Abstract
Dolichols are long-chain unsaturated polyisoprenoids with multiple cellular functions, such as serving as lipid carriers of sugars used for protein glycosylation, which affects protein trafficking in the endoplasmic reticulum. The biological functions of dolichols in plants are largely unknown. We isolated an Arabidopsis thaliana mutant, lew1 (for leaf wilting1), that showed a leaf-wilting phenotype under normal growth conditions. LEW1 encoded a cis-prenyltransferase, which when expressed in Escherichia coli catalyzed the formation of dolichol with a chain length around C80 in an in vitro assay. The lew1 mutation reduced the total plant content of main dolichols by ∼85% and caused protein glycosylation defects. The mutation also impaired plasma membrane integrity, causing electrolyte leakage, lower turgor, reduced stomatal conductance, and increased drought resistance. Interestingly, drought stress in the lew1 mutant induced higher expression of the unfolded protein response pathway genes BINDING PROTEIN and BASIC DOMAIN/LEUCINE ZIPPER60 as well as earlier expression of the stress-responsive genes RD29A and COR47. The lew1 mutant was more sensitive to dark treatment, but this dark sensitivity was suppressed by drought treatment. Our data suggest that LEW1 catalyzes dolichol biosynthesis and that dolichol is important for plant responses to endoplasmic reticulum stress, drought, and dark-induced senescence in Arabidopsis.
INTRODUCTION
Dolichols are long-chain unsaturated polyisoprenoids present in all eukaryotic cells; they consist of 15 to 23 isoprenic units depending on the species (Chojnacki and Dallner, 1988; Swiezewskaa and Danikiewiczb, 2005). In animals, dolichols are broadly distributed in different cells. They are localized within the two halves of the phospholipid bilayer, with high contents in the plasma membranes, peroxisomes, Golgi vesicles, and lysosomes. Dolichols exist as free alcohols or products derived by phosphorylation, esterification, or glycosylation (Chojnacki and Dallner, 1988; Swiezewskaa and Danikiewiczb, 2005). The biosynthesis of dolichols takes place mainly in the endoplasmic reticulum (ER) and to a lesser extent in peroxisomes (Chojnacki and Dallner, 1988). This biosynthesis is initiated by the formation of farnesyl diphosphate (FPP), primarily through the mevalonate pathway (Chojnacki and Dallner, 1988; Swiezewskaa and Danikiewiczb, 2005). The formation of longer chain polyprenyl diphosphate is catalyzed by enzymes called cis-prenyltransferases, which sequentially add isopentenyl diphosphates (IPPs) onto FPP (Grabinska and Palamarczyk, 2002). The number of IPPs added is species-dependent. Polyprenyl diphosphate is further converted to dolichol and dolichyl phosphate by a series of reactions (Grabinska and Palamarczyk, 2002). Dolichyl phosphate serves as a glycosyl carrier lipid in the biosyntheses of protein C- and O-mannosylation, in glycosylphosphatidylinositol anchors, and is one of the rate-limiting factors in N-linked protein glycosylation in yeast and mammalian cells (Burda and Aebi, 1999; Jones et al., 2005).
The first eukaryotic cis-prenyltransferase–encoding gene, RER2, which is responsible for dolichol synthesis, was cloned from the yeast mutant rer2 (Sato et al., 1999). The multicopy suppressor of rer2, STR1, encodes a protein similar to RER2, but with different physiological roles during cell growth (Sato et al., 2001). rer2 shows many phenotypes, including temperature and hygromycin sensitivity, slow growth, defects in N- and O-glycosylation, and abnormal accumulation of ER and Golgi membranes (Sato et al., 1999). By contrast, the str1 mutant does not have noticeable phenotypes (Sato et al., 2001). Through comparison of cis-prenyltransferase sequences with plant genomic sequences, a homologous gene (AT2G23410) was cloned from Arabidopsis thaliana (Cunillera et al., 2000; Oh et al., 2000). Expression of this gene in yeast rer2 complements its thermosensitive phenotype (Cunillera et al., 2000). A recombinant protein produced from AT2G23410 shows dehydrodolichyl diphosphate synthase activity, and dolichols could be synthesized in an in vitro assay (Cunillera et al., 2000; Oh et al., 2000). It should be noted that Escherichia coli cis-prenyltransferase synthesizes undecaprenyl diphosphate (C55), which functions as a carrier lipid in cell wall polysaccharide synthesis (Kato et al., 1999). However, no dolichol-deficient mutants have been isolated from any multicellular organisms, including plants, and the physiological roles of dolichols in plants are largely unknown.
N-Glycosylation of secreted or membrane proteins occurs, shortly after synthesis, in the lumen of the ER. Inhibition of N-glycosylation often reduces the folding efficiency, enhances incorrect folding, and accelerates the degradation of the hypoglycoprotein. Chaperone proteins such as BiP (for BINDING PROTEIN) play crucial roles in assisting protein folding during ER stress, which can result from N-glycosylation defects (Wilson, 2002). Increases in secretory activity and in the accumulation of unfolded proteins induce the transcripts of chaperones through the unfolded protein response (UPR) pathway (Wilson, 2002). Recently, two BASIC DOMAIN/LEUCINE ZIPPER (bZIP) transcription factor genes, bZIP60 and bZIP28, were isolated, both of which have been shown to activate BiP expression, probably through ER stress response element–like sequences (Iwata and Koizumi, 2005; Liu et al., 2007a). bZIP60 and bZIP28 are membrane proteins that are anchored to the ER membrane under normal conditions. Upon ER stress, bZIP28, and probably also bZIP60, is cleaved and the bZIP domain translocates to the nucleus to activate the expression of BiP (Iwata and Koizumi, 2005; Liu et al., 2007a).
Drought stress is one of the most damaging environmental conditions that limit plant growth and distribution. Drought stress induces the accumulation of the phytohormone abscisic acid, which, as an endogenous signal, plays crucial roles in mediating plant responses in order to adjust the water deficit (Xiong et al., 2002; Christmann et al., 2006; Shinozaki and Yamaguchi-Shinozaki, 2007). During drought stress, many genes are upregulated. Expression of some of these genes is mediated by abscisic acid, while others are abscisic acid–independent (Xiong et al., 2002; Christmann et al., 2006; Shinozaki and Yamaguchi-Shinozaki, 2007). The expression of BiP was found to be induced by water stress in soybean (Glycine max) (Cascardo et al., 2000), and overexpression of BiP in transgenic tobacco (Nicotiana tabacum) improves plant drought tolerance (Alvim et al., 2001). However, the upregulation of BiP is not necessarily related to increased drought tolerance (Koiwa et al., 2003). Gene expression profiling of soybean leaves shows that some genes are induced by both osmotic and UPR stresses, suggesting that the two pathways may converge in plants (Irsigler et al., 2007). These results suggest complex crosstalk between drought stress responses and the UPR pathway (Alvim et al., 2001; Koiwa et al., 2003; Irsigler et al., 2007).
Through a genetic screen, we isolated the Arabidopsis mutant lew1 (for leaf wilting1). Here, we show that lew1 is altered in a gene encoding a cis-prenyltransferase for dolichol synthesis. lew1 mutant plants showed a leaf-wilting phenotype under normal growth conditions due to impaired membrane integrity. lew1 mutant plants were hypersensitive to tunicamycin and had reduced levels of protein glycosylation. Drought stress induced higher expression of the UPR genes BiP and bZIP60 as well as earlier expression of the abiotic stress–responsive genes COR47 and RD29A in lew1 than in the wild type. Furthermore, drought treatment suppressed dark-induced senescence in lew1. These data implicate LEW1 as having crucial roles in dolichol synthesis, the UPR pathway, and the abiotic stress response in Arabidopsis.
RESULTS
lew1 Mutant Plants Exhibit a Leaf-Wilting Phenotype under Normal Growth Conditions Despite Increased Hydraulic Conductivity in Roots
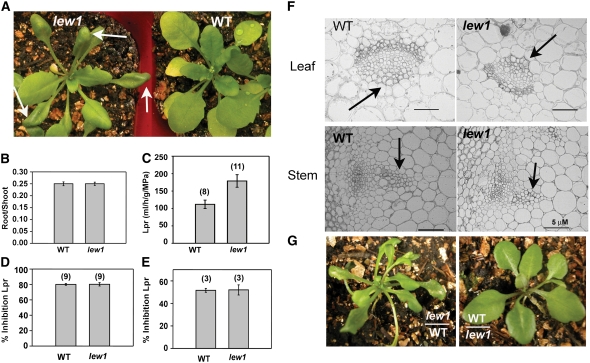
The lew1 mutant was isolated during a genetic screen for leaf-wilting phenotypes (Figure 1A) in an ethyl methanesulfonate–mutagenized Arabidopsis M2 population (Chen et al., 2005). We found that lew1 plants showed a clear leaf-wilting phenotype even under well-watered normal growth conditions. The wilting in lew1 was observed mainly in the margins of old leaves but not in young leaves. This differs from leaf wilting normally caused by water deficit, in that it was not seen in all leaves and was not associated with changed water status of the seedlings. The overall size of lew1 mature plants was smaller than that of wild-type plants (Figure 1A). There was no apparent difference in flowering time between lew1 and wild-type plants (Figure 2A).
Figure 1.
The Phenotypes of the lew1 Mutant.
(A) Wild-type and lew1 mutant plants grown in soil. Left, lew1 (arrows point to wilted parts of leaves); right, the wild type.
(B) Comparison of root:shoot ratio. Data are means ± se (n = 41).
(C) Lpr of wild-type and lew1 roots. The numbers in parentheses indicate the numbers of plants analyzed. Data are means ± se.
(D) and (E) Effects of aquaporin inhibitors on Lpr of wild-type and lew1 plants: azide at 1 mM, 30 min (D) and mercury at 50 μM, 60 min (E). Data are means ± se. The numbers in parentheses indicate the numbers of plants analyzed.
(F) Structure of xylem in leaves (top row) and stems (bottom row) of wild-type and lew1 plants. Arrows point to xylem. Bars = 5 μm.
(G) Grafting experiments. Left, lew1 shoot with wild-type root; right, wild-type shoot with lew1 root.
Figure 2.
Analysis of Drought Resistance and Related Parameters in lew1.
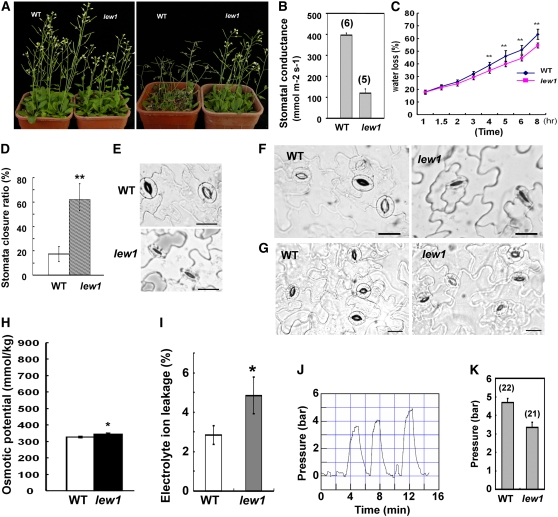
(A) Comparison of wild-type and lew1 plant growth under normal (left) and drought stress (right) conditions. For drought treatment, 3-week-old seedlings were subjected to water withholding for 2 weeks.
(B) Stomatal conductance in the wild type and lew1. Data are means ± se. The numbers in parentheses indicate the numbers of analyzed plants. Two to four leaves were used per plant for the wild type, and one to two leaves were used per plant for lew1.
(C) Water loss from detached leaves. Three independent experiments were performed, and 15 leaves per genotype were used in each experiment. Data are means ± se (Student's t test; ** statistically significant difference [P < 0.01]).
(D) Stomata closure rates of the wild type and lew1. For lew1 plants, stomata from wilting parts were measured. About 40 stomata were measured in each of three experiments for both the wild type and lew1. Data are means ± se (Student's t test; ** statistically significant difference [P < 0.05]).
(E) Stomata comparison between the wild type and lew1. The stomata on the wilting part of a leaf are shown. Bars = 20 μm.
(F) Comparison of stomata in the wild type and lew1 under conditions that promote full stomatal opening. The stomata from the wilting part of a lew1 leaf were not open under conditions when all stomata in the wild-type leaf were open. Bars = 20 μm.
