Abstract
Heart failure (HF) is characterized by a skeletal muscle myopathy with increased expression of fast myosin heavy chains (MHCs). The skeletal muscle-specific molecular regulatory mechanisms controlling MHC expression during HF have not been described. Myogenic regulatory factors (MRFs), a family of transcriptional factors that control the expression of several skeletal muscle-specific genes, may be related to these alterations. This investigation was undertaken in order to examine potential relationships between MRF mRNA expression and MHC protein isoforms in Wistar rat skeletal muscle with monocrotaline-induced HF. We studied soleus (Sol) and extensor digitorum longus (EDL) muscles from both HF and control Wistar rats. MyoD, myogenin and MRF4 contents were determined using reverse transcription-polymerase chain reaction while MHC isoforms were separated using polyacrylamide gel electrophoresis. Despite no change in MHC composition of Wistar rat skeletal muscles with HF, the mRNA relative expression of MyoD in Sol and EDL muscles and that of MRF4 in Sol muscle were significantly reduced, whereas myogenin was not changed in both muscles. This down-regulation in the mRNA relative expression of MRF4 in Sol was associated with atrophy in response to HF while these alterations were not present in EDL muscle. Taken together, our results show a potential role for MRFs in skeletal muscle myopathy during HF.
Keywords: heart failure, myogenic regulatory factors, myosin heavy chain, skeletal muscle, Wistar rats
Heart failure (HF) is characterized by a reduced tolerance to exercise due to early fatigue and dyspnea; this may in part be due to skeletal muscle myopathy, with atrophy and shift from type I ‘slow’ to type II ‘fast’ fibres (Lipkin et al. 1988; Sullivan et al. 1990; Mancini et al. 1992; De Sousa et al. 2000). HF induces skeletal muscle myosin heavy chain (MHC) isoform expression towards the fast isoform (Simonini et al. 1996; Vescovo et al. 1998), which is related to HF severity (Spangenburg et al. 2002; Carvalho et al. 2003).
Different pathways regulate skeletal muscle MHC expression (Allen et al. 2001), including myogenic regulatory factors (MRFs), a family of transcriptional factors that control the expression of several skeletal muscle-specific genes. The family has four members: MyoD, myogenin, Myf5 and MRF4. MRFs form dimers with ubiquitous E proteins (e.g. E12 or E47), resulting in heterodimeric complexes that bind to the E-box consensus DNA sequence (5′-CANNTG-3′) that is found in the regulatory region of many muscle-specific genes (Murre et al. 1989), including the MHCIIB gene (Wheeler et al. 1999). During embryogenesis, MRFs are critical for establishing myogenic lineage and controlling terminal differentiation of myoblasts (for a review, see Parker et al. 2003). Previous studies have suggested that myogenin and MyoD may also be involved in establishing and maintaining slow and fast mature muscle fibre phenotype; myogenin is expressed at higher levels compared with MyoD in slow muscles, whereas the opposite is true for fast muscles (Hughes et al. 1993; Voytik et al. 1993). Similarly, MyoD is associated with the expression of fast type IIX and IIB MHC isoforms (Hughes et al. 1993; Hughes et al. 1997; Kraus & Pette 1997; Mozdziak et al. 1998; Seward et al. 2001). Therefore, it is reasonable to suggest that MRFs contribute to mechanisms controlling skeletal muscle MHC expression during HF.
This investigation was undertaken to examine MRF mRNA expression and its potential relationships with changes in MHC composition in young Wistar rat skeletal muscle with monocrotaline (MCT)-induced HF. To compare muscles with different fibre-type composition and function, we performed analyses in the fast phasic extensor digitorum longus (EDL) and slow postural soleus (Sol) muscles.
