Abstract
In this study, we have created a conditional deletion of AP-2α in the developing mouse lens (Le-AP-2α mutants) to determine the cell-autonomous requirement(s) for AP-2α in lens development. Embryonic and adult Le-AP-2α mutants exhibited defects confined to lens placode derivatives, including a persistent adhesion of the lens to the overlying corneal epithelium (or lens stalk). Expression of known regulators of lens vesicle separation, including Pax6, Pitx3, and Foxe3 was observed in the Le-AP-2α mutant lens demonstrating that these genes do not lie directly downstream of AP-2α. Unlike germ-line mutants, Le-AP-2α mutants did not exhibit defects in the optic cup, further defining the tissue specific role(s) for AP-2α in eye development. Finally, comparative microarray analysis of lenses from the Le-AP-2α mutants vs. wild-type littermates revealed differential expression of 415 mRNAs, including reduced expression of genes important for maintaining the lens epithelial cell phenotype, such as E-cadherin.
Keywords: activating protein-2alpha, lens development, E-cadherin, epidermal growth factor receptor
Introduction
Development of the vertebrate ocular lens arises from a series of interactions and cumulative inductive signals between the surface ectoderm (SE) of the head, the neural plate, mesoderm and foregut endoderm, and later between the optic vesicle (OV) and neural crest-derived mesenchyme (Chow and Lang, 2001; Fisher and Grainger, 2004; Lovicu and McAvoy, 2005). The head ectoderm is predetermined or biased to form lens by early signals from the underlying neural plate. The presumptive lens ectoderm is then stimulated by the OV, an out-pocketing of the forebrain, to proliferate and thicken into the lens placode. The lens placode later invaginates to form the lens pit, which eventually separates from the surface ectoderm to form the lens vesicle, ultimately giving rise to the mature lens.
As with the development of other tissues and organs, lens development relies on a complex genetic hierarchy of multiple regulators. These regulators often act in combination to activate or repress downstream effectors that control determination and morphogenetic movements of cells and tissues. The genetic cascade controlling vertebrate lens development has been extensively studied. Previous work indicates that the homeobox containing transcription factors Pax6 and Six3, along with Bmp and Fgf signaling growth factors, are essential for lens ectoderm competence and growth (Wawersik et al., 1999; Faber et al., 2001; Liu et al., 2006). Furthermore, these upstream components are required for the activation of several downstream regulators required for proper lens development, some of which include, Prox1, c-Maf, Sox2/3, Pitx3, and FoxE3 (Brownell et al., 2000; Sakai et al., 2001; Chauhan et al., 2002; Lengler et al., 2005). Of interest, mutations in several of these genes have been shown to result in comparable lens phenotypes indicative of their related pathways yet in many cases the nature of these genetic pathways and the interaction between these genes remains unknown.
Activating Protein-2alpha (AP-2α) has been shown to be an important regulator of lens development, with germ-line mutant mice exhibiting lens defects reminiscent of mutants in the Pax6 regulatory pathway. There are five members of the AP-2 family, AP-2α, AP-2β, AP-2γ, AP-2δ, and AP-2∈, each encoded by separate genes (Tcfap2a, Tcfap2b, Tcfap2c, Tcfap2d, and Tcfap2e, respectively) with overlapping and distinct expression patterns in the developing embryo (Williams et al., 1988; Moser et al., 1995; Chazaud et al., 1996; Zhao et al., 2001; Tummala et al., 2003). The first AP-2 family member to be cloned, AP-2α, is expressed in multiple tissues contributing to the developing eye including the SE, forebrain, and neural crest cells and their derivatives. At embryonic day (E) 9.5, AP-2α and AP-2β are coexpressed in cells of the lens placode. However, as the lens separates from the surface ectoderm at E11, AP-2α becomes uniquely expressed in the lens epithelium (West-Mays et al., 1999b). Previous investigations of Tcfap2a germ-line (null) and chimeric mice have shown a requirement for AP-2α in development of the vertebrate eye and lens (Nottoli et al., 1998; West-Mays et al., 1999b). For example, Tcfap2a null mice exhibited a range of abnormal ocular phenotypes, including anophthalmia (lack of eyes) and persistent adhesion of the lens to the overlying surface ectoderm, forming a lens stalk. In addition to the lens, defects in the optic cup (OC), including the conversion of the dorsal retinal pigmented epithelium (RPE) into neural retina (NR) and aberrant retinal lamination, were observed in the Tcfap2a null mice (Zhang et al., 1996; West-Mays et al., 1999b). Because AP-2α is expressed in both the developing lens placode and the developing NR, tissues that provide critical signals to each other during development, it remained unclear whether or not the ocular defects observed in the Tcfap2a null mouse were due to the intrinsic loss of AP-2α expression. Furthermore, due to its broader developmental expression pattern, deletion of AP-2α in mice also led to additional developmental defects in the head, including excencephaly and failure of the anterior neural tube to close, as well as a dysmorphogenesis of the developing face (Zhang et al., 1996; West-Mays et al., 1999b). Thus, these defects may have also contributed, in a non–cell-autonomous manner, to the ocular defects observed in the Tcfap2a mutants. These findings combined with the scarcity in number of null mutants to examine (owing to embryonic lethality) and variability in ocular phenotypes, has hampered the genetic screening analyses needed to further understand the regulatory roles AP-2α in lens and eye development.
Using Cre-loxP technology, we have created a conditional deletion of Tcfap2a in the developing lens placode (Le-AP-2α mutants) to determine the cell-autonomous requirement(s) for AP-2α in lens development. Embryonic and adult Le-AP-2α mutants exhibited defects confined to lens placode derivatives, including a persistent adhesion of the lens to the overlying corneal epithelium (or lens stalk). The expression of Pax6, Pitx3, and Foxe3, was observed in the Le-AP-2α mutant lens epithelium demonstrating that, while mutations (or deletions) in the genes encoding these transcription factors are associated with lens separation defects, they do not lie downstream of AP-2α. Unlike Tcfap2a germ-line mutants, the Le-AP-2α mutants did not exhibit defects in the OC, further defining the tissue specific role(s) for AP-2α in eye development. Finally, comparative microarray analysis of lenses from Le-AP-2α mutants vs. wild-type littermates revealed differential expression of 415 mRNAs, including reduced expression of genes important for maintaining the lens epithelial cell phenotype, such as E-cadherin. Together, data from the current study has revealed cell-autonomous roles for AP-2α in lens vesicle separation and maintenance of the lens epithelial cell phenotype.
Results
Targeted Deletion of AP-2α Results in Failure in Lens Vesicle Separation
To reveal the cell-autonomous requirement(s) for AP-2α in lens development, a line of mice possessing a conditional knockout of AP-2α in the lens placode (Le-AP-2α) was created using the Cre/loxP recombination approach (Gu et al., 1994). Polymerase chain reaction (PCR) analysis confirmed the Cre-mediated deletion of the Alflox allele, and creation of the Floxdel allele in tissues derived from the lens placode, including the eyelids and lens (Fig. 1A). In contrast, no Cre-mediated recombination occurred in non–placodal-derived tissues, such as the ear and retina (Fig. 1A). AP-2α protein expression was also examined in the Le-AP-2α mutant lens at multiple stages. A representative section at mid-gestation, E15.5, demonstrated that AP-2α protein expression was absent in the lens epithelium as compared to its nuclear expression in the lens epithelium of wild-type littermates (Fig. 1B,C). The targeted deletion of AP-2α in the lens placode also resulted in deletion of AP-2α expression in lens placode derivatives, including the corneal epithelium (Fig. 1B,C), the margins of the epidermis of the eyelids (Fig. 1D,E), as well as in a ridge of facial epidermis extending from the eyelids toward the snout (Fig. 1F,G).
Fig. 1.
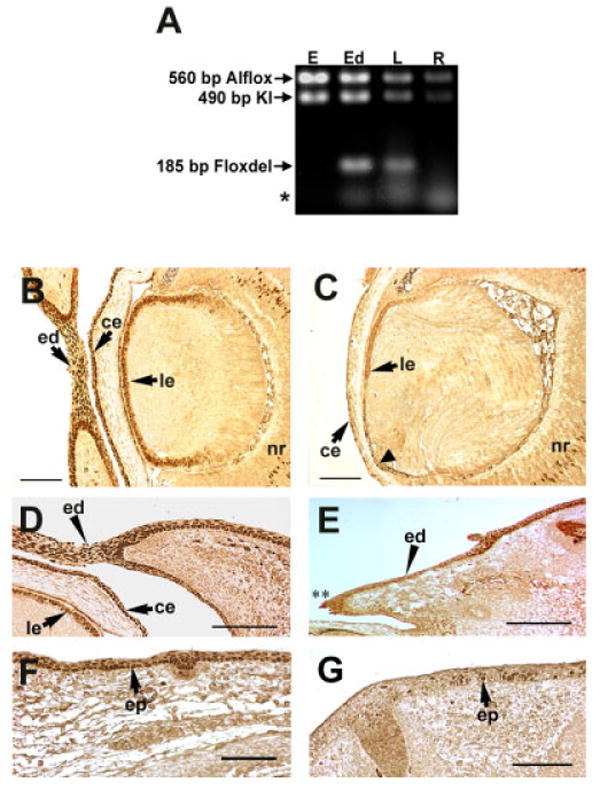
Conditional knockout of AP-2α within lens placode derivatives. A: Polymerase chain reaction (PCR) analysis of the Le-Cre-mediated deletion of the Alflox allele. All Le-AP-2α mouse tissues (E, ear; Ed, eyelid; L, lens; R, retina) contain the AP-2α:LacZ KI allele (490 bp). In addition, these tissues also contain the undeleted Alflox allele (560 bp). The deleted conditional allele (Floxdel; 185 bp) band is only detected in lens placode-derived tissues in the presence of Le-Cre. Asterisk denotes primer dimers. B-G: Immunohistochemical analysis of AP-2α protein expression within lens placode derivatives. All panels show paraffin-embedded sections of E15.5 embryos labeled with anti–AP-2α antibodies. There is an absence of AP-2α protein expression within all lens placode derivatives of Le-AP-2α mice (C,E,G) as compared with control mice (B,D,F), including the developing lens epithelium (compare B and C, arrowhead in C), the developing eyelid epidermis (compare D and E, arrowhead; ** region of open eyelid), and the developing facial epidermis (compare F and G, arrow). ed, eyelid; ce, corneal epithelium; le, lens epithelium; nr, neural retina; ep, epidermis. Scale bars = 100 μm.
