Abstract
A 21-mer peptide that can be used to covalently introduce synthetic molecules into proteins has been developed. Phage displayed peptide libraries were subjected to reaction-based selection with 1,3-diketones. The peptide was further evolved by addition of a randomized region and reselection for improved binding. The resulting 21-mer peptide had a reactive amino group that formed an enaminone with 1,3-diketone and was used as a tag for labeling of maltose binding protein. Using this peptide tag and 1,3-diketone derivatives, a variety of molecules such as reporter probes and functionalities may be covalently introduced into proteins of interest.
Introduction
Development of systems for labeling of proteins with synthetic molecules or small molecules is of considerable interest (1), because labeling systems can be used to address fundamental biological questions and can be used for the preparation of novel therapeutics. For example, labeling of proteins with fluorescent molecules allows tracking of the proteins within native environs (2, 3, 4). Whereas GFP variants have a given fluorescence, use of a tag-fusion protein allows for labeling with a variety of synthetic fluorescent molecules that differ in fluorescence. In addition, modifications to a tag-fusion protein are not limited to introduction of fluorescent molecules; other molecules can be introduced to the tag-fusion protein. Proteins conjugated with carbohydrates or lipids are tools for exploring interactions among these biomolecules on the cell surface and inside cells (5). Drug-protein conjugates, including antibody conjugates with cytotoxic molecules or radioisotopes, are often safer and more effective therapeutics than drug or protein alone (6, 7, 8, 9). Proteins labeled with polymers are more stable than the corresponding unlabeled proteins in serum and are useful for therapeutic applications (10). Labeling reactions of proteins with synthetic molecules can also be used for covalently attaching proteins to the surfaces of microchips or microarrays (11, 12, 13).
In order to label a protein of interest selectively and specifically, several strategies and methods have been developed. For example, using a tag protein or peptide (either enzyme-derived or designed) that selectively reacts with designed synthetic molecules provides selective and specific labeling. Methods using a covalently-modifiable tag fusion include labeling of O6-alkylguanine-DNA alkyltransferase with O6-benzylguanine derivatives (3, 14), Sfp phosphopantetheinyl transferase-catalyzed labeling of a peptide excised from a nonribosomal peptide synthetase with an adenosine 3′-monophosphate derivative (11, 15), phosphopantetheine transferase-catalyzed labeling of the acyl carrier protein with an adenosine 3′-monophosphate derivative (4), biotin ligase-catalyzed labeling of the acceptor peptide with a biotin-mimic derivative (16), labeling of a serine esterase cutinase with phosphate derivatives (13), and labeling of a tetracysteine α-helix motif with biarsenical ligands (17, 18). Other useful covalent labeling methods include intein-mediated labeling (19, 20, 21), incorporation of an unnatural amino acid into a protein and labeling of the unnatural amino acid (5) and introduction of a N-terminal cysteine and its labeling with aldehyde derivatives (22). When serine or threonine is introduced at N-terminus of a protein, oxidation of the N-terminal serine or threonine to form aldehyde group followed by oxime or hydrazone formation reaction is another useful chemical modification method (23, 24, 25). Labeling of fusion proteins with noncovalent, but tight-binding compounds has also been reported. Examples include labeling of a dihydrofolate reductase (DHFR)-fusion with methotrexate conjugates (26, 27), labeling of an FKBP12 mutant (F36V)-fusion with its ligand derivatives (28), labeling of a single chain antibody fusion with its hapten-conjugated molecules (29), and labeling of avidin fusion with biotin-conjugates (30).
For labeling of fusion proteins, a smaller peptide tag is preferred as the fusion partner because a larger protein tag may affect the function of the protein of interest and because production of larger proteins is often more difficult. When multiple labeling of a protein of interest with different molecules is required, one convenient solution is the introduction of different tags into the protein at the same time followed by specific labeling of each tag of the fusion. For multiple labels, smaller fusion partners allow for the fusion protein to be of reasonable size. When small peptides are used, noncovalent binding is unlikely to provide sufficient hydrogen bonds and/or charge and hydrophobic interactions for tight binding. In contrast, a covalent bond is sufficient to retain the label and numerous noncovalent interactions are not necessary. Thus small peptides that form covalent bonds with designed compounds should be suitable for conjugation of the fusion proteins with synthetic molecules. In addition, it is preferred that labeling methods do not require additional catalysts or toxic metal reagents. Here we report the development of a small 21-mer peptide that forms an enaminone with 1,3-diketone derivatives. We have demonstrated that when fused to a protein of interest, the peptide can be used to introduce synthetic molecules into the fusion protein without the requirement of additional catalysts or reagents.
Experimental Procedures
Selection of peptides from initial libraries
Ph.D.-C7C, Ph.D.-7, and Ph.D.-12 phage display peptide libraries (New England Biolabs, NEB) were used for the selection. Wells of a microtiter plate (Costar 3690) were coated with 1-bovine serum albumin conjugate (1-BSA) (2 μg/PBS 25 μL/well) at 4 °C overnight, washed with H2O two times, and blocked with 3% BSA/PBS (170 μL/well) at room temperature for 1.5 h (PBS is 10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4). Blocking solution was removed and the library phage were added. After 2 h, the wells were washed 10 times with 0.5% Tween 20/PBS (PBST) to remove unbound phage. The bound phage were eluted by trypsin digestion (Difco Trypsin 1:250, 10 mg/mL in TBS, 50 μL/well) at 37°C for 30 min, added to E. coli 2537 cells in LB medium, and grown using the procedures recommended by NEB. After four rounds of selection using each phage displayed peptide library separately, panned libraries were combined and additional three rounds of selection were performed. The DNA sequences of the clones from the seventh round were analyzed using procedures recommended by NEB. Individual clones were analyzed by ELISA for binding to 1-BSA immobilized on wells of a microtiter plate using anti-M13 antibody-horseradish peroxidase conjugate (Amersham) and the peroxidase substrates 2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) and hydrogen peroxide.