Methods
Heart failure induction
Seventeen weaned male Wistar rats (3–4 weeks old; 80–100 g) were obtained from the Central Animal House at São Paulo State University. HF was experimentally induced in 10 rats (HF group) by a single intraperitoneal (i.p., 30 mg/kg) injection of MCT, a widely accepted HF model (Vescovo et al. 1998; Dalla Libera et al. 1999; Dalla Libera et al. 2001; Leineweber et al. 2002). MCT is a pyrrolizidine alkaloid that induces pulmonary vascular disease with severe right ventricle hypertrophy and failure (Vescovo et al. 1989; Reindel et al. 1990) without itself producing changes in skeletal muscle MHC composition (Vescovo et al. 1998). Preliminary experiments revealed that 30 mg/kg i.p. was an appropriate dose of MCT for our animals with regard to survival and HF induction. MCT-treated rats were allowed to eat freely from a supply of standard rat cubes. Seven control rats (CT group) were injected with saline and were given the same quantity of food as consumed on the previous day by the treated rats. HF and CT rats were studied 22 days after MCT administration when the HF group had developed overt HF. After anaesthesia with i.p. sodium pentobarbital (50 mg/kg), the animals were killed; body weight (BW), Sol weight and EDL weight were evaluated. The Sol/BW and EDL/BW ratios were used as indexes of muscle atrophy. Muscles were immediately frozen in liquid nitrogen and stored at −80 °C. Left ventricle weight (LVW), right ventricle weight (RVW) and atrium weight normalized by BW (LVW/BW, RVW/BW and ATW/BW, respectively) were used as indexes of heart hypertrophy. This experiment was approved by Ethics Committee of Instituto de Biociências, UNESP, Botucatu, SP, Brazil.
Semi-quantitative reverse transcription-polymerase chain reaction analyses of mRNA for MRF genes
Total RNA was extracted from Sol and EDL muscles with TRIzol Reagent (Invitrogen Life Technologies, Carlsbad, CA, USA), which is based on the guanidine thiocyanate method. Frozen muscles were mechanically homogenized on ice in 1 ml of ice-cold TRIzol reagent. Total RNA was solubilized in Rnase-free H2O, incubated in Dnase I (Invitrogen Life Technologies) to remove any DNA present in the sample and quantified by measuring the optical density (OD) at 260 nm. RNA purity was ensured by obtaining a 260/280 nm OD ratio of approximately 2.0. Two micrograms of RNA was reverse-transcribed with random hexamer primers and Superscript II reverse transcription (RT) in a total volume of 21 µl, according to standard methods (Invitrogen Life Technologies). Control ‘No RT’ reactions were performed by omitting the RT enzyme. These reactions were then polymerase chain reaction (PCR)-amplified to ensure that DNA did not contaminate the RNA. One microlitre of cDNA was then amplified using 1 µm of each primer (Table 1), ×1 PCR buffer minus Mg, 5 mm MgCl2, 1 mm deoxyribonucleotide triphosphates and 2 units of Platinum® Taq DNA Polymerase (Invitrogen Life Technologies, São Paulo, SP, Brazil) in a final volume of 25 µl. Primer pairs for MyoD were designed from a sequence published in GenBank; myogenin and MRF4 primer sequences were those used by Smith et al. (1994). Preliminary experiments were conducted with each gene to determine the number of PCR cycles that represented the linear range of amplification. All PCR products were verified by restriction digestion or by sequencing. The cDNA from each muscle for both CT and HF groups were amplified simultaneously using aliquots from the same PCR mixture. After the PCR amplification, 10 µl of each reaction underwent electrophoresis on 1.0% agarose gels and stained with ethidium bromide. Images were captured and the bands corresponding to each gene were quantified by densitometry as Integrated Optical Density. PCR products were run in duplicate on different gel for each gene, and results averaged. The size (the number of base pairs) of each band corresponded to the size of processed mRNA. The PCR signals were normalized to the housekeeping gene cyclophilin (Alway et al. 2002a).
Table 1.
Oligonucleotide primers used for polymerase chain reaction (PCR) amplification of reverse transcribed RNA
| Product | Accession number | Sequence | Start position | TA (°C) | Cycles | PCR length (bp) | Restriction enzyme | Restriction products (bp) |
|---|---|---|---|---|---|---|---|---|
| MyoD | M84176 | 5′-GACGGCTCTCTC TGCTCCTT | 259 | 60 | 32 | 544 | Sequenced | |
| 3′-GTCTGAGTCGCCGCTGTAGT | 782 | |||||||
| Myogenin | M24393 | 5′-TGCCACAAGCCAGACTACCCACC | 827 | 63 | 31 | 246 | Pst I | 234,80,65 |
| 3′-CGGGGCACTCACTGTCTCTCAA | 1050 | |||||||
| MRF4 | M30499 | 5′-AGAGACTGCCCAAGGTGGAGATTC | 491 | 63 | 32 | 272 | Pst I | 118,96,57 |
| 3′-AAGACTGCTGGAGGCTGAGGCATC | 1344 | |||||||
| Cyclophylin | M19533 | 5′-ACGCCGCTGTCTCTTTTC | 9 | 57.7 | 32 | 440 | HindII | 40,400 |
| 3′-TGCCTTCTTTCACCTTCC | 431 |
Accession number, GenBank accession number; TA, annealing temperature.