(G) Comparison of stomata in the wild type and lew1. Stomata on nonwilting parts in the leaf base of lew1 are shown. Bars = 20 μm.
(H) Osmotic potential in the wild type and lew1 under normal growth conditions. Values are means ± se, n = 3 (Student's t test; * statistically significant difference [P < 0.05]).
(I) Comparison of leaf electrolyte leakage between lew1 and the wild type. Values are means ± se, n = 3 (Student's t test; * statistically significant difference [P < 0.05]).
(J) Typical recording trace showing measurements of turgor in epidermal cells of a wild-type leaf. Following successful impalement of a cell with the pressure probe, the intracellular recording was continued until a stationary turgor was reached. The trace shows successive recordings in three individual cells.
(K) Mean (±se) epidermal cell turgor in areas of leaves of wild-type and lew1 plants showing no obvious sign of wilting (wild type, n = 22 cells; lew1, n = 21 cells). There is a statistically significant difference (P = 0.0005) between the two genotypes.
In principle, leaf wilting in plants may be caused by (1) reduced water uptake by roots, (2) low water transport through the xylem, (3) high transpiration, (4) low osmotic potential in leaf cells, or (5) impaired cell or membrane integrity. We observed that wild-type and lew1 plants showed similar root:shoot ratios (Figure 1B). This suggested that no major imbalance between water uptake and water loss was created by gross morphological differences between the two genotypes. The intrinsic capacity of roots to take up water (i.e., their hydraulic conductivity [Lpr]) was determined from measurements of pressure-dependent sap flow in excised root systems (Boursiac et al., 2005). As shown in Figure 1C, lew1 plants exhibited a significantly greater (+64%) Lpr than wild-type plants. Previous studies have shown that aquaporin activity contributes to most of the Lpr in Arabidopsis (Tournaire-Roux et al., 2003). In order to understand whether the enhanced Lpr in lew1 was due to enhanced root aquaporin activity or to enhanced flow through the cell walls, we tested the effects of two independent aquaporin inhibitors, mercury and azide. Mercury is the most commonly described aquaporin blocker, but azide is even more potent, as it can reversibly block Lpr of wild-type plants by up to 80% (Tournaire-Roux et al., 2003). The results showed that wild-type and lew1 plants had similar levels of inhibition, particularly in the case of azide (Figures 1D and 1E), which meant that most of the increase in Lpr shown by lew1 (+64%) could be accounted for by enhanced aquaporin activity. These results disprove a possible hydraulic limitation of roots in the leaf-wilting phenotype. They further suggest that the lew1 mutation increases the efficiency of water transport across root cells.
We previously showed that leaf wilting in lew2 is caused by impeded water transport due to a collapsed xylem (Chen et al., 2005). To investigate a possible xylem defect, we examined cross sections of vascular bundles in the xylem of stems and leaves and found no difference between lew1 and wild-type plants (Figure 1F). We also performed grafting experiments to test whether leaf wilting in lew1 might have been due to an overall defect of water supply to the shoot. When lew1 shoots were grafted onto wild-type roots, leaf wilting still occurred. By contrast, when grafted onto lew1 roots, wild-type shoots did not show any wilting (Figure 1G).
In conclusion, the leaf-wilting phenotype of lew1 was not caused by defects in root water uptake or xylem transport.
The lew1 Mutation Impairs Cell Turgor in Leaves by Causing Increased Electrolyte Leakage and Reduces Leaf Transpiration
In contrast to the increased Lpr, increased transpiration could cause leaf wilting. Surprisingly, we found that stomatal conductance, as measured using a porometer, was reduced by ∼75% in lew1 (Figure 2B). Consistent with this, spontaneous water loss from detached leaves was slower in lew1 than in the wild type (Figure 2C). Importantly, lew1 plants grown in soil were more resistant to drought stress than were wild-type plants (Figure 2A).
A closer inspection of leaf epidermis showed that, under normal growth conditions, 60% of stomata were closed in the wilted parts of lew1 leaves but only ∼15% of stomata were closed in wild-type leaves (Figures 2D and 2E). Even under conditions (e.g., high light) that promote stomatal opening, when all stomata of wild-type leaves were fully open, most stomata in the wilted parts of lew1 leaves remained partly closed (Figure 2F). By contrast, no apparent difference in stomatal opening was found in the unwilted parts of lew1 and wild-type leaves (Figure 2G). These results suggested that, in the wilted parts of lew1 leaves, guard cells and possibly surrounding cells were not able to establish a normal turgor. To assess further the basis of the apparent leaf-wilting phenotype of lew1, leaf cell turgor was investigated using the cell pressure probe technique. Because lew1 leaves could show signs of early senescence, the measurements were performed in young, growing leaves that did not show any of these symptoms. Yet, local zones of wilting, mostly in the most distal part of the blade, could be observed in lew1. Among all leaf cell types investigated, the large fusiform epidermal cells located at the edge of the blade yielded the most stable micropipette impalements and turgor recordings. In wild-type plants, cell turgor measurements could be performed all along the leaf edge, yielding a mean stationary turgor value of P0 = 4.70 ± 0.21 bars (n = 22 cells) (Figure 2J). By contrast, impalement of the wilted area in lew1 leaves did not allow any stable turgor recording. Measurements performed in other areas of the leaf provided a mean turgor value of P0 = 3.34 ± 0.29 bars (n = 21 cells), which was significantly lower (P = 0.0005) than the value in wild-type leaves. Thus, leaf cells of lew1 showed a significantly reduced turgor, even in areas showing no clear sign of wilting.
Under normal growth conditions, the osmotic potential of lew1 leaves was slightly higher than that of wild-type leaves (Figure 2H). This was probably due to a slight osmotic compensation in the wilted lew1 cells. We then examined cell membrane integrity by measuring electrolyte leakage. As shown in Figure 2I, lew1 leaves had a higher electrolyte ion leakage than did wild-type leaves, suggesting that the lew1 mutation impaired membrane integrity.
In conclusion, these results suggest that the leaf-wilting phenotype of lew1 is due to increased solute leakage caused by cell membrane lesions. The reduced cell turgor would lead to partial stomatal closure and less transpiration in lew1.
Mapping-Based Cloning of the LEW1 Gene
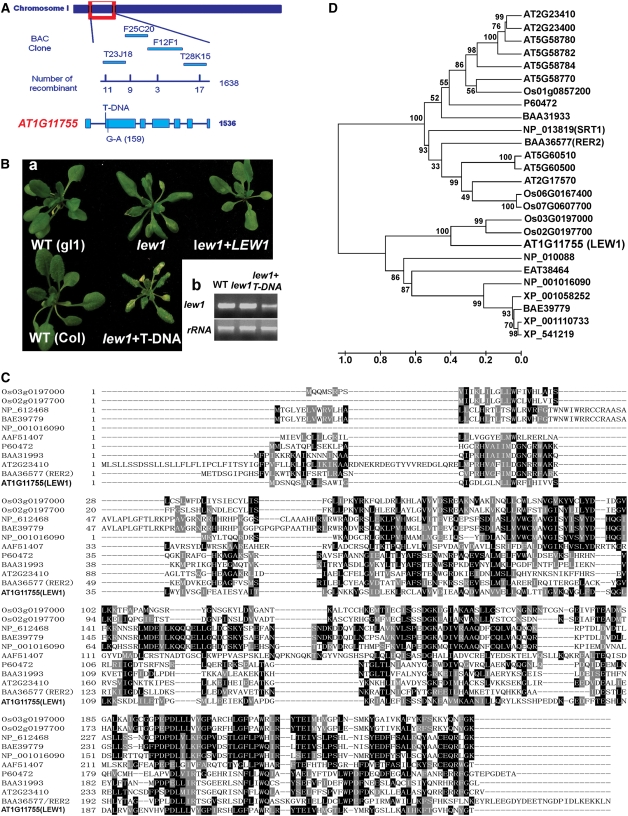
To clone the LEW1 gene, lew1 plants in the Columbia gl1 accession were crossed with wild-type Landsberg plants, and F2 progeny was grown in soil under normal conditions. lew1 plants with a leaf-wilting phenotype were selected for genetic mapping. LEW1 was initially mapped to the upper arm of chromosome 1. Fine-mapping narrowed the lew1 mutation to a region covered by four BAC clones, T23J18, F25C20, F12F1, and T28K15 (Figure 3A). Further fine-mapping delimited lew1 on BAC clones F25C20 and F12F1. We sequenced all of the open reading frames from the lew1 mutant in this region and found a single G-to-A mutation in the open reading frame of AT1G11755, which contains seven exons (Figure 3A). The mutation occurred in the second exon and changed GGT (encoding Gly-21) to GAT (encoding Asp-21) in lew1 (Figure 3A).
Figure 3.
Position Cloning of LEW1 and Sequence Analysis of Long-Chain cis-Prenyltransferases.
(A) Positional cloning of LEW1. The LEW1 locus was mapped to between two polymorphic markers on BAC clones T23J18 (recombinants, 11 of 1638) and T28K15 (recombinants, 17 of 1638). Further mapping delimited the LEW1 locus to a region within BACs F12F1 (recombinants, 3 of 1638) and T28K15. All candidate open reading frames were sequenced in this region, and only one mutation (G159A) was found in AT1G11755.
(B) (a) Complementation of the lew1 mutant. Shown are the wild type (gl1), the lew1 mutant, the lew1 mutant transformed with 35S-LEW1 (cDNA), the wild type (Columbia [Col]), and a heterozygous plant of lew1 crossed with a line in which the LEW1 gene was disrupted by a T-DNA insertion. The T-DNA was inserted at position 205 counting from the first putative ATG of AT1G11755. (b) RT-PCR analysis of LEW1 transcripts in the wild type, lew1, and a heterozygous plant of lew1 crossed with the T-DNA line. rRNA is shown as a control for loading.
(C) Alignment of long-chain cis-prenyltransferases from different organisms: rice, Os02g0197700 and Os03g0197000; human, NP_612468; mouse, BAE39779; Xenopus, NP_001016090; fruit fly, AAF51407; E. coli, P60472; Micrococcus luteus, BAA31993; Saccharomyces cerevisiae, BAA36577 (RER2); Arabidopsis, AT2G23410 and AT1G11755 (LEW1).
(D) Phylogenetic tree of LEW1 and related proteins from different organisms: Arabidopsis, At2g23400, At5g58780, At5g58782, At5g58784, At5g58770, At5g60510, At5g60500, and At2g17570; rice, Os01g0857200, Os06g0167400, and Os07g0607700; monkey, XP_001110733; dog, XP_541219; Aedes aegypti, EAT38464; rat, XP_001058252; S. cerevisiae, NP_013819/SRT1 and NP_010088; the others are the same as in (C). The phylogenetic tree was constructed on the alignment using MEGA (version 4.0) (see Supplemental Data Set 1 online). Bootstrap values were calculated from 1000 trials and are shown at each node. The extent of divergence according to the scale (relative units) is shown at bottom.
The cDNA of AT1G11755 was amplified and overexpressed in lew1 and wild-type plants under the control of the cauliflower mosaic virus 35S promoter. We obtained 25 independent transgenic lew1 lines and found that the leaf-wilting phenotype was complemented in 21 of these lines (Figure 3B shows a representative plant of lew1+LEW1). In parallel to this, an Arabidopsis line (SALK_032276) carrying a T-DNA insertion in the second exon of AT1G11755 was obtained from the Arabidopsis stock center. Our failure to obtain homozygous T-DNA insertion plants suggested that complete disruption of LEW1 is lethal. Plants heterozygous for the T-DNA insertion were crossed with the lew1 mutant. The F2 progeny segregated into three different phenotypes, including typical wild-type and lew1 phenotypes, as expected. We also observed a third class of plants that were smaller, showed earlier senescence, and produced fewer seeds than lew1 or wild-type plants (Figure 3Ba). Genetic and DNA sequence analysis showed these plants to be heterozygous for both the T-DNA insertion and the lew1 mutation. RT-PCR analysis indicated that LEW1 transcripts were present in these heterozygous plants, perhaps at reduced levels (Figure 3Bb).