Construction of the second and third libraries and peptide selections
Synthesized oligonucleotide library was amplified by PCR using 5′-primer SfiI-f (5′-GAGGAGGAGGAGGCCCAGGCGGCC-3′) and 3′-primer SfiI-b (5′-GAGGAGGAGGAGGCCGGCCTGGCC-3′). The PCR conditions as were follows: SfiI-f, 1 μg; SfiI-b, 1 μg; oligonucleotide library, 50 ng; 0.2 mM each dNTP; 10 μL of 10× PCR buffer; 2.5 units of Ampli Taq DNA polymerase (Roche) in a total volume of 100 μL. A program of 94 °C 1 min; 94 °C 15 sec, 52 °C 15 sec, 72 °C 30 sec (25 times); 72 °C 10 min was used for amplification. The PCR products were purified, digested with SfiI, and ligated to SfiI-digested pComb3X using T4 DNA ligase (NEB). The following oligonucleotides were used (N = A, C, G, or T; K = G or T): PhD-C7C-NNK6 (5′-GGCCCAGGCGGCCTGTCATAATCATCAGAAGGCTACGTGCNNKNNKNNKNNKNNKNNKGGCCAGGCCGGCC-3′), PhD7-2-NNK6-f (5′-GGCCCAGGCGGCCGCTGCTGTGGCTAAGCCGCCGNNKNNKNNKNNKNNKNNKGGCCAGGCCGGCC-3′), NNK6-PhD7-2-f (5′-GGCCCAGGCGGCCNNKNNKNNKNNKNNKNNKGCTGCTGTGGCTAAGCCGCCGGGCCAGGCCGGCC-3′), PhD7-6-NNK6-f (5′-GGCCCAGGCGGCCGTGTCGGTGCAGACTAAGTATNNKNNKNNKNNKNNKNNKGGCCAGGCCGGCC-3′), NNK6-PhD7-6-f (5′-GGCCCAGGCGGCCNNKNNKNNKNNKNNKNNKGTGTCGGTGCAGACTAAGTATGGCCAGGCCGGCC-3′), PhD7-14-NNK6-f (5′-GGCCCAGGCGGCCAATCCGCTGGGTGCTAAGTTGNNKNNKNNKNNKNNKNNKGGCCAGGCCGGCC-3′), NNK6-PhD7-14-f (5′-GGCCCAGGCGGCCNNKNNKNNKNNKNNKNNKAATCCGCTGGGTGCTAAGTTGGGCCAGGCCGGCC-3′), PhD12-3-NNK6-f (5′-GGCCCAGGCGGCCGCTGCTATGGATGCTAAGAATTCTCCTGCTTCTGCTNNKNNKNNKNNKNNKNNKGGCCAGGCCGGCC-3′), NNK6-PhD12-3-f (5′-GGCCCAGGCGGCCNNKNNKNNKNNKNNKNNKGCTGCTATGGATGCTAAGAATTCTCCTGCTTCTGCTGGCCAGGCCGGCC-3′), PhD12-9-NNK6-f (5′-GGCCCAGGCGGCCGATTTGCCTATTCCTACTACGAAGCTTGGGCGGTCTNNKNNKNNKNNKNNKNNKGGCCAGGCCGGCC-3′), and NNK6-PhD12-9-f (5′-GGCCCAGGCGGCCNNKNNKNNKNNKNNKNNKGATTTGCCTATTCCTACTACGAAGCTTGGGCGGTCTGGCCAGGCCGGCC-3′).
Construction of the third libraries was performed using the same procedures as used for construction of the second libraries. The following oligonucleotides were used: PhD-C7C-1319-NNK6 (5′-GGCCCAGGCGGCCTGTCATAATCATCAGAAGGCTACGTGCACGGCGCAGGCGCAGAGTNNKNNKNNKNNKNNKNNKGGCCAGGCCGGCC-3′), PhD-C7C-1320-NNK6 (5′-GGCCCAGGCGGCCTGTCATAATCATCAGAAGGCTACGTGCCGGAGGATGCGGTCTAGGNNKNNKNNKNNKNNKNNKGGCCAGGCCGGCC-3′), PhD-C7C-1322-NNK6 (5′-GGCCCAGGCGGCCTGTCATAATCATCAGAAGGCTACGTGCTCGTGGGTTCTTGTGCCGNNKNNKNNKNNKNNKNNKGGCCAGGCCGGCC-3′), PhD-C7C-1326-NNK6 (5′-GGCCCAGGCGGCCTGTCATAATCATCAGAAGGCTACGTGCCCGAGGGATAGGCATGGGNNKNNKNNKNNKNNKNNKGGCCAGGCCGGCC-3′), PhD7-14-2-NNK6 (5′-GGCCCAGGCGGCCAATCCGCTGGGTGCTAAGTTGGCGGGTGTGTTTTGGTGGNNKNNKNNKNNKNNKNNKGGCCAGGCCGGCC-3′), and PhD7-14-10-NNK6 (5′-GGCCCAGGCGGCCAATCCGCTGGGTGCTAAGTTGGCTATGGCTATGTGGGGGNNKNNKNNKNNKNNKNNKGGCCAGGCCGGCC-3′). The library size of the second and third libraries was between 2.3 × 107–3.1 × 108.
Ligation mixtures were ethanol precipitated and transformed into E. coli ER 2537 cells by electroporation. Immediately after transformation, 5 mL of SOC (2% trypton, 0.5% yeast extract, 0.05% NaCl, 2.5 mM KCl, 10 mM MgCl2, 20 mM glucose) was added to each reaction and the cells were grown at 37 °C. After 1 h, 10 mL of super broth (SB, 3% trypton, 2% yeast extract, 1% Mops) and carbenicillin (to 20 μg/mL) were added. After 1 h, additional carbenicillin was added to increase the final concentration to 50 μg/mL. After 1 h, helper phage VCS-M13 (∼1013 pfu) were added and the culture volume was increased to 100 mL by the addition of SB including carbenicillin (at 50 μg/mL). After 1.5 h, kanamycin (for a final concentration of 70 μg/mL) was added and the culture was incubated at 37 °C over night. The cells were removed by centrifugation (4000 rpm, 30 min). Phage were purified by precipitation: Poly(ethylene glycol) 8000 (final concentration 4% w/v) and NaCl (final concentration 3% w/v) were added to the supernatant, and, after 30 min on ice, the mixture was centrifuged at 9000 rpm for 20 min. The phage precipitate was resuspended in 1% BSA/PBS (2 mL) and filtered (0.2 μm).
Selection was performed using 1-BSA immobilized on wells in a microtiter plate as described above. The bound phage eluted by trypsin were added to E. coli ER2537 cells (20 mL in SB) and the culture was shaken at 37 °C. After 15 min, carbenicillin (for a final concentration of 20 μg/mL) was added. After 45 min, helper phage VCS-M13 (∼1012 pfu) was added, and the culture was diluted to the total volume 100 mL with SB containing carbenicillin (50 μg/mL). After 1.5 h, kanamycin (final concentration of 70 μg/mL) was added and the culture was incubated at the same temperature overnight. Phage were purified as described above and were used for further panning.