Electrophoretic separation of MHC
MHC isoform analysis was performed by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE). Six to 10 serial cross sections (12 µm thick) were placed in 450 µl of a solution containing 10% (wt/vol) glycerol, 5% (v/v) 2-mercaptoethanol, 2.3% (wt/vol) SDS and 0.9% (wt/vol) Tris/HCl (pH 6.8) for 10 min at 60 °C. Small amounts of the extracts (6 µl) were loaded on a 7–10% SDS-PAGE separating gel with a 4% stacking gel, run overnight (19–21 h) at 120 V and silver stained. MHC isoforms were identified according to molecular mass, and their relative percentages were quantified by densitometry.
Statistical methods
Data are expressed as mean ± SE. The comparisons between groups were performed using the Student's unpaired t-test and multivariate statistical analysis when appropriate. The level of significance was P < 0.05.
Results
Presence of heart failure in the monocrotaline-treated rats (Table 2)
Table 2.
Anatomical data of CT and HF groups
| Experimental groups | ||
|---|---|---|
| CT (n = 7) | HF (n = 10) | |
| BW (g) | 179.7 ± 2.8 | 182.2 ± 3.9 |
| LVW (g) | 0.42 ± 0.02 | 0.48 ± 0.01* |
| RVW (g) | 0.14 ± 0.01 | 0.37 ± 0.01*** |
| ATW (g) | 0.06 ± 0.00 | 0.10 ± 0.01** |
| LVW/BW (mg/g) | 2.32 ± 0.07 | 2.63 ± 0.08* |
| RVW/BW (mg/g) | 0.77 ± 0.06 | 2.03 ± 0.07*** |
| ATW/BW (mg/g) | 0.32 ± 0.02 | 0.54 ± 0.03** |
| RV W/D | 4.16 ± 0.06 | 4.68 ± 0.03*** |
| LV W/D | 4.13 ± 0.04 | 4.53 ± 0.02*** |
| AT W/D | 3.53 ± 0.23 | 5.24 ± 0.23** |
| Liver W/D | 3.37 ± 0.05 | 3.64 ± 0.07* |
| Lung W/D | 4.57 ± 0.27 | 4.79 ± 0.17 |
Values are means ± SE; n, number of animals; CT, control group; RCT, ration control group; HF, heart failure group; BW, body weight; LVW, left ventricle weight; RVW, right ventricle weight; ATW, atrium weight; W/D, wet-to-dry weight.
P < 0.05 statistical significance vs. control group.
P < 0.001 statistical significance vs. control group.
P < 0.0001 statistical significance vs. control group.
After 22 days, all 10 MCT-treated rats showed HF at post-mortem, confirmed by atrium and right ventricular hypertrophies (RVW/BW > 0.80), pleural and pericardial effusions and congested liver. No alterations were found in the CT rats.
There was no significant difference in BW between HF and CT groups. Heart weight was increased in HF group compared with CT group, as demonstrated by LVW, RVW and ATW and by the heart hypertrophy indexes (LVW/BW, RVW/BW and ATW/BW).
There was no significant difference in lung wet/dry ratio between CT and HF groups. However, RV, LV, AT and liver wet/dry ratio were greater in HF group than in CT group.
Muscle weight and indexes of muscle atrophy (Table 3)
Table 3.
Sol and EDL weight of CT and HF groups
| Experimental groups | ||
|---|---|---|
| CT (n = 7) | HF (n = 10) | |
| Sol (mg) | 98.7 ± 5.1 | 86.0 ± 3.4* |
| Sol/BW (mg/g) | 0.55 ± 0.03 | 0.47 ± 0.01* |
| EDL (mg) | 81.0 ± 7.7 | 85.3 ± 3.2 |
| EDL/BW (mg/g) | 0.45 ± 0.04 | 0.47 ± 0.01 |
Values are mean ± SE; n, number of animals; CT, control group; HF, heart failure group; BW, body weight; Sol, soleus weight; EDL, extensor digitorum longus weight.
P < 0.05 statistical significance vs. control group.
Sol weight and Sol/BW were significantly decreased in HF group compared with CT group. Both EDL weight and EDL atrophy index (EDL/BW) did not differ between CT and HF groups.