Examination of the external phenotypes of the Le-AP-2α mice revealed that deletion of AP-2α expression within lens placode derivatives resulted in the formation of ectodermal defects that persisted throughout adulthood. For example, in comparison to their wild-type littermates, at postnatal day (P) 0 and P21, Le-AP-2α mutant mice possessed an epidermal cleft that originated at the edge of the nasal aspect of the cornea and extended toward the snout (Fig. 2A,B). Additionally, these animals exhibited open eyelids at birth (data not shown). External examination of the corneal surface revealed the presence of pigment granules positioned nasally in both adult and embryonic mutants (Fig. 2C,D). Further defects in corneal epithelial stratification were observed and has been described elsewhere (Dwivedi et al., 2005).
Fig. 2.
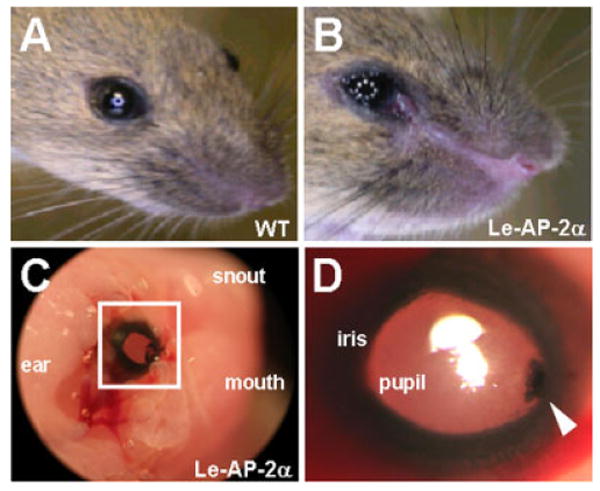
Le-AP-2α mutant mice possess ectodermal defects. A,B: Compared to control littermates (A) at postnatal day (P21), conditional inactivation of AP-2α within lens placode derivatives resulted in a distinct epidermal cleft that extended from the nasal aspect of the cornea toward the snout (B). C: Pigment granules, which were positioned nasally, were also observed on the corneal surface of P0 and adult (not shown) Le-AP-2α mutant mice. D: Increased magnification of the boxed area in C.
Histological analyses of Le-AP-2α mutant mice at multiple developmental stages (E9.5 to E18.5) revealed defects in lens development that were first evident at E12 (Fig. 3). In all mutants examined, a failure of the lens vesicle to separate from the overlying surface ectoderm was observed, yielding an anterior lens epithelium that was continuous with the surface ectoderm, referred to as the lens stalk (Fig. 3B,D,F). Of interest, the lens stalk consistently formed toward the nasal aspect of the cornea in all of the Le-AP-2α mutants. In addition to the defect in lens separation, at E15.5 pigment granules were observed in cells of the mutant lens stalk indicative of aberrant differentiation (Figs. 3D, 5D). In some mutants, the pigmentation of cells had expanded outside of the stalk and into cells lining the surface of the cornea, likely corresponding to the pigment detected on the surface of the corneas (Fig. 2C,D). In adult Le-AP-2α mutants, adhesions between the lens and cornea were also evident and in some cases, an adhesion between the lens and iris was observed (Fig. 3G).
Fig. 3.
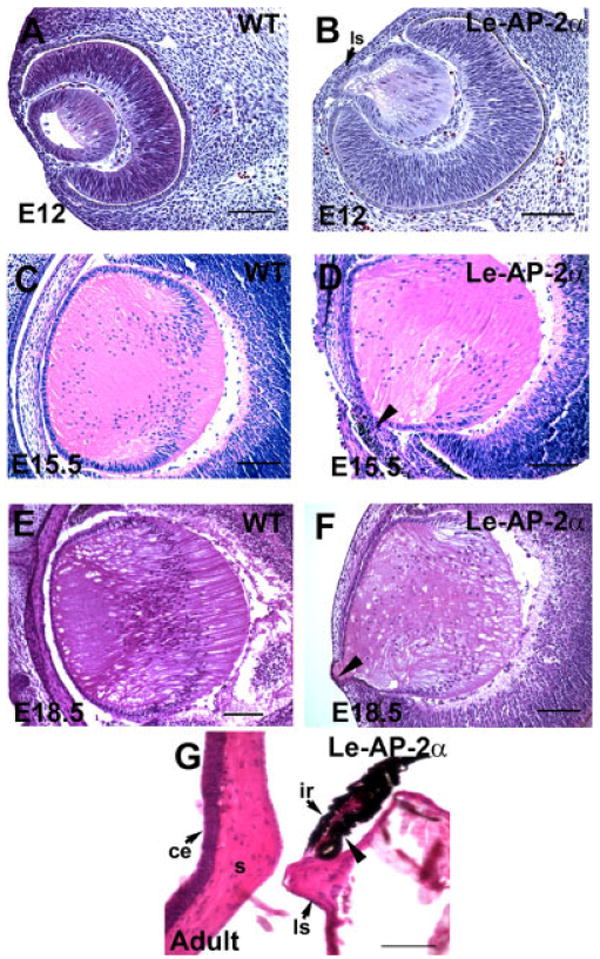
Histological analysis of Le-AP-2α mutant lenses. All panels show hematoxylin and eosin staining of paraffin-embedded sections. A,B: Control (A) and Le-AP-2α (B) mouse eyes at embryonic day (E12) showing that the lens vesicle in Le-AP-2α mice has failed to separate from the surface ectoderm resulting in the formation of a persistent lens stalk. C,D: At E15.5, control eyes (C) develop normally compared to their Le-AP-2α littermates (D) in which pigmentation granules appear in a proportion of cells located within the lens stalk (arrowhead). E,F: At E18.5, control mice (E) develop normally, whereas a lens stalk is observed in Le-AP-2α mice (F, arrowhead). G: Additionally, a proportion of adult Le-AP-2α mutant mice exhibit fusion of the lens to the iris (arrowhead). ls, lens stalk; ce, corneal epithelium; s, corneal stroma; ir, iris. Scale bars = 100 μm.
Fig. 5.

Expression of lens epithelial cell markers in Le-AP-2α mice. All panels show paraffin-embedded mouse embryos at embryonic day (E15.5). A,B: A characteristic Pax6 expression pattern is observed in wild-type (A) mouse embryos as well as in Le-AP-2α mice (B) with distinct staining in the lens stalk (B, inset). C,D: In situ hybridization using a Foxe3 specific digoxigenin (DIG) -labeled riboprobe indicates no overall change in Foxe3 gene expression between control (C) and Le-AP-2α (D) mice. However, cells within the lens stalk of Le-AP-2α embryos fail to express Foxe3 mRNA (D, inset [arrowhead]). Positive Foxe3 mRNA expression (purple stain in D and inset) does not extend into this stalk region, while observable pigment granules (brown stain) are present. E,F: Immunofluorescent staining indicates no overall change in Pitx3 protein expression between control (E) and Le-AP-2α (F) mouse embryos. However, cells within the lens stalk of Le-AP-2α embryos fail to express Pitx3 protein (F, inset (arrowhead). ed, eyelid; ce, corneal epithelium; le, lens epithelium; ls, lens stalk. Scale bars = 100 μm.
In our previous study of the AP-2α germ-line mutants, the lens defects were accompanied by defects in the OC, including a conversion of the RPE into NR on the dorsal aspect of the cup and lack of retinal lamination (West-Mays et al., 1999b). Because the lens is known to influence development of the OC, it was surmised that these defects may have occurred secondary to the defects in the lens. We, therefore, examined the OC morphology in the Le-AP-2α mutants to further determine whether the loss of AP-2α expression in the lens had influenced development of the OC. Although the overall size of the OC was reduced in the Le-AP-2α mutants, histological analysis at E15.5 and adult stages revealed no alteration in the morphology of the RPE or NR as compared with control littermates (Fig. 4A). Corroborating this result, similar retinal expression patterns for Pax6, syntaxin and calretinin were observed in adult Le-AP-2α mice as compared with wild-type littermates (Fig. 4B). Thus, the effects of the conditional deletion of AP-2α in the lens did not appear to have an impact on RPE development or retinogenesis.
Fig. 4.
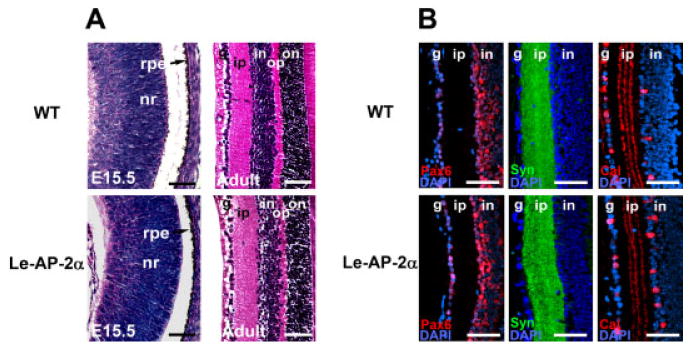
Retinogenesis in Le-AP-2α mutant mice. A: At embryonic day (E15.5), analysis of wild-type (top panels) and Le-AP-2α (bottom panels) embryos indicates no significant change in retinal development, while examination of adult eyes indicates no change in retinal lamination between control and Le-AP-2α mice. B: Immunofluorescent staining of paraffin-embedded sections for the retinal cell markers, Pax6 (red), syntaxin-1 (green), and calretinin (red) revealed no significant alteration in protein expression within the adult retinas of Le-AP-2α mutant mice compared to control littermates. Nuclei are counterstained with 4′,6-diamidine-2-phenylidole-dihydrochloride (DAPI, blue). nr, neural retina; rpe, retinal pigmented epithelium; g, ganglion cell layer; ip, inner plexiform layer; in, inner nuclear layer; op, outer plexiform layer; on, outer nuclear layer. Scale bars = 100 μm.
Expression Patterns of Regulators Associated With Lens Separation Defects Are Maintained in the Le-AP-2α Mutant Lens
To determine the position of AP-2α in previously established genetic pathways regulating lens separation, we examined the expression patterns of Pax6, Pitx3, and Foxe3, mutations and/or deletions of which have been shown to result in phenotypes consisting of a persistent adhesion of the lens and corneal epithelium, similar to that observed in the Le-AP-2α mutants. Strong immunoreactivity for Pax6 was observed in the lens epithelium and the corneal epithelium of the Le-AP-2α mutant lens similar to that observed in the wild-type littermates, and expression was also localized to cells lining the lens stalk (Fig. 5A,B). In situ hybridization using a Foxe3 specific antisense probe revealed that Foxe3 mRNA was expressed in the lens epithelial compartment of the both the wild-type and Le-AP-2α mutant lens. However, its expression was lost in the cells lining the stalk of the mutant lens (Fig. 5C,D). Of interest, many of the cells lacking the Foxe3 signal also exhibited pigment granules. Pitx3 was also detected in the epithelial compartment of the embryonic Le-AP-2α and wild-type lens, but like Foxe3 its expression was lost in cells lining the lens stalk (Fig. 5E,F). That the expression of all three regulators, Pax6, Pitx3, and Foxe3, was detected in the majority of epithelial cells of the Le-AP-2α mutant lens demonstrates that their expression is not dependent on that of AP-2α.
