson HJ. Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc. 2001;123:2970–8. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- 66.Wishart DS, Case DA. Use of chemical shifts in macromolecular structure determination. Methods Enzymol. 2001;338:3–34. doi: 10.1016/s0076-6879(02)38214-4. [DOI] [PubMed] [Google Scholar]
- 67.Lin JC, Barua B, Andersen NH. The helical alanine controversy: an (Ala)6 insertion dramatically increases helicity. J Am Chem Soc. 2004;126:13679–84. doi: 10.1021/ja047265o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andersen NH, Cao B, Chen C. Peptide/protein structure analysis using the chemical shift index method: upfield alpha-CH values reveal dynamic helices and alpha L sites. Biochem Biophys Res Commun. 1992;184:1008–14. doi: 10.1016/0006-291x(92)90691-d. [DOI] [PubMed] [Google Scholar]
- 69.Dyson HJ, Rance M, Houghten RA, Lerner RA, Wright PE. Folding of immunogenic peptide fragments of proteins in water solution. I. Sequence requirements for the formation of a reverse turn. J Mol Biol. 1988;201:161–200. doi: 10.1016/0022-2836(88)90446-9. [DOI] [PubMed] [Google Scholar]
- 70.Dyson HJ, Rance M, Houghten RA, Wright PE, Lerner RA. Folding of immunogenic peptide fragments of proteins in water solution. II. The nascent helix. J Mol Biol. 1988;201:201–17. doi: 10.1016/0022-2836(88)90447-0. [DOI] [PubMed] [Google Scholar]
- 71.Blanco FJ, Serrano L, Forman-Kay JD. High populations of non-native structures in the denatured state are compatible with the formation of the native folded state. J Mol Biol. 1998;284:1153–64. doi: 10.1006/jmbi.1998.2229. [DOI] [PubMed] [Google Scholar]
- 72.Osterhout JJ, Jr, Baldwin RL, York EJ, Stewart JM, Dyson HJ, Wright PE. 1H NMR studies of the solution conformations of an analogue of the C-peptide of ribonuclease A. Biochemistry. 1989;28:7059–64. doi: 10.1021/bi00443a042. [DOI] [PubMed] [Google Scholar]
- 73.Bundi A, Wuthrich K. 1H NMR titration shifts of amide proton resonances in polypeptide chains. FEBS Lett. 1977;77:11–4. doi: 10.1016/0014-5793(77)80182-8. [DOI] [PubMed] [Google Scholar]
- 74.Bundi A, Wuthrich K. Use of 1H-NMR Titration Shifts for Studies of Polypeptide Conformation. Biopolymers. 1979;18:299–311. [Google Scholar]
- 75.Szyperski T, Antuch W, Schick M, Betz A, Stone SR, Wuthrich K. Transient hydrogen bonds identified on the surface of the NMR solution structure of Hirudin. Biochemistry. 1994;33:9303–10. doi: 10.1021/bi00197a034. [DOI] [PubMed] [Google Scholar]
- 76.Baxter NJ, Williamson MP. Temperature dependence of 1H chemical shifts in proteins. J Biomol NMR. 1997;9:359–69. doi: 10.1023/a:1018334207887. [DOI] [PubMed] [Google Scholar]
- 77.Silva Elipe Maria Victoria, M RT, Bednarek Maria A, Arison Byron H. 1H-NMR Studies on a Potent and Selective Antagonist at Human Melanocortin Receptor 4 (hMC-4R) Biopolymers. 2003;68:512–527. doi: 10.1002/bip.10340. [DOI] [PubMed] [Google Scholar]
- 78.Higashijima T, Tasumi M, Miyazawa T. H nuclear magnetic resonance studies of melanostatin: dependence of the chemical shifts of NH protons on temperature and concentration. FEBS Lett. 1975;57:175–8. doi: 10.1016/0014-5793(75)80710-1. [DOI] [PubMed] [Google Scholar]
- 79.Kenakin T. Principles: receptor theory in pharmacology. Trends Pharmacol Sci. 2004;25:186–92. doi: 10.1016/j.tips.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 80.Evans PD, Robb S, Cheek TR, Reale V, Hannan FL, Swales LS, Hall LM, Midgley JM. Agonist-specific coupling of G-protein-coupled receptors to second-messenger systems. Prog Brain Res. 1995;106:259–68. doi: 10.1016/s0079-6123(08)61222-4. [DOI] [PubMed] [Google Scholar]
- 81.Robb S, Cheek TR, Hannan FL, Hall LM, Midgley JM, Evans PD. Agonist-specific coupling of a cloned Drosophila octopamine/tyramine receptor to multiple second messenger systems. Embo J. 1994;13:1325–30. doi: 10.1002/j.1460-2075.1994.tb06385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spengler D, Waeber C, Pantaloni C, Holsboer F, Bockaert J, Seeburg PH, Journot L. Differential signal transduction by five splice variants of the PACAP receptor. Nature. 1993;365:170–5. doi: 10.1038/365170a0. [DOI] [PubMed] [Google Scholar]
- 83.Floyd PD, Li L, Rubakhin SS, Sweedler JV, Horn CC, Kupfermann I, Alexeeva VY, Ellis TA, Dembrow NC, Weiss KR, Vilim FS. Insulin prohormone processing, distribution, and relation to metabolism in Aplysia californica. J Neurosci. 1999;19:7732–41. doi: 10.1523/JNEUROSCI.19-18-07732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li L, Moroz TP, Garden RW, Floyd PD, Weiss KR, Sweedler JV. Mass spectrometric survey of interganglionically transported peptides in Aplysia. Peptides. 1998;19:1425–33. doi: 10.1016/s0196-9781(98)00094-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1: Calculated pKas for all pH dependent resonances in all peptides examined by NMR pH titration and calculated proportion contributions for each pKa to each pH dependant resonance.
S2: NMR chemical shift assignments for all peptides examined by NMR