The LEW1 Gene Encodes a Novel cis-Prenyltransferase Protein
The LEW1 gene encodes a polypeptide of 254 amino acids with a predicted molecular mass of 28.52 kD. Because the genes encoding cis-prenyltransferases form a large family with an overall low level of sequence similarity, we selected for comparison only those candidates that were the most closely related to LEW1 (Figure 3C). The previously identified AT2G23410 and yeast RER2 were included in this analysis. Figure 3D shows that in Arabidopsis, LEW1 (AT1G11755) has only one copy, whereas in rice (Oryza sativa), there are two homologs (Os 03G0197000 and Os 02G0197700). The product of AT2G23410, RER2, and a RER2 homolog in E. coli (P60472) belong to another homology subgroup (Sato et al., 1999; Cunillera et al., 2000; Oh et al., 2000). The analysis indicates that LEW1 is a new member of the cis-prenyltransferase family.
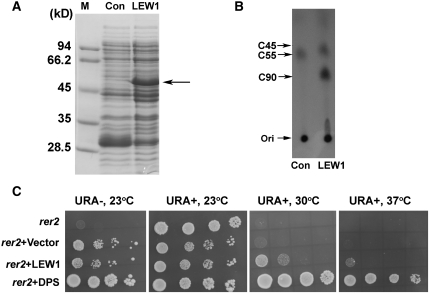
In order to confirm that LEW1 encodes a functional cis-prenyltransferase, we expressed LEW1 in E. coli (Figure 4A) and analyzed its enzyme activity in vitro. The proteins extracted from E. coli carrying LEW1 or an empty vector were incubated with 14C-labeled IPP and FPP, and the reaction products were analyzed by normal-phase thin layer chromatography (TLC) (Figure 4B). The authentic solanesols with chain lengths of C45 and C90 were used as standard compounds (Figure 4B). Because E. coli is able to catalyze the synthesis of dolichol with a chain length of C55, the labeling of C55 products was equally detected in both reactions, as expected. However, larger reaction products were detected in the extract from E. coli expressing LEW1. These products migrated slightly faster than the C90 solanesol standards and therefore must be somewhat smaller (Figure 4B). These results indicate that LEW1 possesses cis-prenyltransferase activity for dolichol synthesis in vitro.
Figure 4.
Biochemical Characterization of LEW1 and Complementation of the Yeast rer2 Mutant.
(A) Ectopic expression of LEW1 in E. coli. Proteins extracted from E. coli cells were analyzed on 12% SDS-PAGE gels and stained with Coomassie blue. The arrow points to the expressed LEW1 protein in E. coli. M, marker; Con, empty vector control; LEW1, isopropylthio-β-galactoside–induced LEW1.
(B) TLC analysis of LEW1 reaction products. [14C]IPP and [14C]FPP were incubated with total proteins isolated from E. coli strains transformed with either the empty vector (Con) or LEW1. At the completion of the reaction, lipid products were extracted and subjected to TLC analysis. Solanesols C45 and C90 were used as standards. A C55 band (representing a product that can be synthesized by the native cis-prenyltransferase in E. coli) was equally detected in E. coli transformed with either empty vector or LEW1, indicating that the reactions were successful. Ori, original spot.
(C) LEW1 partially complemented the temperature-sensitive phenotype of the yeast rer2 mutant. rer2 cells transformed with an empty vector, LEW1, or DPS (AT2G23410) were grown overnight. Serial decimal dilutions were spotted onto plates of synthetic dropout medium with all amino acids (URA+) or excluding URA (URA−). URA is a selection marker of the transformed clone. Plates were incubated at the indicated temperatures and photographed after 4 d.
A previous study indicated that Arabidopsis AT2G23410 was able to complement the temperature-dependent growth phenotype of yeast mutant rer2 (Cunillera et al., 2000). We expressed LEW1 and AT2G23410 (as a positive control) in the rer2 mutant and compared growth at 23, 30, and 37°C. As shown in Figure 4C, all of the tested yeast strains grew very well at 23°C in uracil (URA)-containing medium. By contrast, the rer2 mutant, either native or expressing the empty vector, was not able to grow at temperatures higher than 30°C. However, when transformed with a plasmid carrying the LEW1 cDNA under control of the glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter, rer2 was able to grow, albeit more slowly than the rer2 strain expressing AT2G23410. These results indicate that LEW1 can partially complement the high temperature–sensitive phenotype of the rer2 mutant.
Dolichol Contents Are Reduced in lew1 Mutant Plants but Are Increased in Transgenic Plants Overexpressing LEW1
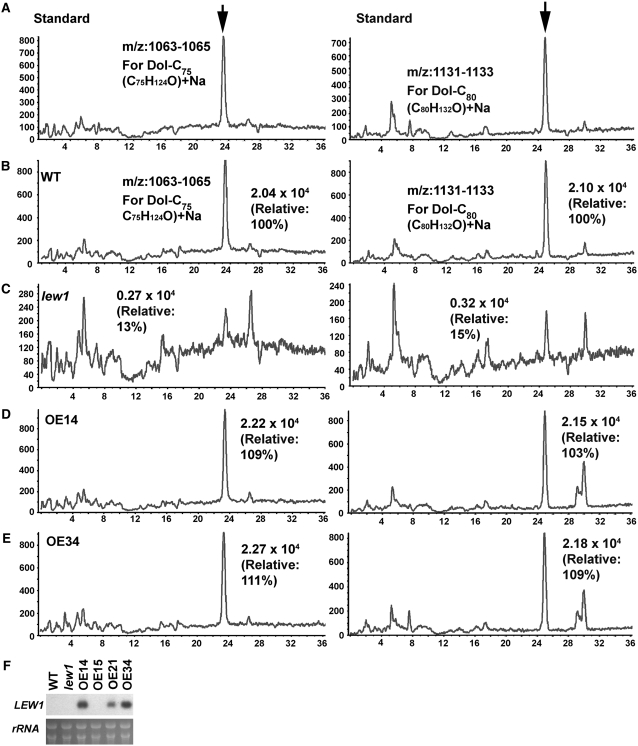
Dolichols with chain lengths of C75 and C80 have been reported to be the most abundant forms in Arabidopsis (Gutkowska et al., 2004). We performed lipid analysis to compare the dolichol contents among wild-type, lew1 mutant, and transgenic plants overexpressing LEW1 (lines OE14 and OE34). Figure 5F shows that LEW1 transcripts increased in the two overexpression lines. The dolichols in Arabidopsis exist as free lipids, lipids covalently bound to proteins, or lipid carriers linked to oligosaccharides for protein glycosylation. The total dolichols were extracted and analyzed by liquid chromatography–mass spectrometry (LC-MS) following previously established methods (Skorupinska-Tudek et al., 2003; Gutkowska et al., 2004). Dol-C65-105 (a mixture of dolichols C65 to C105) was purchased and used as a standard compound. Because we did not have pure standard samples for each dolichol to quantify the exact amounts, here we only show the relative dolichol contents in each sample. As indicated in Figure 5A, we detected Dol-C75 and Dol-C80 at different retention times (Dol-C75, 25 min, mass analysis of m/z 1063 to 1065; Dol-C80, 26 min, m/z 1131 to 1133). Based on this information, we compared the peak areas of Dol-C75 and Dol-C80 of wild-type (Figure 5B), lew1 (Figure 5C), OE14 (Figure 5D), and OE34 (Figure 5E) plants. Compared with wild-type levels, the relative contents of Dol-C75 and Dol-C80 were 13 and 15% in lew1, 109 and 103% in line OE14, and 111 and 109% in line OE34, respectively. These results strongly suggest that LEW1 catalyzes the biosynthesis of Dol-C75 and Dol-C80 in Arabidopsis.
Figure 5.
Analysis of the Relative Dolichol Contents in Wild-Type, lew1, and LEW1 Overexpression Plants by LC-MS.
The peak area in the wild type was taken as 100%, and the relative contents in other plants were compared with that in the wild type.
(A) The dolichol standards (left, Dol-C75; right, Dol-C80). Arrows indicate the peaks.
(B) The wild type.
(C) lew1 mutant.
(D) Overexpression line 14 (OE14).
(E) Overexpression line 34 (OE34).
(F) RNA gel blot analysis of LEW1 expression in OE14 and OE34. Due to its low expression, LEW1 transcript could not be detected in the wild type and the lew1 mutant by RNA gel blot under the conditions used. OE15 and OE21, two other transgenic lines not used in this study, are also included. The bottom panel shows an RNA gel as a loading control.
The lew1 Mutation Impairs Protein N-Glycosylation
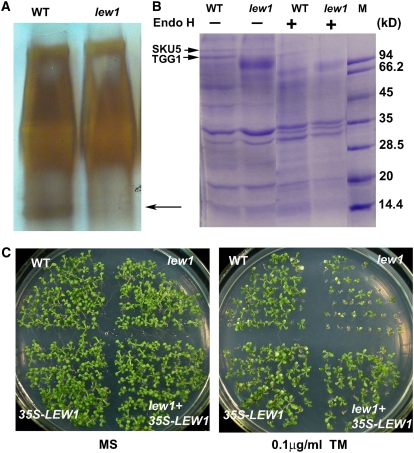
Dolichol acts as a lipid carrier during protein glycosylation, and mutations in the yeast RER2 lead to defects in protein glycosylation (Sato et al., 1999). First, we used periodic acid Schiff (PAS) staining to examine the glycoprotein levels (Nanjo et al., 2006) in wild-type and lew1 plants. In total protein extracts separated by SDS-PAGE and stained by PAS (Figure 6A), several relatively small bands were found to be much less abundant in lew1 than in the wild type, indicating that protein glycosylation levels were reduced by the mutation.
Figure 6.
lew1 Impairs Protein N-Glycosylation, and lew1 Plants Are Hypersensitive to Tunicamycin.
(A) Comparison of protein glycosylation patterns between the wild type and lew1 by PAS staining. Thirty micrograms of total protein were resolved by SDS-PAGE and detected by PAS staining. The arrow points to the bands missing from lew1.
(B) Concanavalin A–Sepharose binding assay for N-glycosylated proteins. Total proteins extracted from the wild type and lew1 were incubated with concanavalin A–Sepharose, and the bound proteins were eluted, treated with (+) or without (−) endoglycosidase H (Endo H), resolved by SDS-PAGE, and stained with Coomassie blue. The arrows at left point to the two protein bands found in the wild type but not in lew1. These two bands were recovered and analyzed by MALDI-TOF MS. The top one corresponds to SKU5, and the bottom one corresponds to TTG1. Endoglycosidase H treatment resolved the proteins found in both the wild type and lew1. M, marker.
(C) lew1 seedlings are sensitive to tunicamycin (TM). Seedlings were germinated on MS medium or MS medium containing 0.1 μg/mL tunicamycin and grown for 10 d in a growth chamber. WT, wild type (gl1); lew1, lew1 mutant; 35S-LEW1, a wild-type line overexpressing LEW1 under the control of the 35S promoter (line 14); lew+35S-LEW1, a lew1 mutant line overexpressing LEW1 under the control of the 35S promoter (line 5).
We then used concanavalin A–Sepharose, which binds to high-mannose-type N-glycans of glycoproteins, to enrich glycoproteins from the total protein extracts (Koiwa et al., 2003). The enriched glycoproteins were separated by SDS-PAGE. As shown in Figure 6B, more bands were detected in the wild type than in lew1. It is also noticeable that the peptide profile from lew1 was smeared, indicating that proteins in lew1 may be abnormally glycosylated. We further treated the enriched proteins with endoglycosidase H to cleave N-linked glycans, and the products were compared by SDS-PAGE. As shown in Figure 6B, similar bands were detected in both the wild type and lew1, suggesting that the differences were due to changes in protein glycosylation.