An enzyme-linked immunosorbent assay (ELISA) of individual clones against 1-BSA immobilized on wells of a microtiter plate was performed using anti-decapeptide antibody-alkaline phosphatase conjugate (Pierce) and p-nitrophenyl phosphate.
Preparation rpf1368-MBP
The SfiI-digested fragment of rpf1368 was ligated to SfiI-digested pComb3-MBP vector (see below) with T4 DNA ligase. The ligation mixture was transformed into E. coli XL1-Blue cells (Stratagene) by electroporation. After transformation, SOC (2 mL) was immediately added and the cells were grown at 37 °C. After 1 h, the culture (100, 10, and 1.0 μL) was plated on LB agar containing carbenicillin (100 μg/mL). Individual colonies were picked from the plates and grown in SB (5 mL) containing carbenicillin (50 μg/mL) at 37 °C for over night. The culture was diluted 1:10 in SB containing 0.2% glucose and carbenicillin (50 μg/mL) and was grown for 2 h at 37 °C. Expression of the rpf1368-MBP genes was induced by addition of IPTG (1 mM) and the culture was incubated for additional 3 h. The culture was centrifuged (3500 rpm) for 20 min and the cells were resuspended in PBS (500 μL) then freeze-thawed five times in a dry ice-ethanol bath and water bath. The lysate sample was spun in a microcentrifuge at maximum speed for 20 min and the supernatant was used for the ELISA with 1-BSA (see below). The sequence of rpf1368-MBP was confirmed by DNA sequencing of peptide rpf1368 region using primer Mal-B (5′-GAATTTCTCTTCCAGTTTATCCG-3′) as well as the positive signal in ELISA with 1-BSA.
For large-scale production of rpf1368-MBP, the following procedures were used: The colony was picked from a plate and grown in SB (5 mL) containing carbenicillin (50 μg/mL) at 37 °C for overnight. A 1 mL aliquot of the culture was added into each of two flasks containing 100 mL of SB containing 0.2% glucose, 20 mM MgCl2, and carbenicillin (50 μg/mL) and was grown for 7 h at 37 °C. Expression of the rpf1368-MBP genes was induced by addition of IPTG (1 mM) and the culture was incubated overnight. The culture was centrifuged (3500 rpm) for 20 min at 4 °C and the cells were resuspended in amylose column buffer (5 mL) composed of 20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA, and 1mM PMSF. The cells were sonicated on ice using Tekmer Sonic Disrupter. The lysate sample was centrifuged (3500 rpm) for 15 min at 4 °C and the supernatant was filtered. The filtrate was applied to the amylose resin column (pre-equilibrated with the amylose column buffer). The column was washed with the column buffer and rpf1368-MBP was eluted with 10 mM maltose in column buffer. The combined eluted fractions were concentrated and the buffer was changed to PBS using Centriprep-10 (Millipore). The protein purity was judged to be >95% by in gel electrophoresis.
Construction of pComb3-MBP
Cloning of the SfiI-digested fragment from the pComb3X (31) into pComb3-MBP allowed the production of the MBP-fusion. Vector pComb3-MBP produces N-terminal MBP-fusion (protein/peptide of interest fused at the N-terminus of MBP) and the N-terminal sequence of protein/peptide of the MBP fusion is identical to that produced from the pComb3X system. Vector pComb3-MBP was prepared by replacing gene III of pComb3X with the MBP gene from pMAL-pIII (32).
PCR was performed using (i) primers Bseq (5′-GTGAGCGAGGAAGCGGAAGAG-3′) and TT-SfiI-b (5′-CATCTGGCCGGCCTGGCCACTAG-3′) and template pComb3X containing anti-tetanus toxoid Fab p313 gene (33) between the SfiI sites, and (ii) primers SfiI-MBP-f (5′-GGCCAGGCCGGCCAGATGAAAATCGAAGAAGGTAAACTG-3′) and MBP-NotI-b (5′-GAGGAGGAGAAGCGGCCGCTTAAATTAATTAGCTAGCTTAAGAGGATCCAAATTCTGAAATCC-3′) which included a silent mutation to destroy an intrinsic EcoRI site in MBP (indicated by underlining) and template pMAL-pIII. The PCR fragments were fused using primers Bseq and MBP-NotI-b. The resulting PCR product was digested with EcoRI and NotI, and purified. The fragment was ligated EcoRI/NotI-digested pComb3X. The correct vector pComb3-MBP was identified by the production of anti-tetanus toxoid Fab fused to MBP and by ELISA against tetanus toxoid using detection of MBP. When gene of a protein is cloned by the SfiI sites into this vector and the protein is overproduced, the protein is delivered into periplasm. Thus in this system disulfide bonds that are conformationally reasonable can form automatically in periplasm when the protein is produced in E. coli cells.
ELISA of rpf1368-MBP
Wells of a microtiter plate (Costar 3690) were coated with 1-BSA (1 μg/PBS 25 μL/well) at 37 °C for 1 h, washed with H2O two times, and blocked with 3% BSA/PBS (170 μL/well) at 37 °C for 1 h. Blocking solution was removed and ELISA sample (cell lysate or purified protein) was added (25 μL/well). After 2 h of incubation at room temperature, the wells were washed 10 times with water. The bound rpf1368-MBP was detected using mouse anti-MBP antibody clone MBP-17 (Sigma), goat anti-mouse antibody-alkaline phosphatase conjugate (Pierce), and the phosphatase substrate p-nitrophenyl phosphate. The resulting yellow color was measured at 405 nm. For the control MBP, MBP2* protein (NEB) was used.
For the ELISA using 1-biotin (Figure 2b), wells of a microtiter plate (Costar 3690) were coated with NeutrAvidin (Pierce) (10 μg/PBS 25 μL/well) at 37 °C for 1 h, washed with H2O two times, and 1-biotin (80 μM/PBS, 25 μL) was added. After 2 h incubation at room temperature, the wells were washed with H2O two times and were blocked with 3% BSA/PBS (170 μL/well) at room temperature for 1 h. Blocking solution was removed and ELISA sample was added. The same procedure was then followed as described for the ELISA with 1-BSA. For background ELISA, the same procedures were used except that 1-biotin was added (i.e., wells coated with NeutrAvidin were used for background binding studies).