Apoptosis is a mechanism proposed to promote tissue separation during development, including that of the lens placode from the SE (Ozeki et al., 2001). Thus, apoptosis-mediated mechanisms were also investigated in the Le-AP-2α mutant using terminal deoxynucleotidyl transferase–mediated deoxyuridinetriphosphate nick end-labeling (TUNEL) analysis. No discernible difference in the number of TUNEL-positive cells was observed between the lenses of embryonic Le-AP-2α mutants and their wild-type littermates (data not shown), suggesting that a decrease in programmed cell death is not associated with the lens separation defect observed in Le-AP-2α mutant mice. Similarly, both proliferating cell nuclear antigen and Ki67 markers were used on the Le-AP-2α mutant sections during the lens separation stage to determine whether proliferation was altered. These studies also showed no overt difference in the numbers of proliferating cells between the mutants and their wild-type littermates (data not shown).
Microarray Analysis of AP-2α-Conditional Knockout Lenses Reveals Alterations in Lens Epithelial Cell Differentiation
To further understand how tissue-specific deletion of AP-2α impacts downstream gene expression during lens vesicle development and maintenance of the lens epithelial cell phenotype, differentially expressed mRNAs within the lenses of Le-AP-2α mutant mice vs. wild-type littermates were identified using an Affymetrix microarray screen. The microarray analysis was performed on RNA isolated from three pairs (R1-R3) of Le-AP-2α mutant and wild-type lenses at P0 (Fig. 6A), a time at which the lens stalk phenotype in the mutants remains intact. The average expression level for each transcript was calculated from the biological triplicates and the average fold change between the Le-AP-2α and wild-type groups was determined. Of the over 14,000 mouse genes represented in the array, 415 genes in the Le-AP-2α mutant group were significantly altered using the t-test or Significance Analysis of Microarrays (SAM) criteria as described in the Experimental Procedures section. The targets identified in the genetic screen were categorized according to Gene Ontology (GO) categories of biological process, molecular function, and cellular compartment (Fig. 6B). This analysis suggests that the majority of genes differentially regulated between wild-type and the Le-AP-2α mutant lens are involved in cellular differentiation, signal transduction, cellular adhesion, and cell–cell signaling. These data are consistent with the processes of lens vesicle separation and lens epithelial/fiber cell differentiation that are disrupted in the Le-AP-2α mutant mice. Furthermore, stratification of these data into molecular function indicated that the majority of genes differentially regulated between normal and mutant lenses lacking AP-2α in the lens, reside in the epidermal growth factor receptor (Egfr) and Janus kinase (Jak) signal transduction cascades, pathways previously implicated in cell differentiation and cellular migration during development (Kumar and Moses, 2001; Andl et al., 2004; Brown et al., 2006).
Fig. 6.

Gene expression profiling of wild-type and Le-AP-2α mutant lenses. A: Biological triplicates (R1 through R3) of mouse lenses were analyzed by Affymetrix GeneChip arrays to identify a set of 415 genes significantly modulated between wild-type (WT) animals and Le-AP-2α mutants. Statistical filtering of array data was performed as described in the Experimental Procedures section. B: Gene Ontology (GO) analyses (GO biological process, molecular function and cellular compartment) of mRNA of the 415 significantly modulated genes. The numbers show percentages of interrogated up-regulated (yellow) or down-regulated (blue) mRNAs, found represented in a specific GO category.
Previous studies have aimed at confirming the reliability of Affymetrix microarray results using various validation techniques (Shi et al., 2006; Strauss, 2006). As such, we did not undergo high-capacity validations of the array results using real-time quantitative polymerase chain reaction (RT-QPCR). However, we did analyze the expression of a subset of genes that were selected based on their role(s) in lens vesicle separation, lens epithelial differentiation and growth. Three genes found to be down-regulated in the Le-AP-2α mutant lens by microarray analysis (Cdh1, Bfsp1, and Etv6) as well as three genes that were up-regulated compared with wild-type (Egfr, Pak1, and L1cam) were selected for RT-QPCR analysis. As shown in Fig. 7, a significant reduction (2.48-fold) in Cdh1 (gene encoding E-cadherin) mRNA expression was observed in Le-AP-2α mutant lenses. Similarly, 1.56-fold and 1.41-fold reductions in gene expression were also observed for Bfsp1 (gene encoding filensin) and Etv6, respectively. Additionally, significant increases in mRNA expression were observed for Egfr (2.53-fold), Pak1 (3.41-fold), and L1cam (4.02-fold), suggesting our microarray screen provided reliable results (see Supplementary Table S1, which can be viewed at http://www.interscience.wiley.com/jpages/1058-8388/suppmat).
Fig. 7.
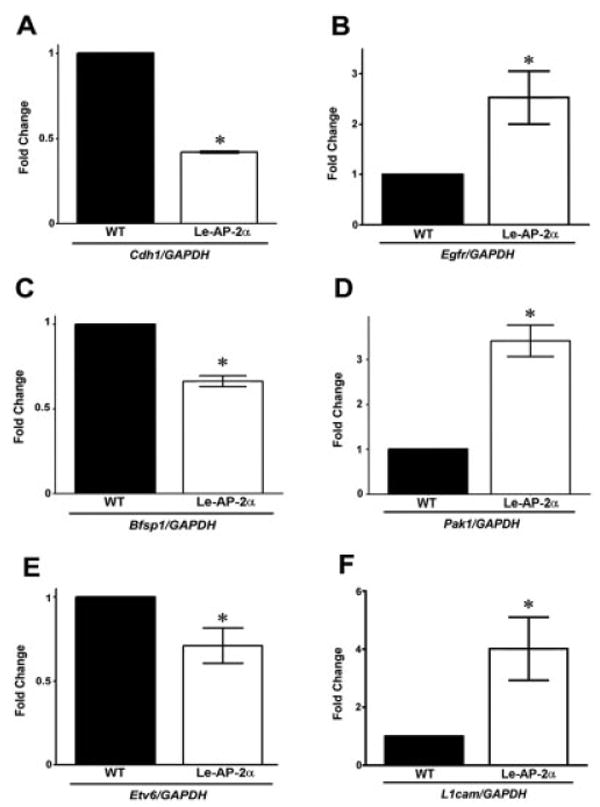
Real-time polymerase chain reaction (RT-PCR) analysis of Le-AP-2α mutant lenses. RT-PCR analysis was performed to confirm some of the genes found to be differentially regulated in Le-AP-2α mutant lenses in the microarray analysis. A–F: A significant reduction in mRNA expression was observed for Cdh1(A), Bfsp1 (C), and Etv6 (E), while a significant increase in mRNA transcripts was observed for Egfr (B), Pak1 (D), and L1cam (F). Data are presented as mean ± SEM. Fold change, relative to control littermates. Asterisks indicate P < 0.05 vs. wild-type (Student's t-test).
AP-2α Directly Interacts With Upstream Sequences of Differentially Regulated Genes in the Mouse Lens
Several genes identified in the array as being differentially expressed in the Le-AP-2α mutant lens vs. wild-type are known to contain potential AP-2α binding sites. To determine whether AP-2α associates in vivo with a subset of genes identified in the genetic screen, we assessed the binding of AP-2α to their promoters in chromatin prepared from newborn mouse lenses using quantitative chromatin immunoprecipitation (ChIP). Specifically, we investigated the promoter region of the mouse filensin, E-cadherin, and Fgf14 genes. Fgf14 was included because it was found to be up-regulated, 6.5-fold, in mutant lenses, and expression of Fgf14 has previously been observed in the mouse embryonic lens (Wang et al., 2000). As a negative control, a genomic region from chromosome 13 was analyzed. Our results (Fig. 8) revealed enrichments of Bfsp1, Cdh1, and Fgf14 promoter sequences, demonstrating the presence of AP-2α in promoter regions of these genes in lens chromatin.
Fig. 8.

AP-2α associates to upstream promoter sequences within the lens. qChIP was used to assess the binding of AP-2α to mouse filensin (Bfsp1), E-cadherin (Cdh1), and Fgf14 promoters in chromatin prepared from newborn wild-type (postnatal day [P] P0) mouse lenses. Enrichments of Bfsp1, Cdh1, and Fgf14 promoter sequences were observed after immunoprecipitation with a polyclonal anti–AP-2α antibody. A genomic region from chromosome 13 served as the negative control. Data are presented as mean ± SEM. Asterisks indicate P < 0.05 vs. control (one-way analysis of variance, Tukey's post hoc test).
Differential Expression of the Cadherins in the Le-AP-2α vs. Wild-Type Lens
Microarray and RT-QPCR analyses confirmed that Cdh1 (cadherin-1) mRNA expression was significantly reduced (58%) within the lenses of Le-AP-2α mutants vs. their wild-type littermates. E-cadherin belongs to a family of calcium-dependent adhesion molecules that have been shown to play critical roles in multiple morphogenetic processes, including tissue separation (Takeichi, 1988; Xu et al., 2002). Thus, to further understand the potential impact of loss of E-cadherin expression, we examined the expression pattern of E-cadherin protein in the Le-AP-2α mutant lens at various embryonic stages (Fig. 9A). At E10.5, a similar pattern of E-cadherin expression was observed within the invaginating lens vesicle and surface ectoderm, in both Le-AP-2α embryos and wild-type littermates. Two days later, at E12.5, expression of E-cadherin was observed in the anterior portion of the lens vesicle in control animals. In contrast, in the Le-AP-2α mutant mouse, E-cadherin was localized to the anterior region of the lens vesicle, yet its expression was also detected within the posterior region of the lens vesicle. At E13.5, reduced expression of E-cadherin was observed throughout the lens epithelium of Le-AP-2α embryos as compared to control counterparts, while almost all E-cadherin expression was lost between the cells of the lens stalk in the mutant tissue. As the lens continued to develop (E15.5), there was a further reduction in total E-cadherin expression in the lens epithelium of mutant mice, while no expression of E-cadherin was observed in the cells comprising the lens stalk. Examining E-cadherin expression postnatally (P0) revealed a further decrease in expression as compared to wild-type embryos, suggesting that the loss in E-cadherin expression in mutant mice occurs in a time-dependent manner. The decrease in E-cadherin protein expression in Le-AP-2α lens, was further confirmed with western blot analyses. Immunoblot analysis on adult lens tissue (4 weeks old) confirmed that there is a significant reduction (49.1%) in E-cadherin protein expression within the lenses of Le-AP-2α mutant mice compared to that of wild-type littermates (Fig. 9B).