Third, we recovered from SDS-PAGE two bands that were present in the total protein extracts from the wild type but not from lew1 (Figure 6B, arrows). Tryptic peptides were resolved by matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) MS (Autoflex II TOF/TOF; Bruker) and analyzed by Mascot at www.Matrixscience.com. The smaller band corresponded to the glycoprotein β-thioglucoside glucohydrolase encoded by TGG1 in Arabidopsis (Xue et al., 1995; Husebye et al., 2002). TGG1 is localized in the vacuole, and underglycosylated TGG1 was also found in the stt3a mutant of Arabidopsis in a previous study (Koiwa et al., 2003). The partial sequence of the larger product was identical to that of a glycoprotein named SKU5 (Sedbrook et al., 2002). SKU5 is localized to both the plasma membrane and the cell wall and is involved in directional root growth in Arabidopsis (Sedbrook et al., 2002). These results further demonstrate that LEW1 plays critical roles in protein glycosylation.
lew1 Mutants Are Hypersensitive to the Glycosylation Inhibitor Tunicamycin
Tunicamycin is a specific inhibitor of UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (EC 2.7.8.15), the first enzyme in the biosynthesis of dolichol-linked oligosaccharides for protein glycosylation modification (Lehrman, 1991; Koizumi et al., 1999). Blocking glycosylation by tunicamycin induces UPR, causing enhanced expression of some UPR pathway genes (Koizumi et al., 1999; Lukowitz et al., 2001; Martinez and Chrispeels, 2003). Because the lew1 mutation impairs protein glycosylation, we tested the sensitivity of lew1 to tunicamycin. As shown in Figure 6C, lew1 plants were more sensitive than wild-type plants to exposure to 0.1 μg/mL tunicamycin. Overexpression of LEW1 in the wild type did not obviously increase the resistance of transgenic plants to tunicamycin, suggesting that LEW1 is not a limiting factor for protein glycosylation.
The lew1 Mutant Is Hypersensitive to Dark Stress
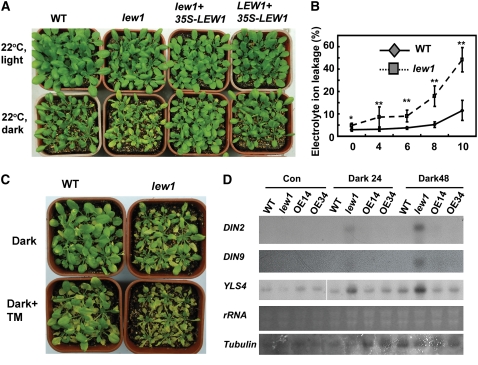
Defects in protein glycosylation greatly affect the biogenesis of membrane and secreted proteins by interfering with their folding, trafficking, degradation, and targeting to their final sites in the cell; therefore, defects in protein glycosylation generally lead to ER stress. We observed that mutant plants with one copy of the T-DNA insertion allele of lew1 and the other copy as the lew1 point mutation allele showed an early-senescence phenotype (Figure 3B). We further tested whether lew1 exhibits a more severe membrane-damage phenotype in the dark, because prolonged dark treatment induces membrane damage and senescence (Fujiki et al., 2000). lew1 and wild-type plants grown under well-watered conditions for 2 weeks were transferred to darkness at 22°C for 10 d. All of the lew1 plants became yellow, but wild-type plants did not. lew1 plants transformed with the LEW1 overexpression construct (35S-LEW1) exhibited similar phenotypes as wild-type plants (Figure 7A). Electrolyte leakage from leaves of lew1 plants was greatly increased and reached 35% after 10 d in the dark, whereas wild-type leaves showed only ∼10% leakage under the same conditions (Figure 7B). These results show that more severe membrane damage occurred in lew1 than in the wild type under prolonged dark treatment.
Figure 7.
lew1 Plants Are Hypersensitive to Dark.
(A) Wild-type and lew1 seedlings (3 weeks old) were grown in light (top row) or dark (bottom row) at 22°C for 10 d. Also shown is a lew1 transgenic line overexpressing LEW1 (line 5) and a wild-type transgenic line overexpressing LEW1 (line 14).
(B) Electrolyte leakage assay of wild-type and lew1 plants in the dark. Two-week-old seedlings were grown in soil in the dark at 22°C for the indicated times, and the leaves were taken for ion leakage assays. Results were from three independent experiments. Data are means ± se (* P < 0.05, ** P < 0.01).
(C) Tunicamycin treatment accelerated dark-induced senescence in lew1. Three-week-old seedlings grown in soil were sprayed with water (Dark) or with 0.1 μg/mL tunicamycin (Dark+TM) and then incubated in the dark for 10 d before photographs were taken.
(D) Expression of dark-inducible genes in lew1 and the wild type. Two-week-old seedlings were grown in the dark for 24 or 48 h, and total RNA was analyzed by RNA gel blot. Also shown are two wild-type transgenic lines (OE14 and OE34) overexpressing LEW1 (35S-LEW1). The two bottom panels show rRNA and tubulin as loading controls.
Because tunicamycin inhibits protein glycosylation, we also investigated the effects of this compound on plant sensitivity to prolonged darkness. Spraying tunicamycin on wild-type leaves accelerated the dark-induced senescence and therefore phenocopied to some extent the dark-sensitive phenotype of lew1 (Figure 7C). In addition, tunicamycin treatment of lew1 leaves rendered their senescence in the dark more pronounced (Figure 7C). The data overall suggest that the lew1 phenotype in the dark is related to a glycosylation defect.
Darkness Stress–Responsive Genes Are Hyperinduced in lew1 by Dark Treatment
We selected two late dark-inducible marker genes, DIN2 (for dark-inducible2) and DIN9, to compare their expression patterns in lew1 and wild-type plants. DIN2 encodes a β-glucosidase, and DIN9 encodes a mannose-6-phosphate isomerase (Fujiki et al., 2000). We also examined a leaf senescence marker gene, YLS4 (for YELLOW-LEAF-SPECIFIC GENE4), which is induced in old leaves by darkness (Fujiki et al., 2005). Two-week-old seedlings were kept under darkness for 24 and 48 h. RNA gel blot analysis revealed DIN2, DIN9, and YLS4 transcripts to be substantially induced in lew1 at 24 and 48 h in the dark, whereas the transcripts were not yet or less detected in the wild type and two transgenic lines overexpressing LEW1 at these time points in the dark (Figure 7D).
Drought or Osmotic Stress Suppresses the Darkness Sensitivity of lew1 Plants, and lew1 Seedlings Are More Tolerant to Osmotic Stress
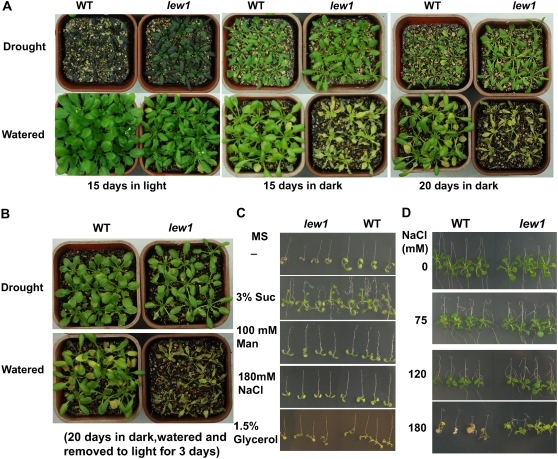
Because lew1 plants are more tolerant to drought stress, we next analyzed the response of lew1 to drought stress in the dark. Ten-day-old seedlings grown in the light were pretreated by withholding water for 7 d prior to the dark treatment, which produced only a modest drought stress. Control plants were watered every third day, and both control and drought-treated plants were kept in the dark or light for various times. After an additional 15 d of drought treatment, wild-type plants grown in the light were seriously wilted (Figure 8A, 15 d in light) and did not recover growth after rewatering. By contrast, lew1 plants, which developed clear drought-tolerant phenotypes in light, were able to regrow after watering (see Supplemental Figure 1 online). These results are consistent with previous data (Figure 2A) and indicate that lew1 is tolerant to drought stress at different developmental stages.
Figure 8.
Drought or Osmotic Stress Improves the Survival of lew1 Plants in the Dark or under Weak Light
(A) Ten-day-old seedlings in soil were subjected to water withholding for 7 d to give a modest drought treatment (Drought) in the light and then incubated in the dark for 15 or 20 d, or in the light for 15 d, without watering. Wild-type but not lew1 plants died from drought treatment for 15 d in the light. All lew1 plants died from drought at 20 d in the light. Plants without drought treatment (Watered) were kept in the light for 15 d or in the dark for 15 or 20 d.
(B) After 20 d, the drought-treated plants in the dark were watered and moved, together with plants without drought treatment in the dark, to light for an additional 3 d. No lew1 plants without drought treatment in the dark survived, but all lew1 plants with drought treatment in the dark survived. No wild-type plants died under the conditions used.
(C) Osmotic stress imposed by mannitol, NaCl, or glycerol increased the survival of lew1 seedlings in MS medium without sucrose under weak light. Four-day-old seedlings grown on MS medium with 3% sucrose were moved to MS medium without sucrose (−), with sucrose (3%), or without sucrose but with 100 mM mannitol, 180 mM NaCl, or 1.5% glycerol and cultured for 7 d. All lew1 seedlings on MS medium without sucrose died, while all wild-type seedlings survived. In other treatment conditions, all lew1 and wild-type seedlings survived.
(D) lew1 seedlings are more tolerant to high-salt stress than wild-type seedlings. Four-day-old seedlings were moved to MS medium containing 3% sucrose with different concentrations of NaCl (0, 75, 120, or 180 mM) and cultured for 10 d. There was no clear growth difference at 0, 75, and 120 mM NaCl between the wild type and lew1. However, at 180 mM NaCl, lew1 seedlings grew better than wild-type seedlings.
lew1 plants growing under well-watered conditions in the dark for 15 d became yellowish and were seriously damaged (Figure 8A, 15 d in dark). However, lew1 plants that had been pretreated by drought were able to survive the dark treatment and were green and healthy after 15 or even 20 d in the dark (Figure 8A). These plants grew very well after being transferred to normal conditions and irrigated in light (Figure 8B, 20 d in drought + dark). These results indicate that drought stress relieved dark-induced senescence in lew1.
We further tested the growth of lew1 mutant plants on medium supplemented with various osmotic stress agents. As controls, both lew1 and wild-type seedlings grew well on Murashige and Skoog (MS) medium containing 3% sucrose (Figure 8C). When no sugar was added to the MS medium, lew1 seedlings grew poorly and died after 10 d in culture. The growth of wild-type seedlings was also inhibited in the absence of sugar in the medium, but in contrast with lew1, all wild-type seedlings survived and still grew (Figure 8C). When grown on MS medium (without sugar) supplemented with 100 mM mannitol, 180 mM NaCl, or 1.5% glycerol for 10 d, all lew1 seedlings survived, as did the wild type (Figure 8C). These results indicate that hyperosmotic stress is able to suppress the sensitivity of lew1 seedlings to conditions of no sugar in culture medium.
We also compared the growth of lew1 and the wild type on MS medium with 3% sucrose and different concentrations of NaCl. As shown in Figure 8D, lew1 seedlings showed similar sensitivity to NaCl concentrations of <120 mM as the wild type. However, lew1 seedlings grew better than wild-type seedlings on 180 mM NaCl. This result suggests that unlike stt3a and other N-glycan defect mutants, which are sensitive to salt stress (Koiwa et al., 2003; Kang et al., 2008), lew1 mutants were more resistant to high NaCl than the wild type.
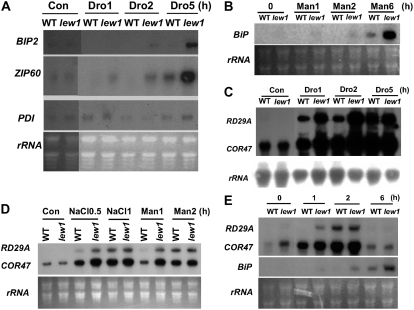
Drought Stress Induces Higher Expression of BiP and bZIP60 in lew1 Than in the Wild Type
Cells have evolved elaborate mechanisms to ensure the accuracy of protein folding and assembly. When unfolded proteins are accumulated to a certain level, UPR signaling is activated to induce the expression of specific proteins to protect cells from the ER stress or to induce cell death if homeostasis cannot be reestablished (Lin et al., 2007). A previous study found that overexpression of BiP improved the drought tolerance of transgenic plants, suggesting that UPR signaling might be involved in drought stress tolerance (Alvim et al., 2001). In order to determine whether drought stress could provoke UPR signaling or not, we compared the transcripts of genes in the UPR pathway in the wild type and lew1 under drought stress conditions. We used a BiP probe that detects the transcripts of both BiP1 and BiP2 because of their high sequence identity. BiP transcripts were hardly detected in the wild type after a 5-h drought treatment under our conditions, whereas the BiP transcripts were clearly detected at 2 h and highly expressed at 5 h in lew1 (Figure 9A). BiP transcripts were also induced by a mannitol treatment at higher levels in lew1 than in the wild type (Figure 9B). bZIP60 is a key bZIP transcription factor responsible for the ER stress response, and its expression is induced by tunicamycin (Iwata and Koizumi, 2005). The expression of bZIP60 was induced to a higher level in lew1 than in the wild type under drought treatment (Figure 9A). The expression of PROTEIN DISULFIDE ISOMERASE (PDI), encoding a disulfide isomerase, is also induced by tunicamycin in Arabidopsis (Martinez and Chrispeels, 2003). However, the level of PDI did not change under our drought stress conditions (Figure 9A). These results indicate that osmotic stress produced from either drought or mannitol treatment can activate the expression of some genes in the UPR pathway, and this response is enhanced by the lew1 mutation.