MRF mRNA levels estimated by semi-quantitative RT-PCR
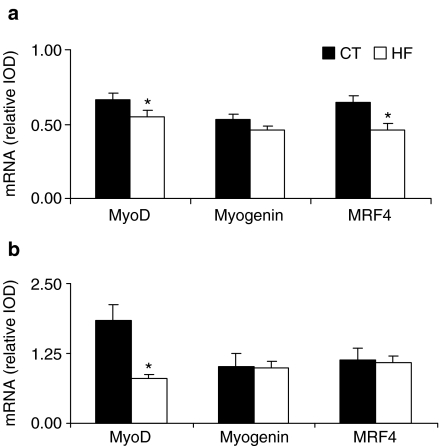
Soleus. MyoD and MRF4 mRNA levels decreased in the HF group compared with its CT group (CT = 0.67 ± 0.04 vs. HF = 0.55 ± 0.04 and CT = 0.64 ± 0.05 vs. HF = 0.46± 0.04, respectively) (P < 0.05). Myogenin mRNA level was similar between HF and CT groups (CT = 0.53 ± 0.04 vs. HF = 0.46 ± 0.03) (Figure 1a).
Figure 1.
MyoD, myogenin and MRF4 mRNA contents estimated by reverse transcription-polymerase chain reaction (RT-PCR) in control (CT) and heart failure (HF) groups from soleus (a) and extensor digitorum longus (EDL) (b) muscles. Data were run in duplicate on different gels for each gene, and the results were averaged. PCR products were visualized with ethidium bromide staining. Quantification of the PCR signal was obtained by densitometric analysis of the product as Integrated Optical Density (IOD). Gene expressions were normalized to the cyclophilin signal from the same RT product. Normalized data are expressed as means ± SE. *Data are significantly different from CT group at P < 0.05.
EDL. MyoD mRNA level decreased in the HF group compared with its CT group (CT = 1.83 ± 0.30 vs. HF = 0.80± 0.08, P < 0.05). Myogenin and MRF4 expressions were similar between HF and CT groups (CT = 1.01 ± 0.24 vs. HF = 0.99 ± 0.12 and CT = 1.14 ± 0.20 vs. HF = 1.09± 0.12, respectively) (Figure 1b).
MHCs electrophoretic pattern
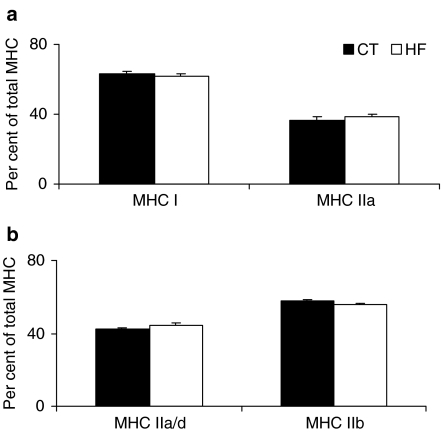
In Sol muscle, two MHC isoforms were separated, MHC1 and MHC2a; their expressions were not different between HF and CT groups (HF = 61.6 ± 1.6% vs. CT = 63.3 ± 1.7% and HF = 38.4 ± 1.6% vs. CT = 36.8 ± 1.7%, respectively) (Figure 2a). In EDL muscle, four MHC isoforms were separated (MHC1, MHC2a, MHC2d and MHC2b). The difference in electrophoretic migration between MHC2a and MHC2d is very small, and it was not possible to quantify them separately. Consequently, we considered the sum (MHC2a/2d) as both isoforms are metabolically similar. In EDL muscle, MHC2a/d and MHC2b were not different between HF and CT groups (HF = 44.5 ± 0.7% vs. CT = 42.5 ± 1.2% and HF = 55.5 ± 0.7% vs. CT = 57.5± 1.2%, respectively) (Figure 2b).
Figure 2.
Percentage distribution of myosin heavy chain (MHC) from soleus (a) and extensor digitorum longus (EDL) (B) muscles from control (CT) and heart failure (HF) groups. Data are expressed as means ± SE.
Discussion
The major finding in this study was that despite there being no change in MHC composition of rat skeletal muscles with MCT-induced HF, the mRNA relative expression of MyoD in Sol and EDL muscles and of MRF4 in Sol muscle were significantly reduced, whereas myogenin was not changed in either muscle.
The pattern of MHC expression during HF from this investigation diverges from previously published data that showed MHC isoform expression towards the fast isoform in chronic HF models such as myocardial infarction in rat (Simonini et al. 1996) and rabbit (Spangenburg et al. 2002), subtotal constriction of the suprarenal abdominal aorta in rabbit (Coirault et al. 1999) and ascending aorta stenosis in rat (Carvalho et al. 2003). The short duration of HF in our model could explain this. We could not push experiments further than 22 days because the severity of the MCT-induced HF in young Wistar rats inevitably leads to death in this time frame. It is noteworthy that the HF period in our study was short and unable to induce change in MHC composition while in other studies, young Sprague–Dawley rats injected with the same dose of MCT as in our experiment (30 mg/kg i.p.) developed HF with consequent changes in MHC composition after 27–30 days (Vescovo et al. 1998; Dalla Libera et al. 1999; Dalla Libera et al. 2001).