Fig. 9.

E-cadherin expression in Le-AP-2α mice. A: Immunofluorescent staining of paraffin-embedded sections at various developmental stages (E 10.5, E12.5, E13.5, E15.5, postnatal day P0) revealed a gradual reduction of E-cadherin protein expression within the lenses of Le-AP-2α mutant mice compared to control littermates. No E-cadherin expression was observed in the cells lining the lens stalk (dotted outline). B: Western blot analysis of adult (4 weeks old) control and Le-AP-2α mouse lenses confirmed a significant decrease in E-cadherin expression in mutant mice. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein expression served as the loading control. Data are presented as relative light units (mean ± SEM) with representative immunoblot (E-cadherin and GAPDH) below. Asterisks indicate P < 0.05 vs. wild-type lens tissue (Student's t-test).
Additional cadherin family members, N-cadherin and P-cadherin, are also known to be temporally and spatially expressed in the developing lens (van Raamsdonk and Tilghman, 2000). Although significant differences in N-cadherin and P-cadherin mRNAs were not revealed in the microarray analyses, we wanted to observe if their protein expression patterns were in any way altered in the Le-AP-2α mutant lens during development. As shown in Figure 10A, at E10.5, there was no overt difference in the N-cadherin expression pattern between Le-AP-2α and control eyes, with normal expression of N-cadherin in the invaginating lens and optic vesicle. However, beginning at E13.5 and continuing onward (E15.5, P0), N-cadherin protein levels were reduced specifically within the lens stalk region of Le-AP-2α mutant mice. Similarly, at E17.5, while normal expression of P-cadherin was observed in the corneal epithelium of Le-AP-2α mutant mice, aberrant P-cadherin expression was evident in cells of the lens stalk (Fig. 10B). In comparison, P-cadherin expression was absent in the lens epithelium of wild-type littermates. In contrast to wild-type littermates, which exhibited positive expression of P-cadherin in the corneal endothelium, P-cadherin immunolocalization within the corneal endothelium was absent in the Le-AP-2α mutants.
Fig. 10.

N-cadherin and P-cadherin expression in Le-AP-2α mice. A: Immunofluorescent staining of paraffin-embedded sections at various developmental stages (embryonic day E10.5, E12.5, E13.5, E15.5, postnatal day P0) revealed no change in N-cadherin protein expression at any of the developmental stages examined. No N-cadherin expression was observed in the cells lining the lens stalk (dotted outline). B: Immunofluorescent staining of frozen sections at E17.5 revealed no alteration in P-cadherin protein expression (red) within the corneal epithelium of wild-type (top panels) and Le-AP-2α mice (middle and bottom panels). While P-cadherin protein expression was maintained within the corneal endothelium of wild-type mice, its expression was discontinuous within the endothelium of Le-AP-2α corneas. Positive P-cadherin immunoreactivity was observed in the cells lining the lens stalk, specifically in cells exhibiting distinct pigmentation (arrowheads in phase-contrast image (bottom-right panel). Bottom panels are increased magnifications of boxes in the middle-right, and nuclei are indicated by 4′,6-diamidine-2-phenylidole-dihydrochloride (DAPI, blue). ce, corneal epithelium; cd, corneal endothelium; ls, lens stalk. Scale bars = 100 μm.
A loss in E-cadherin expression accompanied by an increase in Egfr expression/signaling has been intimately linked to the process known as epithelial to mesenchymal transition (EMT; Hay, 1995), in which epithelial cells undergo alterations in general cellular architecture and signaling capabilities. One hallmark feature of this transformation is the up-regulation of the contractile element α-smooth muscle actin (α-SMA). Although, we observed approximately 2.4-fold differential expression of α-SMA (Acta2) in the Le-AP-2α mutant lenses compared with controls at the P0 stage examined (data not shown), our stringent statistical filtering of the microarray data failed to include the gene (t-test; P = 0.38) into the final list of 415 genes (Supplementary Table S1). Therefore, to confirm that the lens epithelium expresses α-SMA in parallel with the eventual loss of E-cadherin protein, α-SMA protein expression was examined within the lens epithelium of adult Le-AP-2α mutants and wild-type littermates. Positive α-SMA immunoreactivity was observed within the lens epithelium of 6-week-old Le-AP-2α mice, while the lens epithelium of wild-type mice did not exhibit positive expression (Fig. 11).
Fig. 11.

The α-smooth muscle actin (α-SMA) expression in Le-AP-2α mice. An induction of α-SMA protein immunoreactivity (green) was observed within the lens epithelium of adult (6 weeks old) Le-AP-2α mice (right panels). This finding contrasted with wild-type littermates (left panels), which displayed normal expression of α-SMA within the iris. Positive α-SMA immunoreactivity within the cytoplasm of mutant lens epithelial cells is indicated by arrowheads in middle panels, while the presence of distinct nuclei within the lens epithelial layer is indicated by the 4′,6-diamidine-2-phenylidole-dihydrochloride (DAPI) stain (blue) in the bottom panels (increased magnification). le, lens epithelium; ir, iris.
Discussion
Conditional deletion of AP-2α using Le-Cre from E9.5 and onward, resulted in abolished expression of AP-2α in the lens placode and its derivatives, including the lens, corneal epithelium, and eyelid epidermis. The Le-AP-2α mutants exhibited lens defects, including a failure in lens vesicle separation from the overlying ectoderm, resulting in a persistent corneal–lenticular adhesion (lens stalk). A lens stalk phenotype had also been observed in the Tcfap2a germ-line and chimeric mutant mice (West-Mays et al., 1999b). AP-2α is expressed in multiple tissues of the eye during critical periods of development. Therefore, it remained uncertain whether the lens phenotype observed in the Tcfap2a germ-line null mouse was caused by a combination of cell-autonomous and non–cell-autonomous effects. This hypothesis can now be eliminated because targeted deletion of AP-2α in the lens placode resulted in an isolated lens vesicle phenotype demonstrating a cell-autonomous requirement for AP-2α in lens vesicle separation.
Unlike the Tcfap2a germ-line mutants, the Le-AP-2α mutants did not exhibit defects in the OC. Because it is well established that OC development is influenced by the developing lens, we had surmised in an earlier investigation that the defects in the NR and RPE in the Tcfap2a germ-line mutants may have been attributed in part to a loss in signals from the mutant lens (West-Mays et al., 1999b). However, as we have shown herein, targeted deletion of AP-2α in the lens did not result in defects in the NR or RPE specification, which would suggest that either intrinsic loss of AP-2α expression in the developing NR or surrounding tissues (other than the lens) caused the OC defects in the germ-line mutants. AP-2α is expressed in cells of the NR, namely, the inner nuclear and ganglion cell layers during earlier stages of development, becoming further restricted to the amacrine cell population in the adult retina (Bassett et al., 2007). The potential role of AP-2α in retinogeneis has recently been examined by our lab through targeted deletion of AP-2α in the developing NR (Bassett et al., 2007). This study showed that retinal-specific deletion of AP-2α did not lead to retinal defects, suggesting that AP-2α alone is not intrinsically required for retinogenesis. However, a closely related family member, AP-2β, was found to be expressed in the developing NR with striking resemblance to that of AP-2α, and may functionally compensate for its loss during retinogenesis. AP-2α is also expressed in the cranial neural crest cell population, which is known to influence RPE development. Thus, further studies are required to determine the cell-autonomous and non–cell-autonomous roles for AP-2α in OC development.
AP-2α and Lens Vesicle Separation
In addition to Tcfap2a, several other genetic mutations in mice and humans have been shown to result in corneal–lenticular adhesions (lens stalk) and in humans this often referred to as Peter's anomaly (Theiler and Varnum, 1981; Hill et al., 1991; Favor et al., 1997; Prosser and van Heyningen, 1998; Blixt et al., 2000; Brownell et al., 2000; Semina et al., 2000; Collinson et al., 2001; Rieger et al., 2001). Some of these genes, such as Pax6, Foxe3, and Pitx3 are known to act in combination or in a linear pathway (Chow and Lang, 2001). For example, Foxe3 expression was found to be absent in Sey/Sey (Pax6 null) mouse mutants, and its expression diminished in Pax6 enhancer null mutants. Thus, it has been placed downstream of Pax6 in lens development (Chow and Lang, 2001). To determine whether AP-2α fits in this pathway, we examined the expression pattern of these three candidates in the Le-AP-2α mutant lens. Our studies revealed that expression of these candidates was not abolished, indicating that AP-2α does not lie directly upstream of Pax6, Foxe3, and Pitx3 in regulating lens separation and development. In previous investigations of Pax6 expression in the Tcfap2a germ-line mutants, a loss in Pax6 expression was observed in later stages of lens development and we had suggested that this finding may have occurred due to the lack of a true lens epithelial cell compartment or the fact that AP-2α is a required regulator of Pax6 expression (West-Mays et al., 1999b). Given the findings of the current study, the absence in Pax6 expression in the germ-line mutant was likely caused by the former hypothesis involving a reduction or loss in the epithelial compartment.
It must also be considered that AP-2α may lie downstream of the known regulators in the Pax6-regulated cascade that mediate lens separation, such as Pax6 or Foxe3. However, examination of AP-2α expression in the lens of both the Small eye (Pax6) and dysgenetic (dyl) lens (Foxe3) mutants has revealed that it is maintained (J.A. West-Mays, unpublished observations), suggesting that this is not the case. An alternative possibility to explain the phenotypic resemblance of the AP-2α mutant lens with that of the Pax6, Foxe3, and Pitx3 mutants is that these regulators act in parallel to influence the expression of common downstream genes involved in lens vesicle separation. Previous evidence suggests that AP-2α and Pax6 cooperate to control downstream gene expression. For example, Pax6 and AP-2α have been shown in cell culture experiments to control the expression of many common downstream genes, including members of the cadherins (Behrens et al., 1991; Chalepakis et al., 1994; van Raamsdonk and Tilghman, 2000), matrix metalloproteinases (Sivak et al., 2004), and interact with other common modulators of cell cycle during lens differentiation, such as the retinoblastoma protein Rb (Batsche et al., 1998; Cvekl et al., 1999). Also, a recent study from our lab has shown that mice double heterozygous for AP-2α and Pax6 (Pax6+/lacZ/Tcfap2a+/−), exhibit a more severe lens phenotype than that of the single heterozygote mice, further suggesting that the AP-2α and Pax6 pathways cooperate during lens development (Makhani et al., 2007). Future studies involving direct comparisons of microarray data obtained from phenotypically similar mutants, such as these, may help to further pinpoint commonly altered downstream targets, and further define critical pathway(s) involved in lens vesicle separation.