Figure 9.
RNA Gel Blot Analysis of Stress-Inducible Genes under Different Stress Conditions.
(A) The transcripts of BiP and bZIP60 were induced to higher levels in lew1 than in the wild type by drought stress. Four-week-old seedlings were removed from soil and placed on a laboratory bench for 1 h (drought 1 [Dro1]). Then, seedlings were covered with a transparent film (to slow water loss) for 1 or 4 h (drought 2 h and 5 h [Dro2 and Dro5, respectively]). Total RNA from each treatment was used for RNA gel blot analysis. RNA from shoots without treatment was used as a control at 0 h (Con). rRNAs stained with ethidium bromide were used as loading controls.
(B) More BiP transcripts accumulated in lew1 than in the wild type under mannitol treatment. Four-week-old seedlings from soil were dipped into solution containing 300 mM mannitol for 1, 2, or 6 h (Man1, Man2, and Man6, respectively), and total RNA was analyzed by RNA gel blot. rRNAs stained with ethidium bromide served as loading controls. RNA from shoots without treatment was used as a control at 0 h (0).
(C) The transcripts of RD29A and COR47 were induced to higher levels in lew1 than in the wild type by drought stress. Drought treatment was done as in (A). rRNA was used as a loading control.
(D) Four-week-old seedlings were treated with 150 mM NaCl for 0.5 h (NaCl0.5) or 1 h (NaCl1) or with 300 mM mannitol for 1 h (Man1) or 2 h (Man2). rRNAs stained with ethidium bromide served as loading controls. RNA from shoots without treatment was used as a control at 0 h (Con).
(E) Two-week-old seedlings grown on MS medium were treated with 0.1 μg/mL tunicamycin for different times, and total RNA was analyzed by RNA gel blot. rRNAs stained with ethidium bromide served as loading controls.
Because of the sensitivity of lew1 to dark stress, we further tested whether the expression of the UPR marker genes BiP and bZIP60 is induced by dark treatment. RNA gel blot analysis indicated that the expression of neither BiP nor bZIP60 was detected in the wild type or lew1 under dark treatment (data not shown). These results suggest that the dark treatment is not able to provoke the expression of UPR pathway genes.
Stress-Responsive Genes Are Induced to Higher Levels and at Earlier Time Points by Drought Stress in lew1 Than in the Wild Type
We further investigated whether the lew1 mutation also alters the expression of drought-responsive genes. In these experiments, 4-week-old seedlings were removed from soil and subjected to different water stress treatments, and the expression of the drought-responsive genes RD29A and COR47 was analyzed by RNA gel blots. Under drought conditions, the transcripts of RD29A and COR47 were induced to higher levels in lew1 than in wild-type plants (Figure 9C, Dro1 and Dro2). In addition, NaCl and mannitol treatments resulted in earlier induction of RD29A and COR47 in lew1 than in the wild type (Figure 9D).
Because tunicamycin treatment causes ER stress, we also tested the expression of RD29A and COR47 under tunicamycin treatment. We detected higher expression levels of RD29A and COR47 in lew1 than in the wild type at 1 h. The expression levels in wild-type and mutant plants became similar at 2 or 6 h (Figure 9E). The expression of BiP was induced at earlier time points and to higher levels by tunicamycin treatment in lew1 than in the wild type (Figure 9E). These results suggest that lew1 plants are more sensitive to tunicamycin treatment than are wild-type plants in the induction of UPR genes.
DISCUSSION
Here, we report on the genetic and functional analyses of LEW1, a gene encoding a long-chain cis-prenyltransferase in dolichol biosynthesis in Arabidopsis. There are 10 cis-prenyltransferases related to the yeast RER2, and LEW1 belongs to a subgroup that is different from the other 9 cis-prenyltransferases in Arabidopsis. LEW1 is an essential gene in Arabidopsis, since plants with a homozygous T-DNA insertion in LEW1 were nonviable. In yeast, RER2 and STR1 encode two similar cis-prenyltransferases catalyzing the biosynthesis of dolichols with different chain lengths: RER2 for C70 to C85 and STR1 for C95 to C105 (Sato et al., 1999, 2001). The rer2 mutant showed various phenotypes that were not observed in the str1 mutant. Although STR1 overexpression could suppress the growth and glycosylation defects of rer2, the biological functions of RER2 and STR1 are considered to be different (Sato et al., 1999, 2001). These results suggest that different cis-prenyltransferases in Arabidopsis might play distinct roles in plant growth and development.
Our in vitro assay indicated that LEW1 is able to catalyze the biosynthesis of dolichol with a chain length of less than C90. Comparison of total dolichol contents between lew1 and the wild type showed that the dolichols with chain lengths of C75 to C80 were reduced to about one-quarter of the wild-type level, suggesting that LEW1 plays a predominant role in catalyzing the biosynthesis of dolichol C75 to C80 in Arabidopsis. In mammals, the chain lengths of dominating dolichyl phosphates used as glycosyl carrier lipids are around C95 to C105 (Schenk et al., 2001). Our results indicate that the chain lengths of dolichols in Arabidopsis are similar to those in yeast. Although LEW1 catalyzed the biosynthesis of dolichols with chain lengths similar to those produced by RER2, overexpression of LEW1 only partially complemented the temperature-sensitive phenotype of rer2. By contrast, AT2G23410, which catalyzes the formation of dolichol with a chain length of C120, was able to complement the rer2 mutant phenotype (Cunillera et al., 2000; Oh et al., 2000). Similarly, a cDNA that encodes a cis-prenyltransferase for catalyzing the production of dolichols with a chain length of C85 to C95 from human brain complemented the growth and other phenotypes of the rer2 mutant (Shridas et al., 2003). Studies on the rer2 mutant overexpressing SRT1 indicated that yeast cells are able to utilize the longer dolichols in protein glycosylation when shorter ones (C70 to C85) are lacking (Sato et al., 2001). Sequence analysis suggested that LEW1 was more diverged from RER2 than AT2G23410. We speculate that LEW1 expressed in yeast might have low catalytic efficiency for dolichol synthesis, possibly because the LEW1 protein may not be folded correctly in yeast or LEW1 may need some specific cofactors or modifications for full activity.
One of the important functions of dolichols is to serve as the lipid carrier of carbohydrates in the assembly of Dol-P–linked oligosaccharides for protein N-glycosylation (Burda and Aebi, 1999). In plants and animals, some genes involved in the dolichol-related N-linked glycosylation pathway have been identified, and defects in the human genes lead to serious diseases called congenital disorders of glycosylation (Koiwa et al., 2003; Kranz et al., 2004, 2007; Leroy, 2006). In this study, we found that the lew1 mutation reduced protein glycosylation levels and identified two specific proteins with altered glycosylation. There are 10 RER2-related cis-prenyltransferase homologs in Arabidopsis, and at least one of them, AT2G23410, was shown to also be able to catalyze the biosynthesis of dolichols in an in vitro assay (Cunillera et al., 2000; Oh et al., 2000). It is not known yet whether all of these genes play some roles in N-linked protein glycosylation as LEW1 does.
More than 70% of all proteins in eukaryotic cells are predicted to be N-glycosylated (Apweiler et al., 1999). Genes encoding some of the enzymes functioning at various steps of the protein glycosylation pathway have been characterized. These include, for example, GNTI (for N-ACETYL GLUCOSAMINYLTRANSFERASE I, encoded by COMPLEX GLYCAN1 [CGL1]) (von Schaewen et al., 1993; Wenderoth and von Schaewen, 2000; Strasser et al., 2005), GMII (an α-mannosidase II, encoded by HYBRID GLYCOSYLATION1 [HGL1]) (Strasser et al., 2006), STT3a (Koiwa et al., 2003), DGL1 (for DEFECTIVE GLYCOSYLATION1), an Arabidopsis homolog of an oligosaccharyltransferase complex subunit (Lerouxel et al., 2005), CYT1 (for CYTOKINESIS-DEFECTIVE1, mannose-1-phosphate guanylyltransferase) (Lukowitz et al., 2001), GCS1/KNF (glucosidase I), and RSW3 (glucosidase II; glucosidases I and II trim the terminal glucose of N-glycans). Mutations in these genes have some overlapping, yet differential, effects on plant growth, development, and stress responses. For instance, a lesion in GNTI caused by the cgl1 mutation impairs the synthesis of Golgi-modified complex N-linked glycans, and cgl1 mutant plants do not show phenotypes under normal conditions but are more sensitive to heat stress or dark treatment (von Schaewen et al., 1993). Similarly, hgl1 mutants do not show any obvious phenotypes under normal conditions (Strasser et al., 2006), whereas dgl1 and cyt1 mutants do (Zablackis et al., 1996; Bonin et al., 1997; Lukowitz et al., 2001; Lerouxel et al., 2005). Interestingly, some mutants, such as dgl1, gcs1/knf, rsw3, and cyt1, show a clear cell wall–defective phenotype, suggesting that N-glycan processing plays crucial roles in plant cell well formation. Mutations in STT3a, a subunit of the oligosaccharyltransferase complex responsible for protein N-glycosylation, may also affect the cell wall, particularly under salt stress (Koiwa et al., 2003). stt3a mutants are hypersensitive to salt and osmotic stress due to cell swelling and cell cycle arrest at the root tip (Koiwa et al., 2003). The stt3a mutations were also reported to cause increased expression of BiP in the root tip (Koiwa et al., 2003). lew1 mutant plants were not hypersensitive to salt or osmotic stress. Rather, the mutant showed increased drought resistance and enhanced survival under high-salt conditions. These results suggest that defects in different steps of the protein N-glycosylation pathway have different consequences on plant stress responses.
Although we compared the amounts of cellulose and lignin from lew1 and wild-type cell walls and did not find a clear difference (see Supplemental Figure 2 online), we cannot exclude the possibility of a subtle change in cellulose or other cell wall components in lew1 that could not be detected in our experiments. Recent studies suggest that the plant cell wall could be important for stress signaling (Ellis et al., 2002; Zhong et al., 2002; Chen et al., 2005; Hernandez-Blanco et al., 2007; Li et al., 2007). Mutations in genes for cell wall biosynthesis or modifications can activate jasmonate, ethylene, and/or abscisic acid signaling, all of which could be connected to sugar responses (Ellis et al., 2002; Zhong et al., 2002; Chen et al., 2005; Hernandez-Blanco et al., 2007; Li et al., 2007). Our results on lew1 suggest possible complex crosstalk among the different signaling pathways, which may regulate the various stress responses observed in this study.
Dolichols are involved in many different cellular processes. In addition to their roles in protein glycosylation and other modifications, free dolichols also have important biological functions as lipids (Chojnacki and Dallner, 1988; Bizzarri et al., 2003). Because it is an unsaturated lipid, dolichol was postulated to function as a free radical scavenger against reactive oxygen species either produced by chemicals or UV light or accumulated upon aging. However, there is no direct evidence for such a function in vivo (Bergamini et al., 1998; Bizzarri et al., 2003; Sgarbossa et al., 2003). Although we did not observe any phenotypes, such as sensitivity to H2O2 treatment (see Supplemental Figure 3 online), we still cannot exclude that the pleiotropic phenotypes observed in lew1 might be a comprehensive outcome of changing lipid peroxidation, plasma membrane characteristics, protein dolichylation, and/or protein glycosylation.