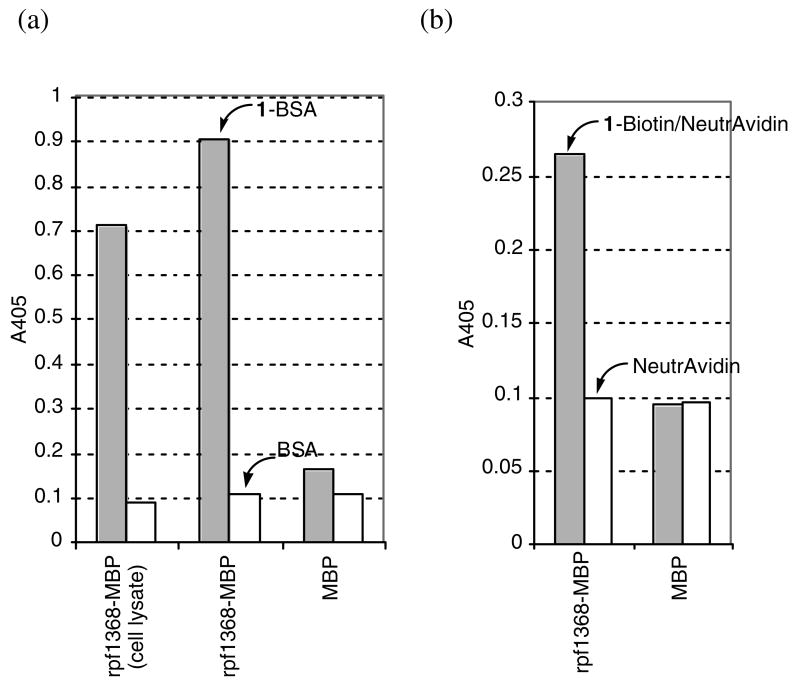
Figure 2.
Binding assays of rpf1368-MBP to diketones. (a) ELISA with 1-BSA. The cell lysate of E. coli cells expressing the fusion protein (labeled as cell lysate) was evaluated, as were purified proteins. Data of binding to 1-BSA and to BSA (background) are shown: 1-BSA (gray), BSA (white). Deviations were within ±10% of the indicated values. (b) ELISA with 1-biotin. Data of binding to 1-biotin supported on NeutrAvidin and to NeutrAvidin (background) are shown: 1-Biotin/NeutrAvidin (gray), NeutrAvidin (white). Deviations were within ±10% of the indicated values.
Whereas 1-BSA possessed ∼30 diketone moieties per BSA molecule, each subunit of NeutrAvidin binds one molecule of biotin. Therefore, ELISA using 1-BSA afforded greater signal (absorption at 405 nm) than that using 1-biotin/NeutrAvidin. Because of the difference in the numbers of the diketone moieties presented on well surfaces, the ELISA signal difference between binding to 1-BSA and to BSA (background) were also greater than that between binding to 1-biotin/NeutrAvidin and to NeutrAvidin (background).
Reaction of rpf1368-MBP with 2,4-pentanedione
Reactions were initiated by adding 2 μL of 2,4-pentanedione (20 mM in CH3CN) to 78 μL of rpf1368-MBP solution in PBS at 25 °C. The final conditions were [rpf1368-MBP] 2 μM, [2,4-pentanedione] 500 μM in 2.5% CH3CN/PBS (total volume 80 μL). Enaminone formation was measured by the increase in absorption at 318 nm using a spectrophotometer.
Peptide rpf1368
Peptide rpf1368 was synthesized on a peptide synthesizer using standard Fmoc solid-phase peptide synthesis chemistry, purified by HPLC to >95% purity, and characterized by mass spectrometry. An intramolecular S-S bond was formed when the peptide was stored in buffer under air at room temperature overnight or during the reaction with diketones. The cyclized form of the peptide was also characterized by mass analysis.
Mass analysis for the reaction of peptide rpf1368 with diketone 2
Reactions were initiated by adding 2.5 μL of diketone 2 (20 mM in CH3CN) to a mixture of 47 μL of 50 mM Na phosphate, pH 7.0 and 0.5 μL of peptide rpf1368 (10 mM in H2O). The final conditions were [rpf1368] 100 μM, [2] 1mM in 5% CH3CN-47 mM Na phosphate, pH 7.0 (total volume 50 μL). After 1 day, MALDI-TOF analysis was performed.
Synthesis of 1-Biotin
See Supporting Information. A mixture of EZ-Link Biotin PEO-LC-Amine (Pierce) (11.2 mg, 0.027 mmol) and diketone N-hydroxysuccinimide ester (N-hydroxysuccinimide ester rather than BSA amide in the structure of 1-BSA shown in Figure 1; 9.3 mg, 0.022 mmol) in CH3CN (0.2 mL)-MeOH (0.2 mL) was stirred at room temperature for 3 h. The mixture was purified by flash silica gel chromatography (EtOAc/MeOH = 4:1 to 2:1) to afford 1-biotin (7.4 mg, 46%). 1H NMR (500 MHz, CD3OD) δ 7.50 (d, J = 8.5 Hz, 2H), 7.19 (d, J = 8.5 Hz, 2H), 4.52 (m, 1H), 4.33 (m, 1H), 3.70-3.62 (m, 8H), 3.59 (t, J = 5.5 Hz, 2H), 3.56 (t, J = 5.5 Hz, 2H), 3.41 (t, J = 5.5 Hz, 2H), 3.38 (t, J = 5.5 Hz, 2H), 3.24 (m, 1H), 2.99-2.86 (m, 2H), 2.92 (t, J = 7.4 Hz, 2H × 0.5), 2.74 (m, 1H), 2.63 (t, J = 7.4 Hz, 2H × 0.5), 2.44 (t, J = 7.4 Hz, 2H), 2.33 (t, J = 7.4 Hz, 2H), 2.25 (t, J = 7.4 Hz, 2H), 2.05 (s, 3H × 0.5), 2.02 (quintet, J = 7.4 Hz, 2H), 1.82-1.43 (m, 6H). MALDI-FTMS calcd for C35H54N5O9S (MH+) 720.3637, found 720.3590.
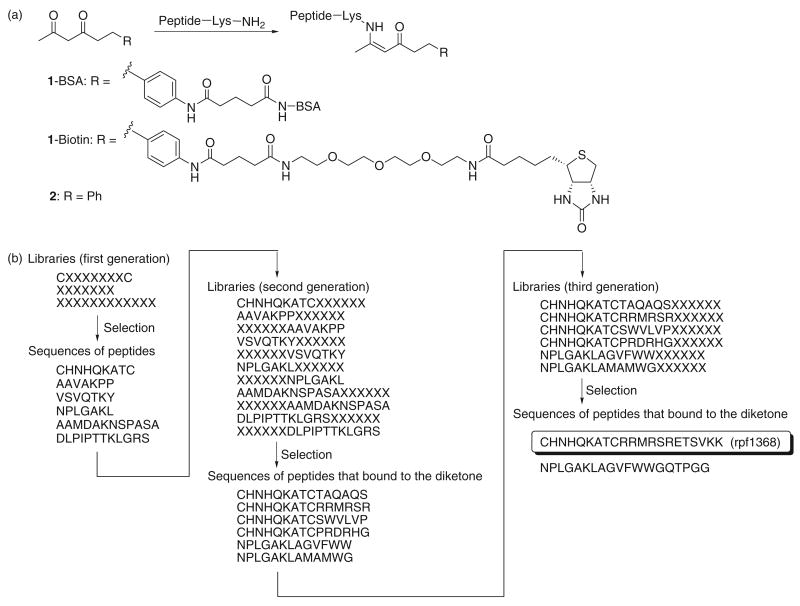
Figure 1.