Although Pitx3 and Foxe3 expression was detected in the Le-AP-2α mutant lens, the expression of both was lost in cells lining the mutant lens stalk. Because Pitx3 and Foxe3 are not expressed in the corneal epithelium, it could be argued that loss in expression of these regulators in the stalk cells may reflect that these cells have adopted a more corneal epithelial cell phenotype. Yet, unlike both corneal and lens epithelial cells, the cells lining the surface of the lens stalk possessed pigment granules, indicative of a more aberrant trans-differentiation event. Of interest, Tcfap2a chimeric mice (derived from a population of Tcfap2a−/− and Tcfap2a+/+ cells) also exhibited pigmentation of cells in the lens stalk region (West-Mays et al., 1999b). The close proximity of the pigmented cells contributing to the developing iris with those of the lens stalk suggests the possibility that these cells could have aberrantly migrated into the stalk region during lens vesicle closure. This explanation has been described for a human patient diagnosed with Peter's anomaly, in which pigment granules observed within the mid-peripheral cornea were thought to be derived from iris tissue (Ohkawa et al., 2003). However, it has been shown that many different types of epithelial cells, such as those in the developing NR, can convert to an RPE phenotype (Gotoh et al., 2004). A recent study has also shown that human lens epithelial cells express components of the melanin synthesis pathway, and are, therefore, equipped to synthesize pigment granules under certain conditions (Wang et al., 2005). Thus, deletion of AP-2α expression in the lens may have rendered cells in the lens stalk cells permissive to signals for trans-differentiation to RPE. Indeed, the cells of the stalk in the Le-AP-2α mutant lens also showed aberrant expression of P-cadherin (Fig. 10B) which is not normally detected in the lens but is expressed by RPE cells.
Spectrum of Genes Altered in the Absence of AP-2α in the Lens
The Affymetrix microarray technology was used in our study to identify the spectrum of genes altered in the Le-AP-2α mutant lens. This genetic screen identified 415 distinct genes (434 transcripts) that were differentially expressed within the lens, including 144 genes that were up-regulated and 271 genes that were found to be down-regulated. The microarray data were subjected to gene ontology mining analysis to implicate specific pathways affected by deletion of AP-2α within the lens. This analysis stratified the array results according to three ontology branches, “biological process,” “molecular function,” and “cellular compartment.” The results suggested that the majority of genes found to be differentially expressed are involved in multiple aspects of cell physiology, ranging from growth and differentiation to migration and adhesion. Some of these genes, which were found to be substantially altered (induced or repressed), do not have any known function in the lens, such as Fgf14, Foxg1, and Olfml3. However, the relevance of these targets in lens development, and whether they are directly related to the observed mutant phenotype, may become more evident after comparisons are made with mutants exhibiting overlapping defects, such as the Sey mutant mice.
Of interest, Tcfap2a, was also found to be up-regulated (∼3.5-fold). This finding was not unexpected, as previous studies have shown the autoregulation of Tcfap2a by AP-2α (Zhang and Williams, 2003). Because no AP-2α protein was detected in the Le-AP-2α mutant lens, induced expression of this transcript likely originated from the 3′-region or other regions of Tcfap2a lying outside of the exon 5–6 region, which is deleted in the mutant lens (Brewer et al., 2004). This explanation is further supported by the position of the Affymetrix Tcfap2a probe set (1421995_at) within exons 7–8 and the 3′-untranslated region of the gene.
The comparative microarray data revealed that the gene encoding for the cell–cell adhesion molecule E-cadherin Cdh1 was significantly reduced in the Le-AP-2α mutant lens as compared to wild-type littermates, and this finding was confirmed by RT-QPCR as a 58% reduction in mRNA. This reduction further resulted in a significant reduction in E-cadherin protein expression, which in adult mutant lenses was reduced by almost half (49%). Because AP-2α had been shown to directly regulate expression of the E-cadherin gene in several different epithelial cell lines by binding to the promoter in multiprotein complexes (Batsche et al., 1998; Decary et al., 2002), we used ChIP analysis to investigate whether this interaction also occurs in lens tissue. These data clearly showed binding of AP-2α to the E-cadherin promoter indicating that AP-2α can act as a direct regulator of E-cadherin mRNA expression in the lens. Together, these experiments are the first to show that AP-2α binds to the endogenous E-cadherin promoter in vivo and that loss of AP-2α results in a corresponding reduction in E-cadherin expression in the affected tissue. These findings further validated our earlier study of the Le-AP-2α mutant cornea in which defects in corneal epithelial stratification coincided with a substantial reduction in E-cadherin protein expression (Dwivedi et al., 2005).
Recent studies of cell adhesion and tissue separation events suggest that the clustering of cells (through adhesion) from two distinct tissues is essential for their normal separation from one another (Takano et al., 2003) and that this cell-sorting event is directly dependent on the types of cadherin molecules expressed at the cell surface (Nose et al., 1988; Steinberg and Takeichi, 1994; Godt and Tepass, 1998; Gonzalez-Reyes and St Johnston, 1998). Specifically in the lens, the switching between E-, N-, and P-cadherin is thought to be important for lens vesicle separation (Xu et al., 2002). Unlike E-cadherin, we did not observe any change in expression of P- or N-cadherin in our microarray analyses. However, we did observe an alteration of P- and N-cadherin protein expression within the lens stalk of the Le-AP-2α mutants. Also, our genetic screen and RT-QPCR analyses indicated a significant increase in expression for the gene encoding the cell adhesion molecule, L1CAM, within the lenses of Le-AP-2α mutant mice. Thus, the cooperative alteration of the cadherins and other adhesion molecules in the developing lens vesicle may have contributed to the observed mutant phenotype. Further investigations involving the deletion of multiple cadherin molecules may help resolve their collective role(s) in lens vesicle separation.
The microarray data also revealed that the expression of the Egfr mRNA transcript was significantly elevated in lenses derived from Le-AP-2α mice as compared to their wild-type littermates. This induction was confirmed by RT-QPCR to show that the induction was 2.53-fold. Multiple AP-2 binding sites are found within the Egfr gene promoter (Johnson et al., 1988), and recent experiments in the epidermis have shown that AP-2α acts as a negative regulator of Egfr expression at the level of transcription (Wang et al., 2006). Our ChIP data further showed that, in the lens, AP-2α can directly bind to the Egfr promoter. Thus, during normal lens development AP-2α may function to repress Egfr expression. Although Egfr activity has been described in normal development of the corneal endothelium and differentiation of lens fiber cells (Ireland and Mrock, 2000; Reneker et al., 2000), little is known about the role of Egfr signaling in early lens development in vertebrates. However, activation of the Egfr signaling in the eye–antennal imaginal disc in Drosophila has been shown to cause transformation of the eye into an antenna (Kumar and Moses, 2001). Further work has shown that Egfr and Notch signaling oppose one another in promoting eye development. Thus, suppression of Egfr expression by AP-2α could potentially be important for promoting normal development of the lens in vertebrates.
Enrichment of the Bfsp1 promoter region was observed in the ChIP analyses performed with AP-2α on normal lenses. Because Bfsp1/filensin is known to be expressed in the fiber cell compartment of the lens, an expression pattern reciprocal to that of AP-2α, the ChIP findings suggest that AP-2α may act as a suppressor of Bfsp1 in the lens epithelium. However, the microarray analysis revealed that Bfsp1 was down-regulated in lenses of Le-AP-2α mutant mice vs. wild-type littermates, suggesting that AP-2α may act to positively regulate Bfsp1. One possible explanation for these contradictory findings is that, while AP-2α has been deleted from the lens epithelium of the Le-AP-2α mutant lens, at P0 there are also defects in lens fiber cells, which may have indirectly contributed to the observed decrease in Bfsp1 gene expression.
A disruption of the lens epithelial cell phenotype in the Le-AP-2α mutant lens was characterized by a loss in E-cadherin expression, presence of pigment granules, and continued expression of P-cadherin. In addition, expression of the contractile filament α-SMA was observed in the adult Le-AP-2α mutant lens epithelium, further suggesting that these cells had lost their epithelial cell phenotype. When epithelial cells acquire α-SMA expression, it is usually an indication that these cells have undergone EMT, and in the lens, this process has been associated with cataractogenesis (de Iongh et al., 2005). The microarray analysis did identify several genes that could contribute to EMT, including p21-activated kinase (Pak1), a Rac-effector protein involved in cytoskeletal reorganization and migration (Manser et al., 1997; Yang et al., 2005), and Egfr, which has been shown to influence EMT through its ability to increase activity of Pak1 (Galisteo et al., 1996). Thus, the loss in epithelial cell phenotype in the Le-AP-2α mutants may be directly related to the loss of AP-2α expression. However, it must also be considered that these epithelial defects may have developed in response to chronic disruption in the lens epithelial cell architecture, including the persistent lens stalk.
In summary, the findings presented in this study clearly demonstrate that AP-2α has cell-autonomous roles in lens vesicle separation and maintenance of the lens epithelial cell phenotype. The comparative microarray data have also revealed downstream genes that are altered in the lens in the absence of AP-2α and included those involved in cell adhesion and maintenance of the epithelial cell phenotype. Comparisons of these microarray data with that from other mutants with overlapping lens phenotypes should help to further determine the relationship between these regulators in the genetic pathways controlling morphogenesis of the lens.