The synthesis of muscle-specific proteins may be associated with myogenin and MyoD mRNA levels. Myogenin is expressed at higher levels compared with MyoD in predominantly slow muscle, whereas MyoD is expressed at higher levels compared with myogenin in predominantly fast muscles of mature animals (Hughes et al. 1993; Voytik et al. 1993). MyoD is associated with the expression of fast type IIX and IIB MHC isoforms (Hughes et al. 1993; Hughes et al. 1997; Kraus & Pette 1997; Mozdziak et al. 1998; Seward et al. 2001). Additionally to the role of myogenin in establishing mature slow muscle fibre phenotype (Hughes et al. 1993; Voytik et al. 1993), it is also involved with oxidative gene expression and metabolic enzyme activity (Coirault et al. 1999; Hughes et al. 1999; Ekmark et al. 2003). Although we did not analyse these metabolic parameters in our study, they probably were not altered because the mRNA relative expression of myogenin was not changed. We also showed that HF induced a reduction in the mRNA relative expression of MyoD in both Sol and EDL muscles suggesting an MHC phenotypic adaptation towards a slower profile in both muscles, as the mRNA relative expression of myogenin was not changed. It is well known that MHC phenotypic adaptation in limb skeletal muscles during HF occurs towards a faster profile (Simonini et al. 1996; Vescovo et al. 1998; Spangenburg et al. 2002; Carvalho et al. 2003).
Interestingly, our results indicate that skeletal muscle fibre phenotype modulation during this syndrome is not exclusively dependent on the MRFs but may also be regulated by distinct pathways, such as those involving activated calcineurin and transcriptional factor nuclear factor of T cells (Chin et al. 1998; McCullagh et al. 2004), calcium-dependent CaM kinase (McKinsey et al. 2000; Wu et al. 2000), peroxisome proliferator-activated receptor-gamma coativator 1 (PGC1α) (Wu et al. 2001; Lin et al. 2002), Ras (Murgia et al. 2000) and the transcriptional complex Six1/Eya1 (Grifone et al. 2004).
What caused the down-regulation in MyoD mRNA expression during HF needs to be determined; however, cytokine activation (Anker et al. 1999) may have been involved. This point has been the subject of considerable debate given that tumour necrosis factor-α (TNF-α) is markedly increased in patients (Levine et al. 1990; McMurray et al. 1991) and Sprague–Dawley rats with HF (Dalla Libera et al. 1999; Dalla Libera et al. 2001). One hallmark of TNF-α is the activation of nuclear factor (NF)κB, a ubiquitous transcription factor normally inactive and sequestered in the cytoplasm through association with IκB (Baeuerle & Baltimore 1988). TNF-α exposure leads to the degradation of IκB, allowing NFκB translocation to the nucleus (Israël 2000) where it down-regulates MyoD mRNA at post-transcriptional level (Guttridge et al. 2000). This mechanism may partially explain the down-regulation of MyoD that we found in both Sol and EDL muscles. However, further studies need to be conducted to demonstrate this mechanism during HF.
The analysis of MRF4 mRNA expression revealed that, during HF, MRF4 mRNA transcripts were down-regulated in the slow Sol muscle but not in the fast EDL. MRF4 is expressed mainly after birth and is likely to have a role in the maintenance of skeletal muscles rather than the development, differentiation and regeneration processes which may be controlled by MyoD, myogenin and Myf5 expressions (Perry & Rudnick 2000). The concept that MRF4 is involved in regulating maturation events and maintaining adult skeletal muscle is further sustained by the fact that MRF4 is not expressed at substantial levels in limb until very late in fetal development (Hinterberger et al. 1991) and is present at higher levels in postnatal muscle (Voytik et al. 1993; Hughes et al. 1997). Interestingly, in this investigation, a down-regulation in the mRNA relative expression of MRF4 in Sol was associated with atrophy in response to HF while these alterations were not present in EDL muscle. This soleus-specific down-regulation of MRF4 was also previously observed in response to atrophy disuse of skeletal muscle in adult rats (Loughna & Brownson 1996) indicating a possible function for MRF4 in muscle atrophy. Additionally, it has been demonstrated that an increase in Id repressor protein expression, which forms heterodimers with MRFs and prevents their DNA binding (Benezra et al. 1990), is related to muscle atrophy in conditions such as disuse (Gundersen & Merlie 1994), hindlimb (Alway et al. 2002a) and ageing (Alway et al. 2002b). Taken together, these results shows that the potential of MRFs in muscle atrophy clearly requires further examination. Furthermore, the knowledge of the mechanisms controlling MRF expression during HF may explain many alterations in skeletal muscle-specific gene and protein expression during this syndrome.