A striking phenotype, which served in the initial isolation of lew1, is leaf wilting even under normal growth conditions. We found that the leaf wilting was not due to a defect in water uptake by roots or in water transport from root to shoot through the xylem. The phenotype was also not caused by excessive transpiration or a low leaf cell osmotic potential. On the contrary, lew1 plants displayed enhanced water permeability in roots and had a reduced stomatal conductance. Our results suggest that the main cause of the leaf-wilting phenotype is probably cell membrane lesions that result in electrolyte leakage and thus impair the ability to maintain cell turgor. The turgor defect may explain, at least partly, the reduced stomatal conductance and enhanced drought resistance of the lew1 mutant. The cell membrane lesions reflect a critical role of dolichol in membrane function and/or its function in protein glycosylation. Because virtually all membrane proteins and secreted proteins are folded and assembled in the ER before being exported and transported to final destinations in the cell, defects in protein glycosylation would greatly affect protein folding, induce ER stress, and eventually reduce the levels and/or activities of certain membrane proteins. Membrane trafficking is known to play crucial roles in abiotic stress responses (Carter et al., 2004). One of the two identified proteins that are affected by lew1, SKU5, is localized in the plasma membrane (Sedbrook et al., 2002). Other plasma membrane proteins, such as H+-ATPases and aquaporins, which are critical in turgor maintenance, may also be altered by the lew1 mutation.
The increased dark sensitivity of lew1 and the early-senescence phenotype exhibited by heterozygous plants carrying lew1 and a T-DNA insertion also support membrane defects in the mutant. The dark-inducible genes DIN2 and DIN9 and the leaf senescence marker YLS4 were induced to higher levels in lew1 than in the wild type by the dark treatment. Leaf senescence is considered to be a type of programmed cell death, with plasma membrane breakage as an earlier symptom (Lim et al., 2007). In yeast, defects in protein N-glycosylation induce apoptosis (Hauptmann et al., 2006). Apoptosis, or programmed cell death, is strongly induced by reduced activities of ER and the Golgi apparatus in plant cells (Crosti et al., 2001; Malerba et al., 2004). The fact that tunicamycin-treated wild-type plants mimicked the leaf-senescence phenotype of lew1 in the dark suggests that protein N-glycosylation defects in lew1 contribute to the increased dark-sensitivity and early-senescence phenotypes of the mutant. Nevertheless, dark treatment did not upregulate UPR pathway genes. Interestingly, drought but not tunicamycin treatment greatly improved the survival of lew1 plants during prolonged dark treatment. This suggests that drought treatment suppresses the dark sensitivity of the mutant by a mechanism independent of the UPR pathway.
The transcriptional upregulation of a large number of genes involved in the UPR pathway is the most prominent strategy for the cell to cope with ER stress (Urade, 2007). BiP is a chaperone protein that is induced by chemicals such as the specific N-glycosylation inhibitor tunicamycin (Koizumi et al., 1999). Previous studies have indicated that UPR pathway genes are important for both biotic and abiotic stress responses (Alvim et al., 2001; Wang et al., 2005; Fujita et al., 2007; Liu et al., 2007b). In this study, we observed that drought stress induced the expression of the UPR pathway genes BiP and bZIP60, and the induction was much stronger in lew1 than in the wild type. On the other hand, tunicamycin treatment was able to induce the expression of the drought-responsive genes RD29A and COR47, with stronger induction in the lew1 mutant. These stress marker genes were also more responsive to drought in the mutant. Taken together, these results suggest that drought stress may cause ER stress and activate UPR responses, and part of the drought responses (e.g., induction of RD29A and COR47) may be mediated by the UPR pathway. The enhanced inducibility of UPR genes and drought-responsive genes in lew1 may contribute to the drought resistance and salt/osmotic tolerance phenotypes of the mutant.
METHODS
Plant Growth Conditions
Arabidopsis thaliana ecotype Columbia (gl1) was grown in long-day conditions (16-h-light/8-h-dark cycle) with forest soil and vermiculite (1:1) or on MS medium (M5519; Sigma-Aldrich) containing 3% (w/v) sucrose and 0.8% (w/v) agar at 22°C. All seeds were kept at 4°C for 3 d before sowing. For drought treatment in soil, we grew Arabidopsis seedlings in short-day conditions (12-h-light/12-h-dark cycle) at 22°C. For drought treatment, 10-d-old seedlings in soil were not watered for 7 d to give a modest drought treatment in the light. Then, the plants were moved to darkness for 15 or 20 d, or continually grown in the light for 15 d, without watering. At the end of treatment, plants were watered and moved to light for an additional 3 d. For osmotic stress, 4-d-old seedlings grown on MS medium with 3% sucrose were moved to MS medium without sucrose, with sucrose (3%), or without sucrose but with 100 mM mannitol, 180 mM NaCl, or 1.5% glycerol and cultured for 7 d. For salt treatment, 4-d-old seedlings were moved to the MS medium containing 3% sucrose with different concentrations of NaCl (0, 75, 120, or 180 mM) and cultured for 10 d.
Isolation of the lew1 Mutant and Genetic Analysis
Ethyl methanesulfonate–mutagenized M2 seeds were sterilized with 0.5% NaClO for 15 min, washed five times with distilled water, sown on MS medium, kept at 4°C for 3 d, and then moved to a plant growth chamber to grow for 5 d. The seedlings were finally transferred to soil in a greenhouse. After growing for 2 weeks, water was withheld from the seedlings. lew1 was chosen because it showed the leaf-wilting phenotype. lew1 was backcrossed with the wild type, and F2 plants showed a segregation of ∼1:3 lew1:wild type (a total 159 seedlings counted, 38 lew1 and 121 wild-type seedlings), indicating that lew1 is recessive and caused by a single nuclear gene. The lew1 mutant was backcrossed with the wild type four times before being used for detailed analysis. We ordered the SALK_065628 heterozygous seeds from the ABRC. We isolated genomic DNA from the progeny of SALK lines and performed PCR analysis using primer pair 5′-CCGAGACGTCTCACACTCCTC-3′ and 5′-GCTTTCCGTCAGGCGCAAC-3′, which covered the T-DNA insertion region, but did not find the homozygous lines, suggesting that the null mutation of LEW1 is lethal. Crossing the heterozygous T-DNA line with lew1, we recovered the heterozygous plants in which the LEW1 gene was impaired by both lew1 mutation and T-DNA insertion. RT-PCR was used to compare the LEW1 transcripts in the wild type, lew1, and a heterozygous plant of lew1 crossed with the T-DNA line. LEW1 transcripts were amplified for 30 cycles with the primers 5′-GCCTGACGGAAAGCGCATTG-3′ and 5′-GGGAAACCGAGGTGGCTCC-3′ using the first-strand cDNA reverse transcribed from mRNAs (M-MLV RTase cDNA synthesis kit, D6130; TaKaRa). Products were visualized on ethidium bromide–stained gels.
Genetic Mapping and Complementation
lew1 mutant plants were crossed with the Landsberg accession, and 1638 mutant plants were chosen from the F2 generation according to the leaf-wilting phenotype. Simple sequence length polymorphism markers were designed according to the information in the Cereon Arabidopsis Polymorphism Collection and used to analyze recombination events (Bell and Ecker, 1994; Jander et al., 2002). The lew1 mutation was first mapped to chromosome 1 between T7I23 and F2J6. To narrow the lew1 mutation, markers in BAC clones F21M12, T23J18, T28K15, F21M12, T20H2, T19E23, and F14M2 were designed. Finally, the lew1 mutation was delimited between BAC clones F25C20 and F12F1. We sequenced all candidate genes in this region from the lew1 mutant and compared the sequences with those in GenBank in order to find the lew1 mutation.
For complementation of the lew1 mutant, LEW1 cDNA was amplified from reverse-transcribed cDNA with the primer pair 5′-CACCATGGATTCGAATCAATCGATGCGGCTCCTC-3′ (added CACC at the 5′ end) and 5′-GATCTCTGAAACTCTGCTCTCTAGTCACCG-3′. The amplified fragment was cloned into Gateway vector pMDC32 (Curtis and Grossniklaus, 2003) with the Gateway Technology system (Invitrogen). Then, the cloned construct was transformed to Agrobacterium tumefaciens and transferred into plants using the floral dip method (Clough and Bent, 1998).
RNA Gel Blot Analysis
Shoots of 4-week-old seedlings from soil were dipped into solution containing 300 mM mannitol or 150 mM NaCl for different times. For drought treatment, the removed shoots were put on a laboratory bench for 1 h, which resulted in the loss of ∼20% of their water (relative air humidity was ∼70%) (drought 1 h), then the seedlings were covered with a transparent film in order to prevent seedlings from quickly losing water for 1 or 4 h (drought 2 h and 5 h). Shoots lost ∼30% of their water at the end of drought treatment for 5 h. For dark treatment, seedlings in soil were moved to a plant growth chamber with no light. Seedlings used for RNA extraction in Figure 5F were grown on MS medium with 3% sucrose for 15 d, and the full length of LEW1 cDNA was used as a probe for hybridization. Total RNA was extracted and hybridized as described previously (Chen et al., 2005). Probes were amplified with the following primer pairs: for RD29A (967 bp), forward, 5′-GACGAGTCAGGAGCTGAGCTG-3′, and reverse, 5′-CGATGCTGCCTTCTCGGTAGAG-3′; for COR47 (413 bp), forward, 5′-GAAGCTCCCAGGACACCACGAC-3′, and reverse, 5′-CAGCGAATGTCCCACTCCCAC-3′; for BiP (652 bp), forward, 5′-ACCAGAAGGACATCAGCAAGGAC-3′, and reverse, 5′-GGTGGGACTCCAGTGAGGTC-3′; for BZIP60 (400 bp), forward, 5′-GAGATGCGGCGGTTAGATCGAG-3′, and reverse, 5′-GGCCTCGAACCCTTACATCTCC-3′; for PDI1 (487 bp), forward, 5′-CTCGCGTGCAGAGAATGCGTC-3′, and reverse, 5′-TCTTGCGCATTTCTCCGTCGAC-3′; for DIN2 (383 bp), forward, 5′-GGAAGATGGGTGTGACGTAAGAGG-3′, and reverse, 5′-ATGAAGATGGGTTCTCTATGCCTTGC-3′; for DIN9 (414 bp), forward, 5′-CCAGAGATCACGGAGCTTGTAGG-3′, and reverse, 5′-GCGATGTTTAGGAGTAAGACCAGCTC-3′; for YLS4 (713 bp), forward, 5′-CTCATACTAGGCGCTGACAGTCC-3′, and reverse, 5′-GATCAGCCATGGCCTTCAGTTCC-3′.
Protein Analysis
Concanavalin A binding proteins were purified using concanavalin A–Sepharose (Sigma-Aldrich) according to the described methods (Koiwa et al., 2003). The isolated concanavalin A binding proteins were resolved on a 12% SDS-PAGE gel and stained with Coomassie Brilliant Blue R 250. Two unique bands in extracts from the wild type were excised and resolved by MALDI-TOF MS (Autoflex II TOF/TOF; Bruker) and then analyzed with the help of Mascot on www.Matrixscience.com. For glycoprotein PAS staining, 30 μg of total soluble proteins was resolved on 12% SDS-PAGE, and glycoprotein levels were detected using the described protocol (Nanjo et al., 2006).