Experimental Procedures
Generation of Mouse Lines
All animal studies were carried out in accordance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research and the Canadian Council on Animal Care guidelines. To generate a conditional knockout of AP-2α in the lens placode, mice heterozygous for the Tcfap2a:LacZ KI allele (maintained on an outbred Black Swiss genetic background), a null allele (Brewer et al., 2002), were bred with heterozygous Le-Cre mice (maintained on an FVB/N genetic background) that expressed Cre-recombinase specifically in cells within the early lens placode (Ashery-Padan et al., 2000). Resultant progeny from this cross, heterozygous for both the Cre transgene and the Tcfap2a:LacZ KI allele, were then bred with mice homozygous for the Alflox allele (maintained on a mixed 129/Black Swiss background), an allele of Tcfap2a in which paired loxP sites flank a region encoding a critical region of the DNA binding domain (Brewer et al., 2004). Noon of the day of the appearance of the vaginal plug was considered day 0.5 of embryogenesis (E0.5). DNA from embryonic or adult mouse tail biopsies was extracted using the DNeasy tissue kit (Qiagen, Valencia, CA). Mice genotypes were determined with PCR using protocols that have been previously described and well established (Ashery-Padan et al., 2000; Brewer et al., 2004). Additionally, biopsies of placodal-derived and non–placodal-derived tissues (ear, eyelids, lens, and retina) were analyzed for an intact or recombined floxed Tcfap2a allele. PCR analysis was performed in the presence of 4 mM magnesium chloride using the following experimental conditions: 1 cycle of 95°C for 2 min, followed by 30 cycles of 95°C for 45 sec, 65°C for 45 sec, 72°C for 1.5 min, and 1 cycle of 72°C for 15 min. The primers Alflp (5′-CCT GCC TTG GAA CCA TGA CCC TCA G-3′), Alflox4 (5′-CCC AAA GTG CCT GGG CTG AAT TGA C-3′), and Alfscsq (5′-GAA TCT AGC TTG GAG GCT TAT GTC-3′), were used to identify the lacZ knock-in allele (490 bp, KI), an intact floxed allele (560 bp, Alflox), and the correctly deleted floxed allele (185 bp, Floxdel). Littermates that contained a wild-type copy of Tcfap2a and/or lacked the Le-Cre transgene were used as normal controls.
Histology and Immunohistochemistry
Animals were killed by CO2 overdose, and either the embryos were dissected from pregnant females or whole eyes were removed. Tissue was either fixed in 10% neutral buffered formalin overnight at 4°C, processed and embedded in paraffin or fixed in 4% paraformaldehyde overnight at 4°C, cryopreserved in a 30% sucrose/phosphate buffered saline solution, and embedded in Tissue-Tek OCT compound. Serial sections were cut at 5 μm in thickness and stained with hematoxylin and eosin (H&E), or used for subsequent experiments. Indirect immunohistochemistry was used to detect AP-2α (3B5; University of Iowa, used neat) and Pax6 (Covance, Princeton, NJ, 1:50) protein expression in embryonic mouse eyes using an indirect biotin/avidin-immunoperoxidase system (Vector Laboratories, Burlingame, CA; West-Mays et al., 1999a, 2002). Indirect immunofluorescence was used to detect Pitx3 (rabbit polyclonal, kindly provided by Dr. Marten Smidt, University Medical Center Utrecht, Utrecht, Netherlands, 1:250; Smidt et al., 2000), Pax6 (Covance), calretinin (Santa Cruz Biotechnology, Santa Cruz, CA, 1:800), syntaxin-1 (Sigma-Aldrich, Oakville, ON, 1:2,000), E-cadherin (BD Transduction Laboratories, Franklin Lakes, NJ, 1:100), N-cadherin (BD Transduction Laboratories, 1:100), P-cadherin (R&D Systems, Minneapolis, MN, 1:50), and α-smooth muscle actin (Sigma-Aldrich, 1:100) protein expression. The locations of these antigens were then revealed using either a fluorescein isothiocyanate (Jackson ImmunoResearch Laboratories, Inc. West Grove, PA) or an Alexa Flour 568 (Invitrogen - Molecular Probes, Burlington, ON) fluorescent secondary antibody followed by mounting with Vectashield mounting medium containing 4′,6-diamidine-2-phenylidole-dihydrochloride (DAPI; Vector Laboratories).
In Situ Hybridization
To detect Foxe3 mRNA expression, digoxigenin (DIG) -labeled antisense RNA strands were generated using a DIG RNA labeling kit (Roche Diagnostics, Laval, PQ, Canada). The template DNA probes were subcloned from open reading frame fragments containing the Foxe3 forkhead box sequence. In situ hybridization using the labeled probe was carried out on paraffin sections as previously described (Blixt et al., 2000). Specificity of hybridization was confirmed using a sense probe in parallel reactions, yielding no significant binding.
Immunoblotting
Whole intact lenses were extracted from 4-week-old control and Le-AP-2α mice, homogenized in a protein lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM ethylenediaminetetraacetic acid, 1% Triton X-100) and measured for total protein according to the protocol developed by Bradford (1976). Equal amounts of protein lysate (20 μg) were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes by electroblotting. Membranes were blocked in a 5% skim milk solution and probed with a monoclonal E-cadherin primary antibody (1:2,500, BD Transduction Laboratories). The signals were detected using an anti-mouse IgG conjugated with horseradish peroxidase and an enhanced chemiluminescence kit (GE Healthcare Life Sciences, Piscataway, NJ). Blots were stripped with a membrane stripping solution and re-probed with a mouse monoclonal primary antibody directed against glyceraldehyde-3-phosphate dehydrogenase (GAPDH, 1:1000, Chemicon International).
RNA Isolation and Reverse Transcription
Whole intact lenses (P0) were extracted from both Le-AP-2α mutant and control mice and stored in an RNAlater stabilizing solution (Ambion, Austin, TX). Total RNA was isolated from lenses using the Qiagen RNeasy Mini Kit. Briefly, lens tissue was hand-homogenized in a homogenizing buffer (Buffer RLT) and centrifuged at maximum speed. Supernatants were collected and incubated in 70% ethanol. RNA was treated with DNaseI and purified using the RNA-easy spin column by first washing and then eluting in RNase-free water. RNA integrity and concentration was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Foster City, CA) according to the manufacturer's recommended protocol.
cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Briefly, between 500 ng and 1 μg of total RNA was reverse transcribed in a 40-μl reaction containing 100 ng of random hexamer primers, 1 mM dNTP mix, and 200 units of SuperScript II reverse transcriptase. All samples were stored at −70°C until further use.
Gene Expression Profiling and Microarray Data Analysis
The microarray expression profiling was performed in triplicate for each experimental condition (three wild-type lens samples and three Le-AP-2α mutant samples). The labeled mRNA/cDNA target population was prepared using Ovation Aminoallyl RNA amplification system (NuGEN, San Carlos, CA). The hybridization to GeneChip Mouse Genome 430A 2.0 arrays and subsequent processing was carried out following the recommendations of the array manufacturer (Affymetrix). Raw GeneChip data were normalized at the probe level by Robust Multichip Average (RMA) algorithm (Irizarry et al., 2003) and further filtered using GeneSpring 7.2 software (Agilent Technologies, Santa Clara, CA). The differentially abundant mRNAs (between the wild-type and mutant groups) were statistically filtered using an intersection of t-test results (with a P value cutoff of 0.05) and SAM with false discovery rate (FDR) set to 5% (Tusher et al., 2001). Gene Ontology classifications were conducted using a web based GO analysis tool, FatiGO of the Babelomics suite (Al-Shahrour et al., 2004).
Quantification of Gene Expression Using RT-QPCR
Gene expression from recovered cDNA was analyzed with RT-PCR using a 96-well TaqMan optical reaction plate format on an ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA). cDNA was normalized to Gapdh expression for each reaction. Each 25-μl PCR reaction (including controls) contained TaqMan Universal Master Mix (Applied Biosystems), gene-specific forward and reverse primers (Applied Biosystems), and probes for target and endogenous control genes (Applied Biosystems). Serial dilutions (one- to fivefold) of standard samples were prepared in separate wells in duplicate for the endogenous control gene, Gapdh, and the gene targets, Cdh1, Egfr, Pak1, L1cam, Bfsp1, and Etv6. Standard and unknown samples were added in a volume of 5 μl. Thermal cycling parameters consisted of the following: 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 sec at 95°C, and 1 min at 60°C. The number of target gene copies was calculated from a standard curve generated in parallel with each batch of samples. A linear relationship was detected over at least 5 orders of magnitude. In all experiments, the correlation coefficient was between 0.998 and 0.999. The normalization of samples was performed by dividing the number of copies of each target gene by the number of copies of Gapdh. PCR reactions for target gene cDNA quantification were performed using standard cDNA dilution curves.
Quantitative ChIPs
Quantitative ChIPs (qChIPs) were performed using chromatin obtained from microdissected P1 mouse lenses and analyzed as described earlier (Stopka et al., 2005; Yang et al., 2007). The formaldehyde cross-linked chromatin was incubated with 2 μg of rabbit anti–AP-2α antibody (C-18, Santa Cruz Biotechnology; McPherson et al., 2002; Schwartz et al., 2007) or the same amount of normal rabbit serum (Oncogene Research Products, San Diego, CA) The immunoprecipitated fragments of DNA were purified by QIAquick columns (Qiagen) for the quantitative analysis by PCR. Primers were designed using Primer3 software to generate amplicons within the promoter regions of Bfsp1 (forward 5′-AAG GAT TTC TGT TAA GTT TGA AGC A-3′ and reverse 5′-AGG AAG ACT GGT GAT ACT GAC CT-3′), Cdh1 (forward 5′-AGG AAG CTG GGA AGT CTT CTA-3′ and reverse 5′-GTG GCA GCC AAG GAA CTG-3′) and Fgf14 (forward 5′-AAT GCG CTG ATA CAC TGT CG-3′ and reverse 5′-AAA GGT TGG CAG ATC TGT GG-3′). Three AP-2α binding sites are located in the −250/+80 fragment of the mouse Bfsp1 promoter (Masaki et al., 1998), and the amplicon generated in this analysis was 82 base pairs (−99/−18), a range that includes this region. AP-2α has also been shown to bind a GC-rich region (−58/−24) of the Cdh1 promoter (Faraldo et al., 1997), and the amplicon generated was 94 base pairs (−147/−54) in length, directly adjacent to this region. Fgf14 transcript information was derived from the Ensembl (ENSMUSG00000025551) database and the amplicon generated was 83 base pairs (−398/−316) in length; two AP-2α consensus binding sites are found within 600 bp of this region. The internal control sequence used was from mouse chromosome 13 (forward 5′-GCT AAG GAA AGA GGG GGA GA-3′ and reverse 5′-AAA AGT GTG TGT GTG TGT GTG TG-3′). A serial dilution of input (0.01%, 0.05%, 0.2%, and 1%) was used to generate a standard curve for each probe in every qChIP experiment. The Ct value of quantitative PCR was normalized to the percentage of input according to the standard curve. The relative enrichments were calculated from at least two independent ChIPs, and every ChIP was tested by QPCR in triplicate. Background from independent nonspecific antibody (IgG) ChIP was subtracted from each probe, and the final signal was normalized and presented vs. 1% input.
Statistical Analysis
Data for immunoblot, RT-QPCR, and qChIP analyses are presented as the mean ± standard error of the mean. Student's t-test was used to compare expression differences between experimental groups (wild-type and Le-AP-2α) for both immunoblot and RT-QPCR data, while statistical analysis of qChIP results was performed using a one-way analysis of variance followed by Tukey's post hoc test. Results were considered significant when P < 0.05. Statistical analysis was performed using Graphpad Prism software (Intuitive Software for Science, San Diego, CA).