Acknowledgments
This study was supported by Fundação de Amparo to Pesquisa do Estado de São Paulo (FAPESP, process n°2004/0156-1), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and CNPq (process n°410942/2003-0). The authors thank José Carlos Georgette for technical assistance. This work is part of the PhD thesis that will be presented by RFC to Universidade Estadual de Campinas – UNICAMP.
References
- Allen DL, Sartorius CA, Sycuro LK, Leinwand LA. Different pathways regulate expression of the skeletal myosin heavy chain genes. J. Biol. Chem. 2001;276:43524–43533. doi: 10.1074/jbc.M108017200. [DOI] [PubMed] [Google Scholar]
- Alway SE, Degens H, Krishnamurthy G, Smith CA. Potential role for Id myogenic repressors in apoptosis and attenuation of hypertrophy in muscles of aged rats. Am. J. Physiol. Cell Physiol. 2002a;283:C66–C76. doi: 10.1152/ajpcell.00598.2001. [DOI] [PubMed] [Google Scholar]
- Alway SE, Degens H, Lowe DA, Krishnamurthy G. Increased myogenic repressor Id mRNA and protein levels in hindlimb muscles of aged rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002b;282:R411–R422. doi: 10.1152/ajpregu.00332.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anker SD, Ponikowski PP, Clark AL, et al. Cytokines and neurohormones relating to body composition alterations in the wasting syndrome of chronic heart failure. Eur. Heart J. 1999;20:683–693. doi: 10.1053/euhj.1998.1446. [DOI] [PubMed] [Google Scholar]
- Baeuerle PA, Baltimore D. I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Carvalho RF, Cicogna AC, Campos GE, et al. Myosin heavy chain expression and atrophy in rat skeletal muscle during transition from cardiac hypertrophy to heart failure. Int. J. Exp. Pathol. 2003;84:201–206. doi: 10.1046/j.1365-2613.2003.00351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin ER, Olson EN, Richardson JA, et al. A calcineurin- dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coirault C, Langeron O, Lambert F, et al. Impaired skeletal muscle performance in the early stage of cardiac pressure overload in rabbits: beneficial effects of angiotensin-converting enzyme inhibition. J. Pharmacol. Exp. Ther. 1999;291:70–75. [PubMed] [Google Scholar]
- Dalla Libera L, Sabbadini R, Renken C, et al. Apoptosis in the skeletal muscle of rats with heart failure is associated with increased serum levels of TNF-alpha and sphingosine. J. Mol. Cell. Cardiol. 2001;33:1871–1878. doi: 10.1006/jmcc.2001.1453. [DOI] [PubMed] [Google Scholar]
- Dalla Libera L, Zennaro R, Sandri M, Ambrosio GB, Vescovo G. Apoptosis and atrophy in rat slow skeletal muscles in chronic heart failure. Am. J. Physiol. 1999;277(5)(Pt 1):C982–C986. doi: 10.1152/ajpcell.1999.277.5.C982. [DOI] [PubMed] [Google Scholar]
- De Sousa E, Veksler V, Bigard X, Mateo P, Ventura-Clapier R. Heart failure affects mitochondrial but not myofibrillar intrinsic properties of skeletal muscle. Circulation. 2000;102:1847–1853. doi: 10.1161/01.cir.102.15.1847. [DOI] [PubMed] [Google Scholar]
- Ekmark M, Gronevik E, Schjerling P, Gundersen K. Myogenin induces higher oxidative capacity in pre-existing mouse muscle fibres after somatic DNA transfer. J. Physiol. 2003;548(Pt 1):259–269. doi: 10.1113/jphysiol.2002.036228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifone R, Laclef C, Spitz F, et al. Six1 and Eya1 expression can reprogram adult muscle from the slow-twitch phenotype into the fast-twitch phenotype. Mol. Cell. Biol. 2004;24:6253–6267. doi: 10.1128/MCB.24.14.6253-6267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen K, Merlie JP. Id-1 as a possible transcriptional mediator of muscle disuse atrophy. Proc. Natl. Acad. Sci. USA. 1994;91:3647–3651. doi: 10.1073/pnas.91.9.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttridge DC, Mayo MW, Madrid LV, Wang CY, Baldwin AS., Jr NF-kappaB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science. 2000;289:2363–2366. doi: 10.1126/science.289.5488.2363. [DOI] [PubMed] [Google Scholar]