Assay of LEW1 Activity and Analysis of LEW1 Reaction Products
Five microliters of each 20 μM primer (5′-GATCCGGTACCGCGGCCGCATGCCATGGTCGACGAGCTCTAGAAGCTTGG-3′ and 5′-AATTCCAAGCTTCTAGAGCTCGTCGACCATGGCATGCGGCCGCGGTACCG-3′) were mixed well and denatured at 100°C for 5 min. When the mixture was cooled to room temperature, it was ligated to pGEX-2T vector that had been excised with BamHI and EcoRI at 16°C overnight. The modified pGEX-2T vector has the multiple clone sites BamHI, KpnI, SacI, NotI, SphI, NcoI, SalI, SacI, XbaI, HindIII, and EcoRI. LEW1 cDNA was amplified with forward primer 5′-CGGGATCCATGGATTCGAATCAATCGATGCGGCTCCTC-3′ and reverse primer 5′-GCTCTAGAAATTGGGAACAGTAGTGGCTGCACTGACTC-3′ and then cloned to a modified vector pGEX-2T in BamHI and XbaI sites. The plasmids were transformed into BL21 Escherichia coli cells, and the bacteria were induced at 37°C with 1 mM isopropylthio-β-galactoside for 4 h, centrifuged at 3200g at 4°C for 15 min, and suspended in the buffer (pH 8.0) containing 50 mM Tris-HCl, 0.25 M NaCl, 1 mM EDTA, and 1 mM DTT. Phenylmethylsulfonyl fluoride to a final concentration of 0.5 mM was added, and the suspended bacteria were broken with an ultrasonic crasher (Sonics and Materials; model vcx500). NaCl to a final concentration of 0.5 mM was added to the broken bacteria, which were then centrifuged at 3200g at 4°C for 60 min. The upper phase was used for LEW1 activity assays as described previously (Sato et al., 1999) with some modifications. The proteins were analyzed on 12% SDS-PAGE gels. One hundred microliter reaction mixtures including 100 μg of crude proteins, 25 mM Tris-HCl (pH 8.5), 5 mM MgCl2, 1.25 mM DTT, 2.5 mM sodium orthovanadate, 1.4 nmol of farnesyl pyrophosphate (F6892; Sigma-Aldrich), and 5 nmol of [1-14C]isopentenyl pyrophosphate (I4143; Sigma-Aldrich) were incubated at 30°C for 1 h, and then 2 mL of chloroform:methanol (2:1) was added to stop the reaction. Next, 0.8 μL of 0.9% NaCl was added to the reaction mixtures to extract the organic (lower) phase, which was washed three times with chloroform:methanol:water (3:48:47) and dried under nitrogen. The dried samples were dissolved with 5 μL of buffer containing 100 mM sodium acetate (pH 4.8), 0.1% (v/v) Triton X-100, and 60% (v/v) methanol. After brief sonication, 1.5 units of potato (Solanum tuberosum) acid phosphatase was added to treat the products at room temperature overnight. Lipid products were extracted three times with 2 mL of n-hexane. All extracts were washed twice with water and dried under nitrogen. The final products were dissolved in 100 μL of n-hexane and analyzed by reverse-phase thin layer chromatography on a Silica Gel G-60 plate (Merck) developed with benzene:ethyl acetate (90:10). The standard solanesols (C45 and C90; Sigma-Aldrich) were marked under 254-nm UV light, and the radiolabeled products were detected with x-ray film for 15 d.
Analysis of Dolichols in Arabidopsis
Dolichol analyses were performed using LC-MS as reported previously (Skorupinska-Tudek et al., 2003; Gutkowska et al., 2004). Briefly, 150 mg of freeze-dried seedlings was ground and extracted three times with 5 mL of CHCl3:methanol (1:1). After centrifugation, 1.5 mL of methanol and then 1.5 mL of 20% (w/v) KOH were added to the organic layer and residues. After incubating at 80°C for 1 h, the lipids were extracted three times with 4 mL of hexane. The extracts were analyzed by HPLC/electrospray ionization-MS. A mixture of dolichols 13 to 21 was purchased from Avanti Polar Lipids and used as a standard compound. The experiments were repeated three times, and similar results were obtained. Here, we show one representative result.
Complementation Study with Yeast and Plants
LEW1 cDNA was excised from pGEX-2T with BamHI and HindIII and cloned into yeast vector p426-GPD. DPS (AT2G23410) cDNA was amplified from RNA-transcribed cDNA product with primer pair 5′-CGGGATCCATGTTGTCTCTTCTTTCTTCTGATTCTTCTCTATTATCAC-3′ and 5′-GCGTCGACTCAAACCCGACAACCAAATCGTCTTTCC-3′. The amplified cDNA was cut by BamHI and SalI and then cloned into p426-GPD. The cloned constructs and empty vector were transformed into yeast mutant strain SNH23-7D (MATalpha rer2-2 mfalpha1∷ADE2 mfalpha2∷TRP1 bar1∷HIS3 ade2 trp1 his3 leu2 ura3 lys2; American Type Culture Collection catalog number MYA-2727) by the lithium acetate procedure (Gietz and Woods, 2002). Selected URA+ transformants were grown at different temperatures, 23, 30, and 37°C, on 2% (w/v) agar plates containing yeast nitrogen base (6.7 g/L) and 2% (w/v) Glc.
Water Transport Measurements
Lpr was measured as described previously (Boursiac et al., 2005). Briefly, the root system of a freshly detopped Arabidopsis plant was inserted and tightly sealed into a pressure chamber filled with nutrient solution. A pressure (P) was then slowly applied to the chamber, using nitrogen gas, and a linear relationship between the rate of sap flow (Jv) exuded from the sectioned hypocotyl and P was established for P values between 160 and 320 kPa. Lpr (in mL·g−1·h−1·MPa−1) was calculated from the slope of a Jv(P) plot divided by the dry weight of the root system. Water transport inhibition experiments were performed essentially as described by Tournaire-Roux et al. (2003). Briefly, an excised root system was equilibrated in a simplified nutrient solution [5 mM KNO3, 2 mM MgSO4, 1 mM Ca(NO3)2, and 10 mM MES, pH 6] during 20 min at 320 kPa. The root was then transferred in a standard solution supplemented with 1 mM NaN3 or 50 μM HgCl2, and the rate of sap flow at 320 kPa was measured during 30 (NaN3) or 60 (HgCl2) min following the treatment. In all cases, the percentage inhibition at the indicated time was deduced from these kinetic curves. Leaf stomatal conductance was measured using a Li-Cor porometer. To measure root and shoot dry weight, all samples were oven-dried at 96°C for 24 to 48 h and weighed. The cross sections of stems and leaves by light microcopy were described previously (Chen et al., 2005).
Leaf Turgor Assay
Turgor measurements were performed on young growing leaves (∼1 cm in length) using a vertically mounted cell pressure probe device as described previously (Gerbeau et al., 2002). The pressure probe was completely filled with silicone oil (type AS4; Wacker) and a few picoliters of water was drawn into the tip of the micropipette, thus forming a meniscus at the oil–water interface. For measurements, a leaf was excised and laid on a filter paper imbibed with water, and epidermal cells located at the edge of the blade were punctured with the probe using a stereomicroscope (magnification ×160). After cell impalement, the meniscus was stabilized at its initial position with the aid of a motor-driven metal rod, allowing a stationary turgor pressure (P0) to be measured. Measurements were performed within 15 min following leaf excision. No time-dependent decay in P0 values measured successively over this period of time was observed, suggesting that the possible decrease in water potential due to leaf transpiration during the measurement period was negligible.
Stomatal Aperture Assays
Epidermal strips from rosette leaves of 4-week-old seedlings of wild-type and mutant plants were brushed to get rid of chlorophyll and examined for stomata opening with a light microscope (B5-223 IEP; Motic China Group) or floated on solutions containing 50 μM CaCl2, 10 mM KCl, and 10 mM MES-Tris, pH 6.15, and exposed to light (20 μmol·m−2·s−l) for 2 h to open stomata and examined with the light microscope.
Water Loss, Osmotic Potential, and Electrolyte Leakage Measurements
Rosette leaves of mutant and wild-type plants growing under normal conditions for 4 weeks were detached and weighed on a piece of weighing paper at designated time intervals in a greenhouse (40% RH, 50 μmol·m−2·s−l light). Three replicates were done. The percentage of water loss was calculated on the basis of the initial weight. Leaf osmotic potential of 4-week-old seedlings was measured as described previously (Ruggiero et al., 2004). Electrolyte leakage was determined using fully developed rosette leaves as described previously (Guo et al., 2002). Three independent experiments were done.
Phylogenetic Analysis
We used the LEW1 animo acid sequence to query the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov). The amino acid sequences were aligned using MEGA (version 4.0) (Tamura et al., 2007) with the default settings. The alignment is shown in Supplemental Data Set 1 online. A phylogenetic tree was constructed with the neighbor-joining method using the default settings of MEGA (version 4.0; http://www.kumarlab.net/publications) (Tamura et al., 2007). Bootstrap values calculated from 1000 replicates are shown.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers NC_003070 (LEW1/AT1G11755) and NM_127905 (AT2G23410).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotypes of lew1 and Wild-Type Plants under Drought Treatment after Rewatering.
Supplemental Figure 2. Acid Detergent Fiber (ADF) Contents of lew1 and Wild-Type Plants.
Supplemental Figure 3. Comparison of lew1 and Wild-Type Plants on H2O2 Medium.
Supplemental Data Set 1. Multiple Alignments of LEW1 Homologs.
Supplementary Material
Acknowledgments
We thank the ABRC for providing the T-DNA lines, Ueli Grossniklaus (University of Zürich) for providing the Gateway vectors, and Youqun Wang (China Agricultural University) for assisting in cross sections of stems and leaves. This work was supported by grants from the National Basic Research Program of China (Grant 2003CB114306) and the National Nature Science Foundation of China to Z.G.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zhizhong Gong (gongzz@cau.edu.cn).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alvim, F.C., Carolino, S.M., Cascardo, J.C., Nunes, C.C., Martinez, C.A., Otoni, W.C., and Fontes, E.P. (2001). Enhanced accumulation of BiP in transgenic plants confers tolerance to water stress. Plant Physiol. 126 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler, R., Hermjakob, H., and Sharon, N. (1999). On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473 4–8. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Bergamini, E., et al. (1998). A proposed mechanism of the antiaging action of diet restriction. Aging (Milano) 10 174–175. [PubMed] [Google Scholar]
- Bizzarri, R., Cerbai, B., Signori, F., Solaro, R., Bergamini, E., Tamburini, I., and Chiellini, E. (2003). New perspectives for (S)-dolichol and (S)-nor dolichol synthesis and biological functions. Biogerontology 4 353–363. [DOI] [PubMed] [Google Scholar]
- Bonin, C.P., Potter, I., Vanzin, G.F., and Reiter, W.D. (1997). The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proc. Natl. Acad. Sci. USA 94 2085–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursiac, Y., Chen, S., Luu, D.T., Sorieul, M., van den Dries, N., and Maurel, C. (2005). Early effects of salinity on water transport in Arabidopsis roots. Molecular and cellular features of aquaporin expression. Plant Physiol. 139 790–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda, P., and Aebi, M. (1999). The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426 239–257. [DOI] [PubMed] [Google Scholar]
- Carter, C.J., Bednarek, S.Y., and Raikhel, N.V. (2004). Membrane trafficking in plants: New discoveries and approaches. Curr. Opin. Plant Biol. 7 701–707. [DOI] [PubMed] [Google Scholar]
- Cascardo, J.C., Almeida, R.S., Buzeli, R.A., Carolino, S.M., Otoni, W.C., and Fontes, E.P. (2000). The phosphorylation state and expression of soybean BiP isoforms are differentially regulated following abiotic stresses. J. Biol. Chem. 275 14494–14500. [DOI] [PubMed] [Google Scholar]
- Chen, Z., Hong, X., Zhang, H., Wang, Y., Li, X., Zhu, J.K., and Gong, Z. (2005). Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J. 43 273–283. [DOI] [PubMed] [Google Scholar]
- Chojnacki, T., and Dallner, G. (1988). The biological role of dolichol. Biochem. J. 251 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann, A., Moes, D., Himmelbach, A., Yang, Y., Tang, Y., and Grill, E. (2006). Integration of abscisic acid signalling into plant responses. Plant Biol. (Stuttg.) 8 314–325. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Crosti, P., Malerba, M., and Bianchetti, R. (2001). Tunicamycin and brefeldin A induce in plant cells a programmed cell death showing apoptotic features. Protoplasma 216 31–38. [DOI] [PubMed] [Google Scholar]