Acknowledgments
We thank the following individuals for their outstanding assistance with the generation of the mouse strains used in this study: Yu Ji, Morgan Singleton, Lori Bulwith, and Jian Huang. We also thank Dr. Marten Smidt (University Medical Center Utrecht, Utrecht, Netherlands) for kindly providing the Pitx3 antibody and the NYU Cancer Institute Genomics Facility for microarray data processing and analysis.
Grant sponsor: National Institutes of Health; Grant number: EY11910 (J.A.W-M.); EY012200 (A.C.); DE-12728 (T.W.); Grant sponsor: Research to Prevent Blindness funding (J.A.W-M.); Grant sponsor: Natural Sciences and Engineering Research Council of Canada (J.A.W-M.).
Footnotes
The Supplementary Material referred to in this article can be found at http://www.interscience.wiley.com/jpages/1058-8388/suppmat
References
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- Andl CD, Mizushima T, Oyama K, Bowser M, Nakagawa H, Rustgi AK. EGFR-induced cell migration is mediated predominantly by the JAK-STAT pathway in primary esophageal keratinocytes. Am J Physiol Gastrointest Liver Physiol. 2004;287:G1227–G1237. doi: 10.1152/ajpgi.00253.2004. [DOI] [PubMed] [Google Scholar]
- Ashery-Padan R, Marquardt T, Zhou X, Gruss P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 2000;14:2701–2711. doi: 10.1101/gad.184000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, Pontoriero GF, Feng W, Marquardt T, Fini ME, Williams T, West-Mays JA. Conditional deletion of activating protein 2alpha (AP-2alpha) in the developing retina demonstrates non-cell-autonomous roles for AP-2alpha in optic cup development. Mol Cell Biol. 2007;27:7497–7510. doi: 10.1128/MCB.00687-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsche E, Muchardt C, Behrens J, Hurst HC, Cremisi C. RB and c-Myc activate expression of the E-cadherin gene in epithelial cells through interaction with transcription factor AP-2. Mol Cell Biol. 1998;18:3647–3658. doi: 10.1128/mcb.18.7.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J, Lowrick O, Klein-Hitpass L, Birchmeier W. The E-cadherin promoter: functional analysis of a G.C-rich region and an epithelial cell-specific palindromic regulatory element. Proc Natl Acad Sci U S A. 1991;88:11495–11499. doi: 10.1073/pnas.88.24.11495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt A, Mahlapuu M, Aitola M, Pelto-Huikko M, Enerback S, Carlsson P. A forkhead gene, FoxE3, is essential for lens epithelial proliferation and closure of the lens vesicle. Genes Dev. 2000;14:245–254. [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brewer S, Jiang X, Donaldson S, Williams T, Sucov HM. Requirement for AP-2alpha in cardiac outflow tract morphogenesis. Mech Dev. 2002;110:139–149. doi: 10.1016/s0925-4773(01)00579-2. [DOI] [PubMed] [Google Scholar]
- Brewer S, Feng W, Huang J, Sullivan S, Williams T. Wnt1-Cre-mediated deletion of AP-2alpha causes multiple neural crest-related defects. Dev Biol. 2004;267:135–152. doi: 10.1016/j.ydbio.2003.10.039. [DOI] [PubMed] [Google Scholar]
- Brown S, Zeidler MP, Hombria JE. JAK/STAT signalling in Drosophila controls cell motility during germ cell migration. Dev Dyn. 2006;235:958–966. doi: 10.1002/dvdy.20709. [DOI] [PubMed] [Google Scholar]
- Brownell I, Dirksen M, Jamrich M. Forkhead Foxe3 maps to the dysgenetic lens locus and is critical in lens development and differentiation. Genesis. 2000;27:81–93. doi: 10.1002/1526-968x(200006)27:2<81::aid-gene50>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Chalepakis G, Wijnholds J, Giese P, Schachner M, Gruss P. Characterization of Pax-6 and Hoxa-1 binding to the promoter region of the neural cell adhesion molecule L1. DNA Cell Biol. 1994;13:891–900. doi: 10.1089/dna.1994.13.891. [DOI] [PubMed] [Google Scholar]
- Chauhan BK, Reed NA, Zhang W, Duncan MK, Kilimann MW, Cvekl A. Identification of genes downstream of Pax6 in the mouse lens using cDNA microarrays. J Biol Chem. 2002;277:11539–11548. doi: 10.1074/jbc.M110531200. [DOI] [PubMed] [Google Scholar]
- Chazaud C, Oulad-Abdelghani M, Bouillet P, Decimo D, Chambon P, Dolle P. AP-2.2, a novel gene related to AP-2, is expressed in the forebrain, limbs and face during mouse embryogenesis. Mech Dev. 1996;54:83–94. doi: 10.1016/0925-4773(95)00463-7. [DOI] [PubMed] [Google Scholar]
- Chow RL, Lang RA. Early eye development in vertebrates. Annu Rev Cell Dev Biol. 2001;17:255–296. doi: 10.1146/annurev.cellbio.17.1.255. [DOI] [PubMed] [Google Scholar]
- Collinson JM, Quinn JC, Buchanan MA, Kaufman MH, Wedden SE, West JD, Hill RE. Primary defects in the lens underlie complex anterior segment abnormalities of the Pax6 heterozygous eye. Proc Natl Acad Sci U S A. 2001;98:9688–9693. doi: 10.1073/pnas.161144098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Kashanchi F, Brady JN, Piatigorsky J. Pax-6 interactions with TATA-box-binding protein and retinoblastoma protein. Invest Ophthalmol Vis Sci. 1999;40:1343–1350. [PubMed] [Google Scholar]
- de Iongh RU, Wederell E, Lovicu FJ, McAvoy JW. Transforming growth factor-beta-induced epithelial-mesenchymal transition in the lens: a model for cataract formation. Cells Tissues Organs. 2005;179:43–55. doi: 10.1159/000084508. [DOI] [PubMed] [Google Scholar]
- Decary S, Decesse JT, Ogryzko V, Reed JC, Naguibneva I, Harel-Bellan A, Cremisi CE. The retinoblastoma protein binds the promoter of the survival gene bcl-2 and regulates its transcription in epithelial cells through transcription factor AP-2. Mol Cell Biol. 2002;22:7877–7888. doi: 10.1128/MCB.22.22.7877-7888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi DJ, Pontoriero GF, Ashery-Padan R, Sullivan S, Williams T, West-Mays JA. Targeted deletion of AP-2alpha leads to disruption in corneal epithelial cell integrity and defects in the corneal stroma. Invest Ophthalmol Vis Sci. 2005;46:3623–3630. doi: 10.1167/iovs.05-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber SC, Dimanlig P, Makarenkova HP, Shirke S, Ko K, Lang RA. Fgf receptor signaling plays a role in lens induction. Development. 2001;128:4425–4438. doi: 10.1242/dev.128.22.4425. [DOI] [PubMed] [Google Scholar]
- Faraldo ML, Rodrigo I, Behrens J, Birchmeier W, Cano A. Analysis of the E-cadherin and P-cadherin promoters in murine keratinocyte cell lines from different stages of mouse skin carcinogenesis. Mol Carcinog. 1997;20:33–47. doi: 10.1002/(sici)1098-2744(199709)20:1<33::aid-mc5>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Favor J, Grimes P, Neuhauser-Klaus A, Pretsch W, Stambolian D. The mouse Cat4 locus maps to chromosome 8 and mutants express lens-corneal adhesion. Mamm Genome. 1997;8:403–406. doi: 10.1007/s003359900456. [DOI] [PubMed] [Google Scholar]
- Fisher M, Grainger RM. Lens induction and determination. In: Lovicu FJ, Robinson ML, editors. Development of the ocular lens. Cambridge, UK: Cambridge University Press; 2004. pp. 27–47. [Google Scholar]
- Galisteo ML, Chernoff J, Su YC, Skolnik EY, Schlessinger J. The adaptor protein Nck links receptor tyrosine kinases with the serine-threonine kinase Pak1. J Biol Chem. 1996;271:20997–21000. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- Godt D, Tepass U. Drosophila oocyte localization is mediated by differential cadherin-based adhesion. Nature. 1998;395:387–391. doi: 10.1038/26493. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, St Johnston D. The Drosophila AP axis is polarised by the cadherin-mediated positioning of the oocyte. Development. 1998;125:3635–3644. doi: 10.1242/dev.125.18.3635. [DOI] [PubMed] [Google Scholar]
- Gotoh N, Ito M, Yamamoto S, Yoshino I, Song N, Wang Y, Lax I, Schlessinger J, Shibuya M, Lang RA. Tyrosine phosphorylation sites on FRS2alpha responsible for Shp2 recruitment are critical for induction of lens and retina. Proc Natl Acad Sci U S A. 2004;101:17144–17149. doi: 10.1073/pnas.0407577101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel) 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Ireland ME, Mrock LK. Differentiation of chick lens epithelial cells: involvement of the epidermal growth factor receptor and endogenous ligand. Invest Ophthalmol Vis Sci. 2000;41:183–190. [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AC, Ishii S, Jinno Y, Pastan I, Merlino GT. Epidermal growth factor receptor gene promoter. Deletion analysis and identification of nuclear protein binding sites. J Biol Chem. 1988;263:5693–5699. [PubMed] [Google Scholar]
- Kumar JP, Moses K. The EGF receptor and notch signaling pathways control the initiation of the morphogenetic furrow during Drosophila eye development. Development. 2001;128:2689–2697. doi: 10.1242/dev.128.14.2689. [DOI] [PubMed] [Google Scholar]
- Lengler J, Bittner T, Munster D, Gawad Ael D, Graw J. Agonistic and antagonistic action of AP2, Msx2, Pax6, Prox1 AND Six3 in the regulation of Sox2 expression. Ophthalmic Res. 2005;37:301–309. doi: 10.1159/000087774. [DOI] [PubMed] [Google Scholar]
- Liu W, Lagutin OV, Mende M, Streit A, Oliver G. Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 2006;25:5383–5395. doi: 10.1038/sj.emboj.7601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovicu FJ, McAvoy JW. Growth factor regulation of lens development. Dev Biol. 2005;280:1–14. doi: 10.1016/j.ydbio.2005.01.020. [DOI] [PubMed] [Google Scholar]
- Makhani LF, Williams T, West-Mays JA. Genetic analysis indicates that transcription factors AP-2alpha and Pax6 cooperate in the normal patterning and morphogenesis of the lens. Mol Vis. 2007;13:1215–1225. [PubMed] [Google Scholar]