- Hinterberger TJ, Sassoon DA, Rhodes SJ, Konieczny SF. Expression of the muscle regulatory factor MRF4 during somite and skeletal myofiber development. Dev. Biol. 1991;147:144–156. doi: 10.1016/s0012-1606(05)80014-4. [DOI] [PubMed] [Google Scholar]
- Hughes SM, Chi MM, Lowry OH, Gundersen K. Myogenin induces a shift of enzyme activity from glycolytic to oxidative metabolism in muscles of transgenic mice. J. Cell Biol. 1999;145:633–642. doi: 10.1083/jcb.145.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SM, Koishi K, Rudnicki M, Maggs AM. MyoD protein is differentially accumulated in fast and slow skeletal muscle fibres and required for normal fibre type balance in rodents. Mech. Dev. 1997;61:151–163. doi: 10.1016/s0925-4773(96)00631-4. [DOI] [PubMed] [Google Scholar]
- Hughes SM, Taylor JM, Tapscott SJ, Gurley CM, Carter WJ, Peterson CA. Selective accumulation of MyoD and myogenin mRNAs in fast and slow adult skeletal muscle is controlled by innervation and hormones. Development. 1993;118:1137–1147. doi: 10.1242/dev.118.4.1137. [DOI] [PubMed] [Google Scholar]
- Israël A. The IKK complex: an integrator of all signals that activate NF-kappaB? Trends Cell Biol. 2000;10:129–133. doi: 10.1016/s0962-8924(00)01729-3. [DOI] [PubMed] [Google Scholar]
- Kraus B, Pette D. Quantification of MyoD, myogenin, MRF4 and Id-1 by reverse-transcriptase polymerase chain reaction in rat muscles – effects of hypothyroidism and chronic low-frequency stimulation. Eur. J. Biochem. 1997;247:98–106. doi: 10.1111/j.1432-1033.1997.t01-1-00098.x. [DOI] [PubMed] [Google Scholar]
- Leineweber K, Brandt K, Wludyka B, et al. Ventricular hypertrophy plus neurohumoral activation is necessary to alter the cardiac beta-adrenoceptor system in experimental heart failure. Circ Res. 2002;91:1056–1062. doi: 10.1161/01.res.0000045088.59360.b7. [DOI] [PubMed] [Google Scholar]
- Levine B, Kalman J, Mayer L, Fillit HM, Packer M. Elevated circulating levels of tumor necrosis factor in severe chronic heart failure. N. Engl. J. Med. 1990;323:236–241. doi: 10.1056/NEJM199007263230405. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Lipkin DP, Jones DA, Round JM, Poole-Wilson PA. Abnormalities of skeletal muscle in patients with chronic heart failure. Int. J. Cardiol. 1988;18:187–195. doi: 10.1016/0167-5273(88)90164-7. [DOI] [PubMed] [Google Scholar]
- Loughna PT, Brownson C. Two myogenic regulatory factor transcripts exhibit muscle-specific responses to disuse and passive stretch in adult rats. FEBS Lett. 1996;390:304–306. doi: 10.1016/0014-5793(96)00681-3. [DOI] [PubMed] [Google Scholar]
- Mancini DM, Walter G, Reichek N, et al. Contribution of skeletal muscle atrophy to exercise intolerance and altered muscle metabolism in heart failure. Circulation. 1992;85:1364–1373. doi: 10.1161/01.cir.85.4.1364. [DOI] [PubMed] [Google Scholar]
- McCullagh KJ, Calabria E, Pallafacchina G, et al. NFAT is a nerve activity sensor in skeletal muscle and controls activity-dependent myosin switching. Proc. Natl. Acad. Sci. USA. 2004;101:10590–10595. doi: 10.1073/pnas.0308035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray J, Abdullah I, Dargie HJ, Shapiro D. Increased concentrations of tumor necrosis factor in ‘cachectic’ patients with severe chronic heart failure. Br. Heart J. 1991;66:356–358. doi: 10.1136/hrt.66.5.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdziak PE, Greaser ML, Schultz E. Myogenin, MyoD, and myosin expression after pharmacologically and surgically induced hypertrophy. J. Appl. Physiol. 1998;84:1359–1364. doi: 10.1152/jappl.1998.84.4.1359. [DOI] [PubMed] [Google Scholar]