Selection of peptides that form enaminones with diketones. (a) 1,3-Diketones and enaminone formation. (b) Peptide libraries and selected peptide sequences. Selection implies selection using phage display with 1-BSA immobilized on wells in a microtiter plate.
Results and Discussions
Design of labeling system using enaminone formation and selection of peptides
Lysine ε-amino groups and N-terminal amino groups in proteins and peptides are not typically nucleophilic enough to form enaminones with 1,3-diketones at neutral pH, but certain enzymes have nucleophilic lysine ε-amino groups that are essential for catalysis (34, 35). 1,3-Diketones act as inhibitors of such enzymes by formation of enaminones with the nucleophilic amino groups (36). Because of the π-conjugation, enaminones are very stable (34, 36, 37). In order for an amino group to be nucleophilic enough to allow for formation of an enaminone with diketones, the amino group must either have an electrostatic interaction with a positively charged residue or be present within a hydrophobic microenvironment (35, 38, 39). Only proteins and peptides that can provide such interactions/microenvironments can form enaminones with 1,3-diketones. Thus a small peptide that forms an enaminone with a 1,3-diketone derivative should provide selective and stable labeling of a protein of interest through the enaminone formation reaction with a diketone-labeling molecule conjugate.
Although peptides possessing lysine residues in α-helical structures whose lysine ε-amino groups are nucleophilic have also been developed by design (40, 41, 42, 43), design of peptides that form an enaminone with 1,3-diketones is generally a difficult task. To efficiently develop proteins and peptides that have nucleophilic amino group(s), we have used selections from libraries based on formation of enaminones with 1,3-diketones. Using the enaminone formation reaction-based selections with 1,3-diketone derivatives from antibody libaries, we have previously developed catalytic antibodies that possess a nucleophilic lysine ε-amino group for catalysis (33, 34, 44). We have also demonstrated that peptides with improved activities for enamine catalysis can be generated by the reaction-based selection with 1,3-diketones using phage displayed peptide libraries (45, 46) and developed an RNase S-peptide-derived small peptide that forms an enaminone, upon interaction with RNase S-protein (47).
Here we have explored reaction-based selection with 1,3-diketone derivatives using peptide libraries that did not initially include helix or sheet structures. Peptide libraries composed of randomized amino acids that are not part of other folded proteins were previously used to identify peptides that noncovalently bind to small molecules (48). Although selections starting from a completely randomized 80-mer protein library have been performed to select ATP binding proteins (49), current methods allow for only a small portion of such libraries to be probed. Further, the results obtained from such libraries have not superceded results obtained with smaller peptide libraries. Thus, we started our selection using peptides containing seven to twelve randomized amino acids displayed on phage. Initially-selected peptides were improved by extending their structure through the attachment of libraries composed of six randomized amino acids followed by the same reaction-based selections. Multiple rounds of peptide growth and selection were performed to access functional peptides without resorting to a longer peptide library where the theoretically diversity of the library would far exceed that examined in the actual experiment.
First, phage displayed peptide libraries that contained seven randomized amino acids flanked by a pair of cysteine residues (CX7C, X = any of the natural 20 amino acids), linear seven-mer peptides (X7), and linear 12-mer peptides (X12) fused via a short spacer to the N-terminus of a minor coat protein (pIII) of the filamentous bacteriophage M13 phage were screened in a covalent selection with a 1,3-diketone derivative (Figure 1). Four rounds of binding selection of each peptide-phage library were performed against diketone 1-bovine serum albumin conjugate (1-BSA) (34) immobilized on wells in a microtiter plate. Phage from each of the selected libraries were combined and three additional rounds of selection were performed. After the final round of selection from the first libraries, individual clones were examined for binding to the diketone in an enzyme-linked immunosorbent assay (ELISA) using 1-BSA. The amino acid sequences of the 20 individual clones that showed positive signals, including those with borderline signals (the ELISA signal was above the background but less than 2-fold of the background) in the ELISA, were determined and six different peptides (total 10 out of the 20) had at least one lysine residue. Since we were more interested in peptides that formed enaminones through lysine ε-amino groups than through the N-terminal amino group, these six peptide sequences were used as the starting points for further improvement. The peptides were elongated to evolve their activity by attachment of six randomized amino acid residues of each of the natural 20 amino acids. The resulting libraries (the second generation libraries) were selected against 1-BSA for four rounds using the monovalent phage display system pComb3X (31). ELISA identified individual clones, from the final round of selection from the second-generation libraries, that bound to 1-BSA and the amino acid sequences of the bound clones were determined. These peptides were further elongated by attachment of six randomized amino acid residues and the same phage selection procedure was used. After the selection from the third generation libraries, a peptide that showed a relatively strong ELISA signal (more than 5-fold of the background) against 1-BSA, named rpf1368 (Figure 1b), was obtained and this peptide was further characterized.
Preparation of the fusion protein and evaluation of diketone labeling
In order to examine whether peptide rpf1368 could be used to label a protein, a fusion of this peptide with maltose binding protein (MBP) was prepared and labeling was analyzed by ELISA with 1-BSA. The culture supernatant and lysates of E. coli cells that produced fusion protein rpf1368-MBP showed a positive signal in the ELISA, while the culture supernatant and lysate of E. coli cells that did not produce rpf1368-MBP or that produced only MBP were negative in the same ELISA. The ELISA results concerning these cell lysate experiments and those performed with the purified protein are shown in Figure 2a. Both the cell lysate containing rpf1368-MBP and the purified fusion protein rpf1368-MBP bound to the diketone conjugate 1-BSA significantly better than to BSA. These results indicate that rpf1368 tagged protein can be labeled with diketone derivatives. Since MBP that was not fused to peptide rpf1368 did not bind to 1-BSA, the peptide rpf1368 portion of rpf1368-MBP is essential for binding to the diketone. Note that MBP has 34 lysine residues per molecule. The ELISA results indicate that peptide rpf1368 has a different chemical reactivity with the 1,3-diketone and is selectively labeled.