- Manser E, Huang HY, Loo TH, Chen XQ, Dong JM, Leung T, Lim L. Expression of constitutively active alpha-PAK reveals effects of the kinase on actin and focal complexes. Mol Cell Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki S, Kamachi Y, Quinlan RA, Yonezawa S, Kondoh H. Identification and functional analysis of the mouse lens filensin gene promoter. Gene. 1998;214:77–86. doi: 10.1016/s0378-1119(98)00230-3. [DOI] [PubMed] [Google Scholar]
- McPherson LA, Loktev AV, Weigel RJ. Tumor suppressor activity of AP2alpha mediated through a direct interaction with p53. J Biol Chem. 2002;277:45028–45033. doi: 10.1074/jbc.M208924200. [DOI] [PubMed] [Google Scholar]
- Moser M, Imhof A, Pscherer A, Bauer R, Amselgruber W, Sinowatz F, Hofstadter F, Schule R, Buettner R. Cloning and characterization of a second AP-2 transcription factor: AP-2 beta. Development. 1995;121:2779–2788. doi: 10.1242/dev.121.9.2779. [DOI] [PubMed] [Google Scholar]
- Nose A, Nagafuchi A, Takeichi M. Expressed recombinant cadherins mediate cell sorting in model systems. Cell. 1988;54:993–1001. doi: 10.1016/0092-8674(88)90114-6. [DOI] [PubMed] [Google Scholar]
- Nottoli T, Hagopian-Donaldson S, Zhang J, Perkins A, Williams T. AP-2-null cells disrupt morphogenesis of the eye, face, and limbs in chimeric mice. Proc Natl Acad Sci U S A. 1998;95:13714–13719. doi: 10.1073/pnas.95.23.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa K, Saika S, Hayashi Y, Tawara A, Ohnishi Y. Cornea with Peters' anomaly: perturbed differentiation of corneal cells and abnormal extracellular matrix in the corneal stroma. Jpn J Ophthalmol. 2003;47:327–331. doi: 10.1016/s0021-5155(03)00072-8. [DOI] [PubMed] [Google Scholar]
- Ozeki H, Ogura Y, Hirabayashi Y, Shimada S. Suppression of lens stalk cell apoptosis by hyaluronic acid leads to faulty separation of the lens vesicle. Exp Eye Res. 2001;72:63–70. doi: 10.1006/exer.2000.0923. [DOI] [PubMed] [Google Scholar]
- Prosser J, van Heyningen V. PAX6 mutations reviewed. Hum Mutat. 1998;11:93–108. doi: 10.1002/(SICI)1098-1004(1998)11:2<93::AID-HUMU1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Reneker LW, Silversides DW, Xu L, Overbeek PA. Formation of corneal endothelium is essential for anterior segment development - a transgenic mouse model of anterior segment dysgenesis. Development. 2000;127:533–542. doi: 10.1242/dev.127.3.533. [DOI] [PubMed] [Google Scholar]
- Rieger DK, Reichenberger E, McLean W, Sidow A, Olsen BR. A double-deletion mutation in the Pitx3 gene causes arrested lens development in aphakia mice. Genomics. 2001;72:61–72. doi: 10.1006/geno.2000.6464. [DOI] [PubMed] [Google Scholar]
- Sakai M, Serria MS, Ikeda H, Yoshida K, Imaki J, Nishi S. Regulation of c-maf gene expression by Pax6 in cultured cells. Nucleic Acids Res. 2001;29:1228–1237. doi: 10.1093/nar/29.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz B, Melnikova VO, Tellez C, Mourad-Zeidan A, Blehm K, Zhao YJ, McCarty M, Adam L, Bar-Eli M. Loss of AP-2alpha results in deregulation of E-cadherin and MMP-9 and an increase in tumorigenicity of colon cancer cells in vivo. Oncogene. 2007;26:4049–4058. doi: 10.1038/sj.onc.1210193. [DOI] [PubMed] [Google Scholar]
- Semina EV, Murray JC, Reiter R, Hrstka RF, Graw J. Deletion in the promoter region and altered expression of Pitx3 homeobox gene in aphakia mice. Hum Mol Genet. 2000;9:1575–1585. doi: 10.1093/hmg/9.11.1575. [DOI] [PubMed] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, Luo Y, Sun YA, Willey JM, Setterquist RA, Fischer GM, Tong W, Dragan YP, Dix DJ, Frueh FW, Goodsaid FM, Herman D, Jensen RV, Johnson CD, Lobenhofer EK, Puri RK, Schrf U, Thierry-Mieg J, Wang C, Wilson M, Wolber PK, Zhang L, Slikker W., Jr The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivak JM, West-Mays JA, Yee A, Williams T, Fini ME. Transcription factors Pax6 and AP-2alpha interact to coordinate corneal epithelial repair by controlling expression of matrix metalloproteinase gelatinase B. Mol Cell Biol. 2004;24:245–257. doi: 10.1128/MCB.24.1.245-257.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidt MP, Asbreuk CH, Cox JJ, Chen H, Johnson RL, Burbach JP. A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci. 2000;3:337–341. doi: 10.1038/73902. [DOI] [PubMed] [Google Scholar]
- Steinberg MS, Takeichi M. Experimental specification of cell sorting, tissue spreading, and specific spatial patterning by quantitative differences in cadherin expression. Proc Natl Acad Sci U S A. 1994;91:206–209. doi: 10.1073/pnas.91.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J. 2005;24:3712–3723. doi: 10.1038/sj.emboj.7600834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss E. Arrays of hope. Cell. 2006;127:657–659. doi: 10.1016/j.cell.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Takano R, Mochizuki A, Iwasa Y. Possibility of tissue separation caused by cell adhesion. J Theor Biol. 2003;221:459–474. doi: 10.1006/jtbi.2003.3193. [DOI] [PubMed] [Google Scholar]
- Takeichi M. The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development. 1988;102:639–655. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- Theiler K, Varnum DS. Development of coloboma (Cm/+), a mutation with anterior lens adhesion. Anat Embryol (Berl) 1981;162:121–126. doi: 10.1007/BF00318098. [DOI] [PubMed] [Google Scholar]
- Tummala R, Romano RA, Fuchs E, Sinha S. Molecular cloning and characterization of AP-2 epsilon, a fifth member of the AP-2 family. Gene. 2003;321:93–102. doi: 10.1016/s0378-1119(03)00840-0. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Raamsdonk CD, Tilghman SM. Dosage requirement and allelic expression of PAX6 during lens placode formation. Development. 2000;127:5439–5448. doi: 10.1242/dev.127.24.5439. [DOI] [PubMed] [Google Scholar]
- Wang Q, McEwen DG, Ornitz DM. Subcellular and developmental expression of alternatively spliced forms of fibroblast growth factor 14. Mech Dev. 2000;90:283–287. doi: 10.1016/s0925-4773(99)00241-5. [DOI] [PubMed] [Google Scholar]
- Wang L, Prescott AR, Spruce BA, Sanderson J, Duncan G. Sigma receptor antagonists inhibit human lens cell growth and induce pigmentation. Invest Ophthalmol Vis Sci. 2005;46:1403–1408. doi: 10.1167/iovs.04-1209. [DOI] [PubMed] [Google Scholar]
- Wang X, Bolotin D, Chu DH, Polak L, Williams T, Fuchs E. AP-2alpha: a regulator of EGF receptor signaling and proliferation in skin epidermis. J Cell Biol. 2006;172:409–421. doi: 10.1083/jcb.200510002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawersik S, Purcell P, Rauchman M, Dudley AT, Robertson EJ, Maas R. BMP7 acts in murine lens placode development. Dev Biol. 1999;207:176–188. doi: 10.1006/dbio.1998.9153. [DOI] [PubMed] [Google Scholar]
- West-Mays JA, Cook JR, Sadow PM, Mullady DK, Bargagna-Mohan P, Strissel KJ, Fini ME. Differential inhibition of collagenase and interleukin-1alpha gene expression in cultured corneal fibroblasts by TGF-beta, dexamethasone, and retinoic acid. Invest Ophthalmol Vis Sci. 1999a;40:887–896. [PubMed] [Google Scholar]
- West-Mays JA, Zhang J, Nottoli T, Hagopian-Donaldson S, Libby D, Strissel KJ, Williams T. AP-2alpha transcription factor is required for early morphogenesis of the lens vesicle. Dev Biol. 1999b;206:46–62. doi: 10.1006/dbio.1998.9132. [DOI] [PubMed] [Google Scholar]
- West-Mays JA, Coyle BM, Piatigorsky J, Papagiotas S, Libby D. Ectopic expression of AP-2alpha transcription factor in the lens disrupts fiber cell differentiation. Dev Biol. 2002;245:13–27. doi: 10.1006/dbio.2002.0624. [DOI] [PubMed] [Google Scholar]
- Williams T, Admon A, Luscher B, Tjian R. Cloning and expression of AP-2, a cell-type-specific transcription factor that activates inducible enhancer elements. Genes Dev. 1988;2:1557–1569. doi: 10.1101/gad.2.12a.1557. [DOI] [PubMed] [Google Scholar]
- Xu L, Overbeek PA, Reneker LW. Systematic analysis of E-, N- and P-cadherin expression in mouse eye development. Exp Eye Res. 2002;74:753–760. doi: 10.1006/exer.2002.1175. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rayala S, Nguyen D, Vadlamudi RK, Chen S, Kumar R. Pak1 phosphorylation of snail, a master regulator of epithelial-to-mesenchyme transition, modulates snail's subcellular localization and functions. Cancer Res. 2005;65:3179–3184. doi: 10.1158/0008-5472.CAN-04-3480. [DOI] [PubMed] [Google Scholar]
- Yang Y, Wolf LV, Cvekl A. Distinct embryonic expression and localization of CBP and p300 histone acetyltransferases at the mouse alphaA-crystallin locus in lens. J Mol Biol. 2007;369:917–926. doi: 10.1016/j.jmb.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Williams T. Identification and regulation of tissue-specific cis-acting elements associated with the human AP-2alpha gene. Dev Dyn. 2003;228:194–207. doi: 10.1002/dvdy.10365. [DOI] [PubMed] [Google Scholar]
- Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon AP, Flavell RA, Williams T. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238–241. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]
- Zhao F, Satoda M, Licht JD, Hayashizaki Y, Gelb BD. Cloning and characterization of a novel mouse AP-2 transcription factor, AP-2delta, with unique DNA binding and transactivation properties. J Biol Chem. 2001;276:40755–40760. doi: 10.1074/jbc.M106284200. [DOI] [PubMed] [Google Scholar]