- Murgia M, Serrano AL, Calabria E, Pallafacchina G, Lomo T, Schiaffino S. Ras is involved in nerve-activity-dependent regulation of muscle genes. Nat. Cell Biol. 2000;2:142–147. doi: 10.1038/35004013. [DOI] [PubMed] [Google Scholar]
- Murre C, Mccaw PS, Vaessin H, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Parker MH, Seale P, Rudnicki MA. Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat. Rev. Genet. 2003;4:497–507. doi: 10.1038/nrg1109. [DOI] [PubMed] [Google Scholar]
- Perry RL, Rudnick MA. Molecular mechanisms regulating myogenic determination and differentiation. Front. Biosci. 2000;5:D750–D767. doi: 10.2741/perry. [DOI] [PubMed] [Google Scholar]
- Reindel JF, Ganey PE, Wagner JG, Slocombe RF, Roth RA. Development of morphologic, hemodynamic, and biochemical changes in lungs of rats given monocrotaline pyrrole. Toxicol. Appl. Pharmacol. 1990;106:179–200. doi: 10.1016/0041-008x(90)90239-q. [DOI] [PubMed] [Google Scholar]
- Seward DJ, Haney JC, Rudnicki MA, Swoap SJ. bHLH transcription factor MyoD affects myosin heavy chain expression pattern in a muscle-specific fashion. Am. J. Physiol. Cell Physiol. 2001;280:C408–C413. doi: 10.1152/ajpcell.2001.280.2.C408. [DOI] [PubMed] [Google Scholar]
- Simonini A, Massie BM, Long CS, Qi M, Samarel AM. Alterations in skeletal muscle gene expression in the rat with chronic congestive heart failure. J. Mol. Cell. Cardiol. 1996;28:1683–1691. doi: 10.1006/jmcc.1996.0158. [DOI] [PubMed] [Google Scholar]
- Smith CK, Janney MJ, Allen RE. Temporal expression of myogenic regulatory genes during activation, proliferation, and differentiation of rat skeletal muscle satellite cells. J. Cell Physiol. 1994;159:379–385. doi: 10.1002/jcp.1041590222. [DOI] [PubMed] [Google Scholar]
- Spangenburg EE, Talmadge RJ, Musch TI, Pfeifer PC, McAllister RM, Williams JH. Changes in skeletal muscle myosin heavy chain isoform content during congestive heart failure. Eur. J. Appl. Physiol. 2002;87:182–186. doi: 10.1007/s00421-002-0615-3. [DOI] [PubMed] [Google Scholar]
- Sullivan MJ, Green HJ, Cobb FR. Skeletal muscle biochemistry and histology in ambulatory patients with long-term heart failure. Circulation. 1990;81:518–527. doi: 10.1161/01.cir.81.2.518. [DOI] [PubMed] [Google Scholar]
- Vescovo G, Ceconi C, Bernocchi P, Ferrari R, Carraro U, Ambrosio GB, Libera LD. Skeletal muscle myosin heavy chain expression in rats with monocrotaline-induced cardiac hypertrophy and failure. Relation to blood flow and degree of muscle atrophy. Cardiovasc. Res. 1998;39:233–241. doi: 10.1016/s0008-6363(98)00041-8. [DOI] [PubMed] [Google Scholar]
- Vescovo G, Jones SM, Harding SE, Poole-Wilson PA. Isoproterenol sensitivity of isolated cardiac myocytes from rats with monocrotaline-induced right-sided hypertrophy and heart failure. J. Mol. Cell. Cardiol. 1989;21:1047–1061. doi: 10.1016/0022-2828(89)90803-1. [DOI] [PubMed] [Google Scholar]
- Voytik SL, Przyborski M, Badylak SF, Konieczny SF. Differential expression of muscle regulatory factor genes in normal and denervated adult rat hindlimb muscles. Dev. Dyn. 1993;198:214–224. doi: 10.1002/aja.1001980307. [DOI] [PubMed] [Google Scholar]
- Wheeler MT, Snyder EC, Patterson MN, Swoap SJ. An E-box within the MHC IIB gene is bound by MyoD and is required for gene expression in fast muscle. Am. J. Physiol. 1999;276(5)(Pt 1):C1069–C1078. doi: 10.1152/ajpcell.1999.276.5.C1069. [DOI] [PubMed] [Google Scholar]
- Wu H, Naya FJ, McKinsey TA, et al. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 2000;19:1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Rothermel B, Kanatous S, et al. Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. EMBO J. 2001;20:6414–6423. doi: 10.1093/emboj/20.22.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]