Labeling of protein rpf1368-MBP was also analyzed using diketone-biotin conjugate 1-biotin in ELISA experiments. Fusion protein rpf1368-MBP bound to 1-biotin supported on a NeutrAvidin, but native MBP did not (Figure 2b). Biotin was introduced into rpf1368-MBP without requirements of the addition of any second catalysts or reagents. These results indicate that peptide rpf1368 functions as a fusion protein and that this peptide can be used as a tag for proteins to introduce compounds conjugated with the diketone.
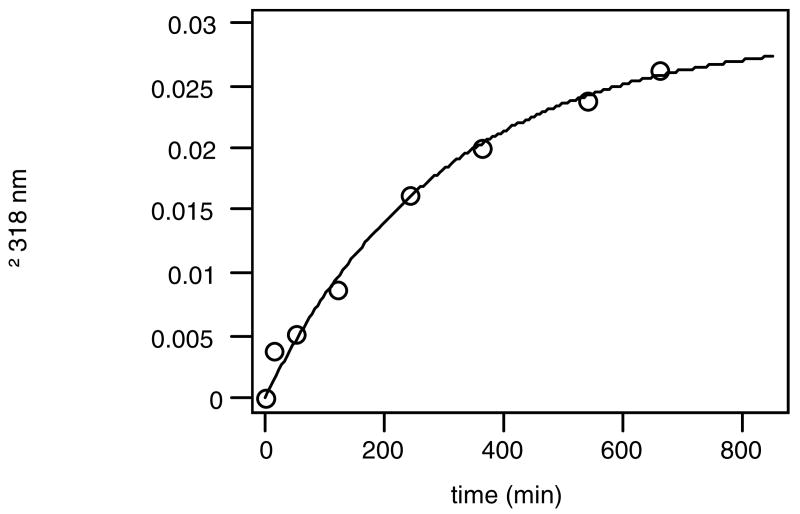
Formation of enaminones can be monitored through the UV absorption characteristic of the enaminone at 318 nm (34), so enaminone formation of rpf1368-MBP was analyzed by monitoring this UV absorption band. When rpf1368-MBP was mixed with 2,4-pentanedione, the smallest compound containing the diketone structure, at pH 7.4, an increase in UV absorption at 318 nm was observed, indicating that rpf1368-MBP covalently bound to this diketone through formation of an enaminone linkage. When 500 μM of 2,4-pentanedione and 2 μM of rpf1368-MBP were mixed in buffer at pH 7.4, the enaminone formation reaction took approximately 10 hours to reach >90% modification as shown in Figure 3. This reaction rate for enaminone formation between rpf1368 and 2,4-pentanedione should be acceptable for labeling. Fusion protein rpf1368-MBP was labeled at a low protein concentration (2 μM). When we studied our previously developed 24-mer peptide FT-YLK3 (46) that possesses nucleophilic lysine residues in a helical structure upon dimerization or oligomerization was fused to MBP, the labeling of FT-YLK3-MBP fusion with 1,3-diketones was not detected at 2 μM fusion protein concentration. At a high concentration, FT-YLK3-MBP formed oligomers, possibly based on oligomerization of the FT-YLK3 portion. Such oligomerization typically is not suitable for many uses of labeled fusion proteins. Thus, as a labeling tag, peptide rpf1368 is superior to our previously developed small peptides containing helical structures where self-association is key for the conformation necessary to provide enaminone forming amino groups.
Figure 3.
Time course of enaminone formation of rpf1368-MBP with 2,4-pentanedione. Increase in absorption at 318 nm, characteristic of the enaminone, was monitored. Conditions: [rpf1368-MBP] 2 μM and [2,4-pentanedione] 500 μM in 2.5% CH3CN/PBS. The second order rate constant k in this reaction {V (M/min) = k[2,4-pentanedione (M)][peptide rpf1368 (M)]} was 6.0 M-1min-1.
Characterization of the labeling reaction using synthetic peptides
In order to characterize the peptide in more detail, the 21-mer peptide rpf1368 was chemically synthesized (with a C-terminal amide, in cyclic form due to the formation of a disulfide bond between cysteines) and evaluated. This synthesized peptide also showed an increase in UV absorption at 318 nm upon addition of 2,4-pentanedione, characteristic of enaminone formation. Enaminone formation was also confirmed by MALDI-TOF mass analysis. A mixture of peptide rpf1368 and diketone 2 gave isotopic mass m/z MH+ of 2725 whereas peptide rpf1368 alone had isotopic mass m/z MH+ of 2553 (See Supporting Information). One molecule of diketone 2 (MW 190) was incorporated per peptide rpf1368 and one molecule of H2O (MW 18) was released, indicating the formation of the enaminone through an amino group of the peptide. Only one amino group of rpf1368 was nucleophilic. Another possible covalent modification, formation of the pyrimidine derivative with a guanidyl group of an arginine was excluded by the mass analysis: If a guanidyl group reacted with diketone 2 and formed a derivative of pyrimidine (50), the mass MH+ should be 2707 because two molecules of H2O are released in the formation of pyrimidine. Unrelated peptides possessing lysine residue(s), ELLELDKWASLWNC, ELKDKWASLWNWFNIT, ELDKWASLWNWFDITGGC, and CDEKSKLQEIYQELTQLKAAVGEL, did not form an enaminone with 2,4-pentanedione; no changes in absorption at 318 nm were observed in the presence of 2,4-pentanedione for these peptides. The single reactive site within peptide rpf1368 is in contrast to our previously developed 24-mer and 35-mer peptides possessing a helical structure (46, 51). Although multiple lysine groups are present in peptide rpf1368, only one amino group was nucleophilic enough to form the enaminone.
The conformation of peptide rpf1368 was critical for the reactivity. An intramolecular disulfide bond between the two cysteines of the peptide formed spontaneously in buffer under air at room temperature. The non-cyclized peptide (prior to disulfide bond formation) did not react with 2,4-pentanedione. The cyclic constraint of rpf1368 by the disulfide bond provided for the formation of favorable electrostatic interactions or a microenvironment that tuned the reactivity of the amino group.