- Cunillera, N., Arro, M., Fores, O., Manzano, D., and Ferrer, A. (2000). Characterization of dehydrodolichyl diphosphate synthase of Arabidopsis thaliana, a key enzyme in dolichol biosynthesis. FEBS Lett. 477 170–174. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, C., Karafyllidis, I., Wasternack, C., and Turner, J.G. (2002). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki, Y., Ito, M., Nishida, I., and Watanabe, A. (2000). Multiple signaling pathways in gene expression during sugar starvation. Pharmacological analysis of din gene expression in suspension-cultured cells of Arabidopsis. Plant Physiol. 124 1139–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki, Y., Nakagawa, Y., Furumoto, T., Yoshida, S., Biswal, B., Ito, M., Watanabe, A., and Nishida, I. (2005). Response to darkness of late-responsive dark-inducible genes is positively regulated by leaf age and negatively regulated by calmodulin-antagonist-sensitive signalling in Arabidopsis thaliana. Plant Cell Physiol. 46 1741–1746. [DOI] [PubMed] [Google Scholar]
- Fujita, M., Mizukado, S., Fujita, Y., Ichikawa, T., Nakazawa, M., Seki, M., Matsui, M., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2007). Identification of stress-tolerance-related transcription-factor genes via mini-scale Full-length cDNA Over-eXpressor (FOX) gene hunting system. Biochem. Biophys. Res. Commun. 364 250–257. [DOI] [PubMed] [Google Scholar]
- Gerbeau, P., Amodeo, G., Henzler, T., Santoni, V., Ripoche, P., and Maurel, C. (2002). The water permeability of Arabidopsis plasma membrane is regulated by divalent cations and pH. Plant J. 30 71–81. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., and Woods, R.A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350 87–96. [DOI] [PubMed] [Google Scholar]
- Grabinska, K., and Palamarczyk, G. (2002). Dolichol biosynthesis in the yeast Saccharomyces cerevisiae: an insight into the regulatory role of farnesyl diphosphate synthase. FEMS Yeast Res. 2 259–265. [DOI] [PubMed] [Google Scholar]
- Guo, Y., Xiong, L., Ishitani, M., and Zhu, J.K. (2002). An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures. Proc. Natl. Acad. Sci. USA 99 7786–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutkowska, M., Bienkowski, T., Hung, V.S., Wanke, M., Hertel, J., Danikiewicz, W., and Swiezewska, E. (2004). Proteins are polyisoprenylated in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 322 998–1004. [DOI] [PubMed] [Google Scholar]
- Hauptmann, P., Riel, C., Kunz-Schughart, L.A., Frohlich, K.U., Madeo, F., and Lehle, L. (2006). Defects in N-glycosylation induce apoptosis in yeast. Mol. Microbiol. 59 765–778. [DOI] [PubMed] [Google Scholar]
- Hernandez-Blanco, C., et al. (2007). Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19 890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye, H., Chadchawan, S., Winge, P., Thangstad, O.P., and Bones, A.M. (2002). Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol. 128 1180–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irsigler, A.S., Costa, M.D., Zhang, P., Braga, P.A., Dewey, R.E., Boston, R.S., and Fontes, E.P. (2007). Expression profiling on soybean leaves reveals integration of ER- and osmotic-stress pathways. BMC Genomics 8 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata, Y., and Koizumi, N. (2005). An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. USA 102 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I.M., and Last, R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J., Krag, S.S., and Betenbaugh, M.J. (2005). Controlling N-linked glycan site occupancy. Biochim. Biophys. Acta 1726 121–137. [DOI] [PubMed] [Google Scholar]
- Kang, J.S., et al. (2008). Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. USA 105 5933–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, J., Fujisaki, S., Nakajima, K., Nishimura, Y., Sato, M., and Nakano, A. (1999). The Escherichia coli homologue of yeast RER2, a key enzyme of dolichol synthesis, is essential for carrier lipid formation in bacterial cell wall synthesis. J. Bacteriol. 181 2733–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwa, H., Li, F., McCully, M.G., Mendoza, I., Koizumi, N., Manabe, Y., Nakagawa, Y., Zhu, J., Rus, A., Pardo, J.M., Bressan, R.A., and Hasegawa, P.M. (2003). The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell 15 2273–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi, N., Ujino, T., Sano, H., and Chrispeels, M.J. (1999). Overexpression of a gene that encodes the first enzyme in the biosynthesis of asparagine-linked glycans makes plants resistant to tunicamycin and obviates the tunicamycin-induced unfolded protein response. Plant Physiol. 121 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz, C., Denecke, J., Lehle, L., Sohlbach, K., Jeske, S., Meinhardt, F., Rossi, R., Gudowius, S., and Marquardt, T. (2004). Congenital disorder of glycosylation type Ik (CDG-Ik): A defect of mannosyltransferase I. Am. J. Hum. Genet. 74 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz, C., et al. (2007). A defect in dolichol phosphate biosynthesis causes a new inherited disorder with death in early infancy. Am. J. Hum. Genet. 80 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman, M.A. (1991). Biosynthesis of N-acetylglucosamine-P-P-dolichol, the committed step of asparagine-linked oligosaccharide assembly. Glycobiology 1 553–562. [DOI] [PubMed] [Google Scholar]
- Lerouxel, O., Mouille, G., Andeme-Onzighi, C., Bruyant, M.P., Seveno, M., Loutelier-Bourhis, C., Driouich, A., Hofte, H., and Lerouge, P. (2005). Mutants in DEFECTIVE GLYCOSYLATION, an Arabidopsis homolog of an oligosaccharyltransferase complex subunit, show protein underglycosylation and defects in cell differentiation and growth. Plant J. 42 455–468. [DOI] [PubMed] [Google Scholar]
- Leroy, J.G. (2006). Congenital disorders of N-glycosylation including diseases associated with O- as well as N-glycosylation defects. Pediatr. Res. 60 643–656. [DOI] [PubMed] [Google Scholar]
- Li, Y., Smith, C., Corke, F., Zheng, L., Merali, Z., Ryden, P., Derbyshire, P., Waldron, K., and Bevan, M.W. (2007). Signaling from an altered cell wall to the nucleus mediates sugar-responsive growth and development in Arabidopsis thaliana. Plant Cell 19 2500–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, P.O., Kim, H.J., and Nam, H.G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58 115–136. [DOI] [PubMed] [Google Scholar]
- Lin, J.H., Li, H., Yasumura, D., Cohen, H.R., Zhang, C., Panning, B., Shokat, K.M., Lavail, M.M., and Walter, P. (2007). IRE1 signaling affects cell fate during the unfolded protein response. Science 318 944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.X., Srivastava, R., Che, P., and Howell, S.H. (2007. a). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19 4111–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.X., Srivastava, R., Che, P., and Howell, S.H. (2007. b). Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 51 897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., Nickle, T.C., Meinke, D.W., Last, R.L., Conklin, P.L., and Somerville, C.R. (2001). Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 98 2262–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malerba, M., Cerana, R., and Crosti, P. (2004). Comparison between the effects of fusicoccin, tunicamycin, and brefeldin A on programmed cell death of cultured sycamore (Acer pseudoplatanus L.) cells. Protoplasma 224 61–70. [DOI] [PubMed] [Google Scholar]
- Martinez, I.M., and Chrispeels, M.J. (2003). Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjo, Y., Oka, H., Ikarashi, N., Kaneko, K., Kitajima, A., Mitsui, T., Munoz, F.J., Rodriguez-Lopez, M., Baroja-Fernandez, E., and Pozueta-Romero, J. (2006). Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-Golgi to the chloroplast through the secretory pathway. Plant Cell 18 2582–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S.K., Han, K.H., Ryu, S.B., and Kang, H. (2000). Molecular cloning, expression, and functional analysis of a cis-prenyltransferase from Arabidopsis thaliana. Implications in rubber biosynthesis. J. Biol. Chem. 275 18482–18488. [DOI] [PubMed] [Google Scholar]
- Ruggiero, B., Koiwa, H., Manabe, Y., Quist, T.M., Inan, G., Saccardo, F., Joly, R.J., Hasegawa, P.M., Bressan, R.A., and Maggio, A. (2004). Uncoupling the effects of abscisic acid on plant growth and water relations. Analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in Arabidopsis. Plant Physiol. 136 3134–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M., Fujisaki, S., Sato, K., Nishimura, Y., and Nakano, A. (2001). Yeast Saccharomyces cerevisiae has two cis-prenyltransferases with different properties and localizations. Implication for their distinct physiological roles in dolichol synthesis. Genes Cells 6 495–506. [DOI] [PubMed] [Google Scholar]
- Sato, M., Sato, K., Nishikawa, S., Hirata, A., Kato, J., and Nakano, A. (1999). The yeast RER2 gene, identified by endoplasmic reticulum protein localization mutations, encodes cis-prenyltransferase, a key enzyme in dolichol synthesis. Mol. Cell. Biol. 19 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, B., Rush, J.S., Waechter, C.J., and Aebi, M. (2001). An alternative cis-isoprenyltransferase activity in yeast that produces polyisoprenols with chain lengths similar to mammalian dolichols. Glycobiology 11 89–98. [DOI] [PubMed] [Google Scholar]
- Sedbrook, J.C., Carroll, K.L., Hung, K.F., Masson, P.H., and Somerville, C.R. (2002). The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgarbossa, A., Lenci, F., Bergamini, E., Bizzarri, R., Cerbai, B., Signori, F., Gori, Z., and Maccheroni, M. (2003). Dolichol: A solar filter with UV-absorbing properties which can be photoenhanced. Biogerontology 4 379–385. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58 221–227. [DOI] [PubMed] [Google Scholar]
- Shridas, P., Rush, J.S., and Waechter, C.J. (2003). Identification and characterization of a cDNA encoding a long-chain cis-isoprenyltransferase involved in dolichyl monophosphate biosynthesis in the ER of brain cells. Biochem. Biophys. Res. Commun. 312 1349–1356. [DOI] [PubMed] [Google Scholar]
- Skorupinska-Tudek, K., Bienkowski, T., Olszowska, O., Furmanowa, M., Chojnacki, T., Danikiewicz, W., and Swiezewska, E. (2003). Divergent pattern of polyisoprenoid alcohols in the tissues of Coluria geoides: a new electrospray ionization MS approach. Lipids 38 981–990. [DOI] [PubMed] [Google Scholar]
- Strasser, R., Schoberer, J., Jin, C., Glossl, J., Mach, L., and Steinkellner, H. (2006). Molecular cloning and characterization of Arabidopsis thaliana Golgi alpha-mannosidase II, a key enzyme in the formation of complex N-glycans in plants. Plant J. 45 789–803. [DOI] [PubMed] [Google Scholar]
- Strasser, R., Stadlmann, J., Svoboda, B., Altmann, F., Glossl, J., and Mach, L. (2005). Molecular basis of N-acetylglucosaminyltransferase I deficiency in Arabidopsis thaliana plants lacking complex N-glycans. Biochem. J. 387 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewskaa, E., and Danikiewiczb, W. (2005). Polyisoprenoids: Structure, biosynthesis and function. Prog. Lipid Res. 44 235–258. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tournaire-Roux, C., Sutka, M., Javot, H., Gout, E., Gerbeau, P., Luu, D.T., Bligny, R., and Maurel, C. (2003). Cytosolic pH regulates root water transport during anoxic stress through gating of aquaporins. Nature 425 393–397. [DOI] [PubMed] [Google Scholar]
- Urade, R. (2007). Cellular response to unfolded proteins in the endoplasmic reticulum of plants. FEBS J. 274 1152–1171. [DOI] [PubMed] [Google Scholar]
- von Schaewen, A., Sturm, A., O'Neill, J., and Chrispeels, M.J. (1993). Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol. 102 1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., Weaver, N.D., Kesarwani, M., and Dong, X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308 1036–1040. [DOI] [PubMed] [Google Scholar]
- Wenderoth, I., and von Schaewen, A. (2000). Isolation and characterization of plant N-acetyl glucosaminyltransferase I (GntI) cDNA sequences. Functional analyses in the Arabidopsis cgl mutant and in antisense plants. Plant Physiol. 123 1097–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, I.B. (2002). Glycosylation of proteins in plants and invertebrates. Curr. Opin. Struct. Biol. 12 569–577. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Schumaker, K.S., and Zhu, J.K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14 (suppl.): S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, J., Jorgensen, M., Pihlgren, U., and Rask, L. (1995). The myrosinase gene family in Arabidopsis thaliana: gene organization, expression and evolution. Plant Mol. Biol. 27 911–922. [DOI] [PubMed] [Google Scholar]
- Zablackis, E., York, W.S., Pauly, M., Hantus, S., Reiter, W.D., Chapple, C.C., Albersheim, P., and Darvill, A. (1996). Substitution of L-fucose by L-galactose in cell walls of Arabidopsis mur1. Science 272 1808–1810. [DOI] [PubMed] [Google Scholar]
- Zhong, R., Kays, S.J., Schroeder, B.P., and Ye, Z.H. (2002). Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14 165–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.