Conclusion
We have developed a small 21-mer peptide that possesses a single reactive amino group using a reaction-based selection with a 1,3-diketone derivative and phage displayed peptide libraries. The libraries did not contain designed helix or sheet structures. We have demonstrated that the peptide can be used as a tag for a protein of interest to covalently label the protein with a synthetic molecule through enaminone formation. No enzyme and no additional reagents were necessary for the labeling of the peptide-protein fusion with the diketone derivatives. Although the reactivity of peptide rpf1368 is still moderate compared to existing covalent labeling tags that are derivatives of natural proteins, this peptide is the smallest peptide tag for covalent labeling with designed synthetic molecules without the requirement of additional catalysts or toxic metal reagents. We have demonstrated that small peptides that covalently react with designer synthetic molecules can be generated. Further improvement of this peptide should provide better small peptide tags for covalent labeling. Using these tags, a variety of molecules such as reporter probes and other functionalities may be conveniently and covalently introduced into proteins of interest.
Supplementary Material
A scheme for synthesis of 1-biotin and mass spectra for the reaction of peptide rpf1368 with diketone 2. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This study was supported in part by The Skaggs Institute for Chemical Biology and by NIH R21GM078447.
References
- 1.Prescher JA, Bertozzi CR. Chemistry in living systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 2.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 3.Keppler A, Pick H, Arivoli C, Vogel H, Johnsson K. Labeling of fusion proteins with synthetic fluorophores in living cells. Proc Natl Acad Sci USA. 2004;101:9955–9959. doi: 10.1073/pnas.0401923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.George N, Pick H, Vogel H, Johnsson N, Johnsson K. Specific labeling of cell surface proteins with chemically diverse compounds. J Am Chem Soc. 2004;126:8896–8897. doi: 10.1021/ja048396s. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z, Gildersleeve J, Yang YY, Xu R, Loo JA, Uryu S, Wong CH, Schultz PG. A new strategy for the synthesis of glycoproteins. Science. 2004;303:371–373. doi: 10.1126/science.1089509. [DOI] [PubMed] [Google Scholar]
- 6.Hamann PR, Hinman LM, Hollander I, Beyer CF, Lindh D, Holcomb R, Hallett W, Tsou HR, Upeslacis J, Shochat D, Mountain A, Flowers DA, Bernstein I. Gemtuzumab ozogamicin, a potent and selective anti-CD33 antibody-calicheamicin conjugate for treatment of acute myeloid leukemia. Bioconjugate Chem. 2002;13:47–58. doi: 10.1021/bc010021y. [DOI] [PubMed] [Google Scholar]
- 7.Lillo AM, Sun C, Gao C, Ditzel H, Parrish J, Gauss GM, Moss J, Felding-Habermann B, Wirsching P, Boger DL, Janda KD. A human single-chain antibody specific for integrin α3β1 capable of cell internalization and delivery of antitumor agents. Chem Biol. 2004;11:897–906. doi: 10.1016/j.chembiol.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 8.Mohsin H, Jia F, Sivagura G, Hudson MJ, Shelton TD, Hoffmann TJ, Cutler CS, Ketring AR, Athey PS, Simon J, Frank RK, Jurisson SS, Lewis MR. Radiolanthanide-labeled monoclonal antibody CC49 for radioimmunotherapy of cancer: Biological comparison of DOTA conjugates and 149Pm, 166Ho, and 177Lu. Bioconjugate Chem. 2006;17:485–492. doi: 10.1021/bc0502356. [DOI] [PubMed] [Google Scholar]
- 9.Antczak C, Jaggi J, LeFave CV, Curcio MJ, McDevitt MR, Scheinberg DA. Influence of the linker on the biodistribution and catabolism of actinium-225 self-immolative tumor-targeted isotope generators. Bioconjugate Chem. 2006;17:1551–1560. doi: 10.1021/bc060156+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng TL, Chen BM, Chan LY, Wu PY, Chern JW, Roffler SR. Poly(ethylene glycol)modification of β-glucuronidase-antibody conjugates for solid-tumor therapy by targeted activation of glucuronide prodrugs. Cancer Immunol Immunother. 1997;44:305–315. doi: 10.1007/s002620050387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin J, Liu F, Li X, Walsh CT. Labeling proteins with small molecules by site-specific posttranslational modification. J Am Chem Soc. 2004;126:7754–7755. doi: 10.1021/ja047749k. [DOI] [PubMed] [Google Scholar]
- 12.Phizicky E, Bastiaens PIH, Zhu H, Snyder M, Fields S. Protein analysis on a proteomic scale. Nature. 2003;422:208–215. doi: 10.1038/nature01512. [DOI] [PubMed] [Google Scholar]
- 13.Hodneland CD, Lee YS, Min DH, Mrksich M. Selective immobilization of proteins to self-assembled monolayers presenting active site-directed capture ligands. Proc Natl Acad Sci USA. 2002;99:5048–5052. doi: 10.1073/pnas.072685299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 15.Yin J, Lin AJ, Golan DE, Walsh CT. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat Protocols. 2006;1:280–285. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- 16.Chen I, Howarth M, Lin W, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 17.Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 18.Martin BR, Giepmans BNG, Adams SR, Tsien RY. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 19.Lesaicherre ML, Lue RYP, Chen GYJ, Zhu Q, Yao SQ. Intein-mediated biotinylation of proteins and its application in a protein microarray. J Am Chem Soc. 2002;124:8768–8769. doi: 10.1021/ja0265963. [DOI] [PubMed] [Google Scholar]
- 20.Wood RJ, Pascoe DD, Brown ZK, Medlicott EM, Kriek M, Neylon C, Roach PL. optimized conjugation of a fluorescent label to proteins via intein-mediated activation and ligation. Bioconjugate Chem. 2004;15:366–372. doi: 10.1021/bc0341728. [DOI] [PubMed] [Google Scholar]
- 21.Muralidharan V, Muir TW. Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat Methods. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 22.Tolbert TJ, Wong CH. New methods for proteomic research: Preparation of proteins with N-terminal cysteines for labeling and conjugation. Angew Chem Int Ed. 2002;41:2171–2174. doi: 10.1002/1521-3773(20020617)41:12<2171::aid-anie2171>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 23.Chelius D, Shaler TA. Capture of peptides with N-terminal serine and threonine: A sequence-specific chemical method for peptide mixture simplification. Bioconjugate Chem. 2003;14:205–211. doi: 10.1021/bc025605u. [DOI] [PubMed] [Google Scholar]
- 24.Dirksen A, Hackeng TM, Dawson PE. Nucleophilic catalysis of oxime ligation. Angew Chem Int Ed. 2006;45:7581–7584. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]
- 25.Dirksen A, Dirlsen S, Hackeng TM, Dawson PE. Nucleophilic catalysis of hydrazone formation and implications for dynamic covalent chemistry. J Am Chem Soc. 2006;128:15602–15603. doi: 10.1021/ja067189k. [DOI] [PubMed] [Google Scholar]
- 26.Miller LW, Sable J, Goelet P, Sheetz MP, Cornish VW. Methotrexate conjugates: A molecular in vitro protein tag. Angew Chem Int Ed. 2004;43:1672–1675. doi: 10.1002/anie.200352852. [DOI] [PubMed] [Google Scholar]
- 27.Miller LW, Cai Y, Sheetz MP, Cornish VW. In vitro protein labeling with trimethoprim conjugates: a flexible chemical tag. Nat Methods. 2005;2:255–257. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- 28.Marks KM, Braun PD, Nolan GP. A general approach for chemical labeling and rapid, spatially controlled protein inactivation. Proc Natl Acad Sci USA. 2004;101:9982–9987. doi: 10.1073/pnas.0401609101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farinas J, Verkman AS. Receptor-mediated targeting of fluorescent probes in living cells. J Biol Chem. 1999;274:7603–7606. doi: 10.1074/jbc.274.12.7603. [DOI] [PubMed] [Google Scholar]
- 30.Wu MM, Llopis J, Adams S, McCaffery JM, Kulomaa MS, Machen TE, Moore HPH, Tsien RY. Organelle pH studies using targeted avidin and fluorescein-biotin. Chem Biol. 2000;7:197–209. doi: 10.1016/s1074-5521(00)00088-0. [DOI] [PubMed] [Google Scholar]
- 31.Rader C, Barbas CF., III Phage display of combinatorial antibody libraries. Curr Opin Biotechnol. 1997;8:503–508. doi: 10.1016/s0958-1669(97)80075-4. [DOI] [PubMed] [Google Scholar]
- 32.Zwick MB, Bonnycastle LLC, Noren KA, Venturini S, Leong E, Barbas CF, III, Noren CJ, Scott JK. The maltose-binding protein as a scaffold for movovalent display of peptides derived from phage libraries. Anal Biochem. 1998;264:87–97. doi: 10.1006/abio.1998.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka F, Fuller R, Shim H, Lerner RA, Barbas CF., III Evolution of aldolase antibodies in vitro: correlation of catalytic activity and reaction-based selection. J Mol Biol. 2004;335:1007–1018. doi: 10.1016/j.jmb.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 34.Wagner J, Lerner RA, Barbas CF., III Efficient aldolase catalytic antibodies that use the enamine mechanism of natural enzymes. Science. 1995;270:1797–1800. doi: 10.1126/science.270.5243.1797. [DOI] [PubMed] [Google Scholar]
- 35.Westheimer FH. Coincidences, decarboxylation, and electrostatic effects. Tetrahedron. 1995;51:3–20. [Google Scholar]
- 36.Guidinger PF, Nowak T. An active-site lysine in avian liver phosphoenolpyruvate carboxykinase. Biochemistry. 1991;30:8851–8861. doi: 10.1021/bi00100a018. [DOI] [PubMed] [Google Scholar]
- 37.Rader C, Sinha SC, Popkov M, Lerner RA, Barbas CF., III Chemically programmed monoclonal antibodies for cancer therapy: Adaptor immunotherapy based on a covalent antibody catalyst. Proc Natl Acad Sci USA. 2003;100:5396–5400. doi: 10.1073/pnas.0931308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu X, Tanaka F, Hu Y, Heine A, Fuller R, Zhong G, Olson AJ, Lerner RA, Barbas CF, III, Wilson IA. Theorigin of enantioselectivity in aldolase antibodies: Crystal structure, site-directed mutagenesis, and computational analysis. J Mol Biol. 2004;343:1269–1280. doi: 10.1016/j.jmb.2004.08.102. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka F, Thayumanavan R, Mase N, Barbas CF., III Rapid analysis of solvent effects on enamine formation by fluorescence: how might enzymes facilitate enamine chemistry with primary amines? Tetrahedron Lett. 2004;45:325–328. [Google Scholar]
- 40.Johnsson K, Allemann RK, Widmer H, Benner SA. Synthesis, structure and activity of artificial, rationally designed catalytic polypeptides. Nature. 1993;365:530–532. doi: 10.1038/365530a0. [DOI] [PubMed] [Google Scholar]
- 41.Perez-Paya E, Houghten RA, Blondell SE. Functionalized protein-like structures from conformationally defined synthetic combinatorial libraries. J Biol Chem. 1996;271:4120–4126. doi: 10.1074/jbc.271.8.4120. [DOI] [PubMed] [Google Scholar]
- 42.Allert M, Baltzer L. A designed folded polypeptide model system that catalyzes the decarboxylation of oxaloacetate. Chem Eur J. 2002;8:2549–2560. doi: 10.1002/1521-3765(20020603)8:11<2549::AID-CHEM2549>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 43.Weston CJ, Cureton CH, Calvert MJ, Smart OS, Allemann RK. A stable miniature protein with oxaloacetate decarboxylase activity. ChemBioChem. 2004;5:1075–1080. doi: 10.1002/cbic.200300805. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka F, Lerner RA, Barbas CF., III Reconstructing aldolase antibodies to alter their substrate specificity and turnover. J Am Chem Soc. 2000;122:4835–4836. [Google Scholar]
- 45.Tanaka F, Barbas CF., III Phage display selection of peptides possessing aldolase activity. Chem Commun. 2001:769–770. [Google Scholar]
- 46.Tanaka F, Fuller R, Barbas CF., III Development of small designer aldolase enzymes: Catalytic activity, folding, and substrate specificity. Biochemistry. 2005;44:7583–7592. doi: 10.1021/bi050216j. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka F, Fuller F. Control of function of a small protein by a protein. Bioorg Med Chem Lett. 2006;16:4059–4062. doi: 10.1016/j.bmcl.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 48.Rozinov MN, Nolan GP. Evolution of peptides that modulate the spectral qualities ofbound, small-molecule fluorophores. Chem Biol. 2001;5:713–728. doi: 10.1016/s1074-5521(98)90664-0. 1998. [DOI] [PubMed] [Google Scholar]
- 49.Keefe AD, Szostak JW. Functional proteins from a random-sequence library. Nature. 2001;410:715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilbert HF, III, O'Leary MH. Modification of arginine and lysine in proteins with 2,4-pentanedione. Biochemistry. 1975;14:5194–5199. doi: 10.1021/bi00694a027. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka F, Barbas CF., III A modular assembly strategy for improving the substrate specificity of small catalytic peptides. J Am Chem Soc. 2002;124:3510–3511. doi: 10.1021/ja0171815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A scheme for synthesis of 1-biotin and mass spectra for the reaction of peptide rpf1368 with diketone 2. This material is available free of charge via the Internet at http://pubs.acs.org.